Abstract
We describe a patient with a novel WT1 pS50X germ line mutation, who developed bilateral Wilms tumours, both with stromal‐type histology. Both tumours showed loss of the wild type WT1 allele (loss of heterozygosity (LOH)) and a tumour specific mutation in catenin beta1 (CTNNB1), S45P in the left and Δ45S in the right tumour. Molecular analysis of microdissected cells from the left tumour revealed the same S45P CTNNB1 mutation in blastema, tubuli, stroma and muscle, and a different CTNNB1 mutation (T41A) in stromal cells isolated from another area of the same slide. Microdissection of two areas of muscle cells from the right tumour revealed the same Δ45S mutation and no CTNNB1 mutation nor LOH of WT1 in normal kidney cells. One year later, the patient developed a new set of bilateral tumours. Both tumours showed LOH of the wild type WT1 allele, but different CTNNB1 mutations as in the first tumours: S45C on the right and S45F on the left side, demonstrating that these developed independently and are not relapses. This case demonstrates the high risk for the development of Wilms tumours in patients with germ line truncation mutations.
Keywords: bilateral Wilms' tumour, wt1 germ line mutation, ctnnb1 mutation, multiple independent tumours
Wilms tumour (WT) or nephroblastoma is the most frequent renal tumour of childhood. WT is thought to be derived from a renal stem cell, with impaired differentiation potential. Most tumours have a mixed histology and are composed of three elements: blastema, epithelia and stroma, recapitulating the development of the normal kidney. Tumours may also contain heterotypic cells not normally found in the kidney such as rhabdomyoblasts, fat, cartilage and bone, probably derived from an abnormal mesenchymal differentiation. This type of differentiation is mainly observed in the stromal‐type variant of WT. Constitutional or somatic mutations in the WT1 gene are found in approximately 10–20% of WT, most occurring in stromal‐type tumours.1,2,3,4 In addition, approximately 75% of WT carry CTNNB1 gene mutations, mostly in or close to amino acids that are important for activity and stabilisation of the protein.5,6,7,8,9
Methods
Mutation and allelotyping analysis
DNA was isolated from the tumour and blood by standard methods. WT1 gene mutation analysis was performed by the DNA‐PCR SSCP method including all 10 exons and flanking intron sequences, as described.2CTNNB1 gene mutation analysis of exon 3 was performed as described.5
To study the mechanism for LOH, microsatellite analysis was performed with CA‐repeat markers from 11q11 (D11S1313), 11p13 (D11S1776) and 11p15.4 (D11S1323). In the PCR one marker was fluorescently labelled with IRD 800 and the products were analysed on an automatic sequencer (Li‐Cor Biosciences, Lincoln, Nebraska, USA).
For the molecular analysis of the different cell types the respective cells were identified after HE staining and the corresponding areas were removed from a consecutive unstained slide using a sterile scalpel. After deparaffinisation DNA was extracted with the Pico Pure DNA extraction buffer according to the manufacturer (Arcturus, Mountain View, California, USA). Each sample served as a template for PCR amplification of exon 1 of WT1 and exon 3 of the CTNNB1 gene.
Case presentation and results
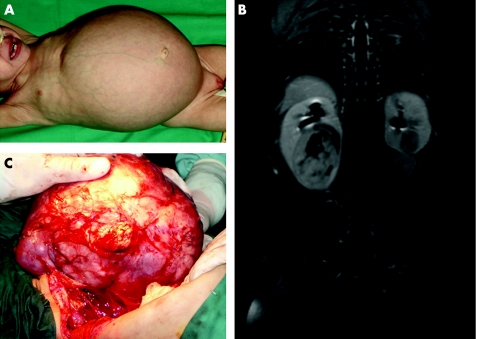
A 10‐month‐old female patient with a 46, XX karyotype presented with large bilateral kidney tumours (fig 1A). She was initially treated in another institution for 8 weeks with empirical chemotherapy including vincristine and actinomycin‐D with no clinico‐radiological response and then referred to the Hospital Sant Joan de Déu in Barcelona where the tumours were completely excised by kidney sparing surgery. The histology was stromal type (rhabdomyomatous) on both sides and she received no cytotoxic therapy after the surgery and did well for 1 year. At this time she developed bilateral renal tumours again (see MRI in fig 1B). At surgery a very large tumour was found in the right kidney, which led to a complete nephrectomy. In the left kidney there was a small round tumour that was totally resected by tumorectomy (fig 1C). Pathology findings showed in the right kidney a large, classical, mixed WT with no capsular infiltration. The left kidney tumour was round and hard, and pathology confirmed the same rhabdomyomatous nature as in the first set of tumours one year earlier. Nephrogenic rests were not observed in any of the tumours nor in the small amount of adjacent normal kidneys removed during surgery.
