Abstract
Background
Eotaxin (CCL11) is a small protein produced in the lungs of patients with asthma, and is a potent chemoattractant for eosinophils.
Aim
To elucidate the role of eotaxin in asthma by an association study of functional and novel eotaxin polymorphisms in case–control and family‐based study designs.
Methods
Eotaxin +67G/A, –384A/G and –426C/T single‐nucleotide polymorphisms and a hexanucleotide (GAAGGA)n repeat 10.9 kb upstream of the gene were genotyped in a cohort of age, sex and ethnically matched patients with asthma (n = 235) and healthy controls (n = 239), and also in a study population of 230 families with asthma recruited from north/northwest India. Total serum IgE (TsIgE) and plasma eotaxin levels were measured using ELISA.
Results
+67G/A polymorphism was found to be significantly associated with asthma in case–control (p = 0.009) and family‐based studies (p = 0.006). Its functional role, as it was correlated with plasma eotaxin levels (p = 0.006), was also demonstrated. Further, –384C/T single‐nucleotide polymorphism was found to be significantly associated with log10 TsIgE (p = 0.016 in case–control and p = 0.018 in families) and eotaxin levels (p = 0.007). Most interestingly, for the first time, a highly significant association of the newly studied (GAAGGA)n hexanucleotide repeat with asthma (p = 3×10−6), log10TsIgE (p = 0.006) and eotaxin levels (p = 0.004) was observed. G_A_C_8 was also identified as an important risk haplotype associated with high TsIgE and plasma eotaxin levels.
Conclusions
This study provides further evidence that eotaxin polymorphisms are associated with the development of asthma by regulating eotaxin levels and reinforces towards the scanning of other chemokine genes present at 17q21 locus for their association with asthma and related phenotypes.
Eosinophils play a major role in the pathogenesis of allergic diseases including asthma by releasing various granular proteins, reactive NO, cytokines and chemokines at the site of inflammation, thus causing tissue damage.1 Interplay between chemokines and their receptors are considered to be crucial for the trafficking of eosinophil and other lymphocytes from the circulation to the bronchoalveolar spaces of the patients with asthma.2 Eotaxin (chemokine, CC motif, ligand; CCL11) is a predominant eosinophil chemoattractant, which binds to chemokine receptor 3.3,4 Chemokine receptor 3 is also present on the Th2 CD4+ lymphocytes, basophils, dendritic cells and mast cells.5,6 Thus, the role of eotaxin in asthma is not confined to eosinophil migration and activation only, but extended to many other effector cells involved in disease pathogenesis. It has also been observed that along with interleukin 5, eotaxin prolongs the viability of eosinophils.7 Eotaxin mRNA and protein are found to be elevated in the induced sputum, bronchial epithelium and airways of the patients with moderate to severe asthma.8,9,10 Bronchoalveolar lavage fluid of the patients with asthma also showed increased levels of eotaxin after allergen inhalation.11 In addition, higher plasma eotaxin levels have been observed in subjects with symptoms of acute asthma and airflow obstruction than subjects with stable asthma. Plasma eotaxin levels have been correlated with severity of asthma even in the presence of steroid treatment.12,13
Eotaxin gene is present on chromosome 17q21.1, where linkage with asthma and related phenotypes has been previously reported in various ethnic populations.14,15 Previous studies on eotaxin gene polymorphisms and asthma remain inconclusive as most of them failed to establish a strong association.16,17,18,19,20 However, in few other studies, eotaxin single‐nucleotide polymorphisms (SNPs) have been correlated significantly with asthma‐associated phenotypes including lung function, serum IgE, circulating blood eosinophils, eosinophil migration and activation, and plasma eotaxin levels.16,17,18,21,22 Studies have also been undertaken to demonstrate the functional implication of various promoter and exonic variants of the eotaxin.20,22 Notably, the variant and the direction of association are inconsistent across different ethnic populations owing to some inherent reasons. However, no such studies have yet been undertaken in an ethnically divergent Indian population.