Figure 1 Clinical presentation of the case. (A) Patient at first presentation after chemotherapy (2004). (B) MRI of recurrent tumours. (C) Second tumour, removed in 2005. Parental informed consent was obtained for publication of this figure.
Analysis of blood DNA revealed an abnormal SSCP banding pattern of exon 1 of WT1. Sequencing identified a heterozygous C>A change at nucleotide position 149, resulting in the creation of a stop codon at Ser50 (S50X) (fig 2A). This mutation has not been described before and represents the most extreme N‐terminal germ line truncation mutation in WT1 to date. Sequencing of the tumour DNA revealed only the mutant allele (fig 2A).
Figure 2 WT1 sequence of exon 1. (a) normal control DNA, patient's blood DNA, patient's tumour DNA, tumour cell line RNA. (b) LOH analysis with markers from 11q11, 11p13 and 11p15. Open circle no loss, filled circle LOH. B, blood DNA; T, tumour DNA.
Different CTNNB1 mutations were found in the tumour DNA from both sides of the initial as well as the second tumour samples. The initial tumours had a S45P on the left side and a ΔS45 on the right side. In the second set of tumours the left side harboured a S45F and the right side a S45C mutation, clearly demonstrating that the second set of tumours occurred independently. Besides the germ line WT1 mutation, present in every cell, each tumour had two additional genetic events: loss of the wild type WT1 allele and a mutation in CTNNB1.
To study whether all different cell types of the tumours, ie, blastemal, stromal, muscle and tubules have the same or different genetic events, we performed manual microdissection. Extracted DNAs were first analysed for CTNNB1 gene mutations. Two different areas from the initial left tumour were used to prepare the A3 and A1 slides. Different cell types from both slides had the same S45P mutation, except one area of stromal cells from the A3 slide, which had a T41A mutation, whereas stromal cells from another part of the same slide had a S45P mutation. From the initial right tumour only one section (C2) was analysed and all tumour cell subtypes contained the same Δ45S mutation.
In normal kidney cells from the right tumour no CTNNB1 mutation was found, confirming by molecular methods that these were indeed normal kidney cells. The same types of analyses were performed with the recurrent bilateral tumours. All cells from two different slides (1E and F1) from the right side had the same S45C mutation except in two normal kidney sections with epithelial morphology, which were wild type for CTNNB1, again confirming that these were normal cells. From the left side two different areas with muscle cells were isolated from one slide and both had a S45F. The CTNNB1 mutations found on both sides were not seen in any cells of the primary set of tumours, demonstrating that the recurrent tumours on both sides occurred independently and had acquired new CTNNB1 gene mutations in addition to loss of wild type WT1. In addition the presence of identical CTNNB1 mutations in the different cell types of each tumour clearly shows that these are derived from one parental transformed cell.
Sequencing of WT1 exon 1 from the microdissected cell types revealed only the mutant allele in all cases that were analysed (for example in table 1, sections A3 and C2 and fig 2A). To study the mechanism which led to loss of the wild type WT1 allele we performed a microsatellite analysis with markers from chromosome 11. For this we first used a tumour cell line that we have established from the left recurrent tumour. The tumour cells had a 46, XX karyotype with two normal appearing copies of chromosome 11. Sequencing of the cell line DNA showed that these were homozygous for the WT1 mutation, and RNA analysis revealed that they expressed the mutant transcript (fig 2A). In addition they carried a S45F CTNNB1 mutation, the same as was observed in the microdissected tumour samples. The DNA from the tumour cell line was heterozygous for the 11q11 marker and homozygous for the 11p13 and 11p15 markers (fig 2B), demonstrating that loss of wild type WT1 was due to a mitotic recombination event between 11q11 and 11p13. Analysis of microdissected blastemal and muscle cells from the second tumour of the right side also showed LOH of 11p13 and 11p15 and no LOH for 11q11, whereas normal appearing epithelial cells did not show LOH of these markers (table 1). Therefore, in both second tumours from the left and right side, a similar mitotic recombination event resulted in loss of wild type WT1.