Importantly, the 17q11–17q21 chromosomal region harbours many other important chemokines including RANTES, MCP1, MCP3 and so on, and also the gene for inducible nitric oxide synthase. Recently, we have reported the association of the gene for inducible nitric oxide synthase microsatellite repeats with asthma and related phenotypes in an Indian population, demonstrating the importance of 17q region in asthma predisposition.23 The results of our previous study and the well‐documented role of eotaxin in asthma prompted us to undertake an association study of eotaxin gene with asthma and associated phenotypes in case–control and family‐based study designs. We have also measured the plasma eotaxin level and attempted to correlate it with eotaxin gene variants.
Methods
Patients, healthy volunteers and family recruitment
In a multicentre‐based asthma‐genetics study programme, unrelated patients were recruited from various collaborating hospitals in northern and northwestern India (n = 235). Ethical approval was obtained from the review board of each hospital. Written informed consent was taken from all individuals (parents in case of minors) participating in the study. Asthma, in the recruited study population was defined by clinical history and later validated by interviews as described previously.23 Patients were diagnosed for asthma on the basis of National Asthma Education and Prevention Program (Expert Panel Report‐2) guidelines and were examined for a self‐reported history of breathlessness and wheezing. Each patient showed airway reversibility as documented by an inhalant bronchodilator‐induced improvement of >15% (using albuterol/salbutamol). Fifteen common environmental allergens (house dust mite, Amaranthus spinosus, Brassica campestris, Cynodon dactylon, Parthenium hysterophorus, Proposis juliflora, Ricinus communis, Alternaria tenuis, Aspergillus fumigatus, cockroach, mosquito, moth, grain dust rice, hay dust and house dust) were used for the skin prick test (SPT). Specific IgE was estimated for the children, where SPT could not be performed. Atopy was defined as having wheal reaction equal to or greater than histamine (3 mm diameter) or high specific IgE (3 times of –ve control) for at least one allergen. Total serum IgE (TsIgE) levels were estimated using ELISA. Only individuals with a family history of asthma/atopy were included in the study. In the interviews conducted, details of environmental factors, geographical region of origin and migration status were also obtained.
Age‐matched and sex‐matched healthy volunteers (referred to as normal controls; n = 239) who answered negatively to a screening questionnaire for respiratory symptoms, and on the basis of the criteria of having no symptoms or history of allergic diseases were recruited from the general population. Pulmonary function tests and SPT were performed, when consent was obtained. Individuals having a history of smoking and parasitic/helminthic infestations in the past 3 years were excluded from the study.
A total of 230 families with asthma (3–12 individuals per family) were recruited by a group of doctors in the follow‐up family visits of the probands recognised by a primary physician.23 Diagnosis of asthma was made, as discussed above, for all the family members after obtaining their consent. Study design was made in such a way that at least one child with asthma along with both the parents were recruited. Families were further extended wherever other members gave their consent to participate. Thus, a total of 918 individuals were recruited with an average size of 3.94 (3–12) individuals per family.
Plasma eotaxin levels measurement
To determine eotaxin levels in the plasma, we used the eotaxin ELISA Kit (R&D Systems, Minneapolis, Minnesota, USA) as per the manufacturer's instructions.
Genomic DNA isolation
DNA was isolated from peripheral white blood cells using the modified salting out method as described,23 and was stored at −20°C until further analysis.
Genotyping of repetitive sequences
Putative repetitive sequences in and around the eotaxin gene (NT_010799) were identified using the RepeatMasker software (http://www.repeatmasker.org/). PCR was carried out in a total volume of 5 μl containing 25 ng of genomic DNA, 0.5 pmol each of a 6‐FAM‐labelled forward primer and a non‐labelled reverse primer, 1.5 mM MgCl2, 0.25 mM of each dNTP, 0.03U/μl of Taq DNA polymerase (Bangalore Genie, Bangalore, India) and the buffer recommended by the manufacturer. Fragment lengths were determined using the GeneMapper V.3.5 (Applied Biosystems, Foster City, California, USA). The number of repeats was determined by sequencing the PCR fragments from individuals (n = 5) homozygous for one allele.