Table 1 Mutations found in microdissected tumour samples.
| Slides | Tissue cell types | CTNNB1 mutation | Nucleotide change | WT1 mutation* | LOH analysis |
|---|---|---|---|---|---|
| Initial left tumour | |||||
| A3 | Tubuli | S45P | TCT>CCT | m/m | |
| A3 | Stroma | T41A | ACC>GCC | m/m | |
| A3 | Blastema | S45P | TCT>CCT | m/m | |
| A3 | Stroma II | S45P | TCT>CCT | nd | |
| A3 | Muscle I | S45P | TCT>CCT | m/m | |
| A3 | Muscle II | S45P | TCT>CCT | nd | |
| A1 | Muscle | S45P | TCT>CCT | nd | |
| A1 | Stroma | S45P | TCT>CCT | nd | |
| A1 | Muscle/fat | S45P | TCT>CCT | nd | |
| Initial right tumour | |||||
| C2 | Tumour pool | ΔS45 | ΔTCT | nd | |
| C2 | Muscle I | ΔS45 | ΔTCT | m/m | |
| C2 | Muscle II | ΔS45 | ΔTCT | nd | |
| C2 | Normal kidney I | wild type | m/w | ||
| C2 | Normal kidney II | wild type | nd | ||
| Recurrence left | |||||
| 3B | Muscle I | S45F | TCT>TTT | nd | |
| 3B | Muscle II | S45F | TCT>TTT | nd | |
| Cell line | S45F | TCT>TTT | m/m | LOH 11p13 + p15 no LOH 11q11 | |
| Recurrence right | |||||
| 1E | Blastema I | S45C | TCT>TGT | nd | LOH 11p13 no LOH 11q11 |
| 1E | Blastema II | S45C | TCT>TGT | nd | |
| 1E | Muscle | S45C | TCT>TGT | nd | |
| 1E | Stroma | S45C | TCT>TGT | nd | |
| F1 | Blastema I | S45C | TCT>TGT | nd | LOH 11p15 |
| F1 | Blastema II | S45C | TCT>TGT | nd | LOH 11p15 |
| F1 | Epithelia I | wild type | nd | no LOH 11p15 | |
| F1 | Epithelia II | wild type | nd | no LOH 11p15 | |
| F1 | Muscle | S45C | TCT>TGT | nd | LOH 11p15 |
Microdissected areas labelled in italics correspond to normal kidney cells by molecular analysis, one WT1 mutant allele as in all cells of the patient and no CTNNB1 mutation.
*WT1 mutation analysis was performed by sequencing, m/m indicates mutant/mutant allele, and m/w one mutant one wild type allele, nd: not done
Discussion
The patient presenting here with a novel WT1 germ line truncation mutation developed four independent WT: bilateral synchronous WT and bilateral synchronous second tumours. The molecular proof for their independent origin was derived from the CTNNB1 mutation analysis as the four tumours were associated with five different mutations and each showed loss of the WT1 wild type allele. The WT1 S50X mutation represents the most extreme N‐terminal truncation of the protein described so far. Another child with a F40fsX90 WT1 germ line mutation who developed a bilateral WT at age 8 months was described as having Denys–Drash syndrome due to a female phenotype but a male karyotype.10 At the time of surgery this child did not have glomerulosclerosis or proteinuria. The authors discuss the possibility that the short WT1 protein acted in a dominant negative fashion to produce some aspects of the Denys–Drash syndrome phenotype. In our patient the tumours showed loss of the wild type allele excluding the possibility of a dominant negative effect for tumour development and suggesting either gain or loss of function as the mechanism involved in tumour formation. As our patient is female, and so far has had no proteinuria, we cannot exclude the possibility that this truncation mutation would act as dominant negative for sex development or nephropathy.
Key points
Germ line mutations in the WT1 gene result in a high risk for Wilms tumour development, often as a synchronous bilateral tumour.
Three genetic events occur in these tumours, (1) a mutation in WT1, (2) loss of wild type WT1 and (3) a mutation in CTNNB1.
The timing of these events is at present not known as all analysed tumour sections harboured all three genetic events.
Patients with germ line WT1 mutations and tumour sparing surgery should be surveyed for the development of independent second tumours.
The different independent tumours harboured the same mitotic recombination event between 11q11 and 11p13 as a mechanism for LOH.
The novel findings of the molecular genetic tumour analysis are the high frequency of mitotic recombination events between 11q11 and 11p13 leading to loss of wild type WT1, and the strong selection pressure for CTNNB1 mutations in cells with a complete loss of a functional WT1 protein. These results suggest that WT1 negative kidney precursor cells may not survive without a CTNNB1 mutation. As a matter of fact in the Wt1 knockout mouse Wt1‐negative mesenchymal cells die by apoptosis during kidney development.11
It will be interesting to study whether the type of the WT1 mutation influences the molecular mechanisms, and therefore the risk for tumour development, as has been observed for the age of tumour onset, with the earliest onset in patients with truncation mutations.4 Also we have observed a higher percentage of bilateral tumours in cases with truncation mutations in the 5′ half of the gene before the nuclear localisation signal.4 In agreement with our earlier observation, this patient has a termination signal at codon 50. The molecular proof that this patient developed independent second sets of bilateral tumours shows the high risk for WT in patients with germ line WT1 mutations and the need for a close clinical tumour surveillance of such patients.
Acknowledgements
We thank Dr Barbara Leube for help with the microsatellite analysis and Dr Barbara Hildebrandt for the cytogenetic analysis.
Abbreviations
CTNNB1 - catenin beta1
LOH - loss of heterozygosity
WT - Wilms' tumour
WT1 - Wilms Tumour 1
Footnotes
Competing interests: None declared
This work was supported by the Elterninitiative Kinderkrebsklinik e.V., Düsseldorf to B R‐P and the Developmental tumour biology laboratory in Hospital Sant Joan de Deu, Barcelona, is supported by a generous gift from Fondo Margarita del Pozo.
Parental informed consent was obtained for the publication of figure 1.
References
- 1.Little M, Wells C. A clinical overview of WT1 gene mutations. Hum Mutat 19979209–225. [DOI] [PubMed] [Google Scholar]
- 2.Schumacher V, Schneider S, Figge A, Wildhardt G, Harms D, Schmidt D, Weirich A, Ludwig.R, Royer‐Pokora B. Correlation of germ‐line mutations and two‐hit inactivation of the WT1 gene with Wilms tumors of stromal‐predominant histology. Proc Natl Acad Sci USA 1997943972–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shibata R, Hashigushi A, Sakamoto J, Yamada T, Umezawa A, Hata J. Correlation between a specific Wilms tumours suppressor gene (WT1) mutation and the histological findings in Wilms tumour (WT). J Med Genet 200239e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Royer‐Pokora B, Beier M, Henzler M, Alam R, Schumacher V, Weirich A, Huff V. Twenty‐four new cases of WT1 germline mutations and review of the literature: genotype/phenotype correlations for Wilms tumor development. Am J Med Genet 2004127249–257. [DOI] [PubMed] [Google Scholar]
- 5.Koesters R, Ridder R, Kopp‐Schneider A, Betts D, Adams V, Niggli F, Briner J, von Knebel Doeberitz M. Mutational activation of the β‐catenin proto‐oncogene is a common event in the development of Wilms' tumors. Cancer Res 1999593880–3882. [PubMed] [Google Scholar]
- 6.Maiti S, Alam R, Amos C I, Huff V. Frequent association of β‐catenin and WT1 mutations in Wilms tumors. Cancer Res 2000606288–6292. [PubMed] [Google Scholar]
- 7.Kusafuka T, Miao J, Kuroda S, Udatsu Y, Yoneda A. Codon 45 of the β‐catenin gene, a specific mutational target site of Wilms' tumor. Int J Mol Med 200210395–399. [PubMed] [Google Scholar]
- 8.Fukuzawa R, Heathcott R W, Sano M, Morison I M, Yun K, Reeve A E. Myogenesis in Wilms' tumors is associated with mutations of the WT1 gene and activation of Bcl‐2 and the Wnt signaling pathway. Pediatr Dev Pathol 20047125–137. [DOI] [PubMed] [Google Scholar]
- 9.Li C ‐ M, Kim C E, Margolin A A, Guo M, Zhu J, Mason J M, Hensle T W, Murty V V V S, Grundy P E, Fearon E R, D'Agati V, Licht J D, Tycko B.CTNNB1 mutations and overexpression of Wnt/β‐catenin target genes in WT1‐mutant Wilms' tumors. Am J Pathol 20041651943–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Little S, Hanks S, King‐Underwood L, Picton S, Cullinane C, Rapley E, Rahman N, Pritchard‐Jones K. A WT1 exon 1 mutation in a child diagnosed with Denys–Drash syndrome. Pediatr Nephrol 20052081–85. [DOI] [PubMed] [Google Scholar]
- 11.Kreidberg J A, Sariola H, Loring J M, Maeda M, Pelletier J, Housman D, Jaenisch R. WT‐1 is required for early kidney development. Cell 199374679–691. [DOI] [PubMed] [Google Scholar]