PCR amplification and genotyping of the SNPs
Two promoter (–426C/T, –384A/G), one exonic (+67G/A) and one intronic (+1599G/A) SNPs were investigated in the study population using primers detailed in supplementary table 1 (available online at http://jmg.bmj.com/supplemental). The –384A/G and +1599G/A polymorphisms were studied using SNaPshot ddNTP Primer Extension Kit (Applied Biosystems). These samples were subsequently electrophoresed using the ABI Prism 3100 Genetic Analyzer (Applied Biosystems) as per the manufacturer's instructions. The –426C/T and +67G/A polymorphisms were assessed using Taqα1 and Bsr1 restriction endonuclease digestion, respectively. Genomic DNA from 40 individuals was also sequenced to verify the genotyping.
Table 1 Characteristics of the patient and the control groups (case–control study)* and the probands (family‐based study).
| Patients (n = 235) | Controls (n = 239) | Probands (n = 230) | |
|---|---|---|---|
| Native place* | North/northwest India | North/northwest India | North/northwest India |
| Mean (SD) age (years) | 34.4 (12.5) | 35.0 (10.6) | 15.6 (11.2) |
| Sex ratio (F:M) | 48:52 | 47:53 | 44:56 |
| Familial history of asthma/ | All | None | All |
| atopy† | |||
| Smoking history‡ | None | None | None |
| Reversibility (%) from baseline FEV1 (after β2 agonist usage) | >15% | ND | >15% |
| Log10 mean (SD) serum total IgE (IU/ml) | 2.84 (0.49) | 2.30 (0.54) | 2.84 (0.63) |
| Atopy/self‐reported history of allergies | All | None | All§ |
FEV1, forced expiratory volume in one second; ND, test not done.
*Patients and controls were recruited from Delhi, Lucknow (UP), Jaipur (Rajasthan), Shimla (HP), Chandigarh (Punjab), Mumbai and Guwahati (Assam).
†Control individuals were subjected to a questionnaire so as to eliminate all individuals having atopic disorders or family history of atopic disorders.
‡Patients and controls known to have experienced smoking in the past, or having parasitic infections, were excluded from the study.
§86% of the probands were found to be SPT +ve or had high specific serum IgE.
Parentheses contain the values for standard deviation (SD).
Statistical analysis
Linkage disequilibrium (LD) between all the identified SNPs and repeat polymorphism was evaluated using EMLD (https://cge.mdanderson.org/˜qhuang/Software/pub.htm). An association analysis, based on the case–control design, was performed using the Armitage trend test following the guidelines given by Sasieni24 as implemented in the FINETTI program. We tested whether the genotype distribution was in HWE using FINETTI. CLUMP2.3 was used to study the association of microsatellite repeat with asthma.25 Analysis of variance (ANOVA) was carried out to test the effect of these polymorphisms on total serum IgE levels and plasma eotaxin levels. Haplotypes of each individual were inferred together for cases and controls using the algorithm (PHASE) developed by Stephens et al.26 Differences in haplotype frequencies in cases and controls were compared using a Monte Carlo approach as implemented in CLUMP2.3.25 Odds ratios (OR) were calculated wherever required.
Family‐based association test (FBAT) (http://www.biostat.harvard.edu/˜fbat/fbat.htm) was used to evaluate the association in the presence of linkage for the binary traits of asthma or atopy using additive and dominant models. Haplotype‐based association testing (HBAT) option was used for the haplotypic association studies. Quantitative trait disequilibrium test (http://www.sph.umich.edu:80/csg/abecasis/QTDT) was used to test the total evidence for association of IgE and various eotaxin variants in families with asthma.
Results
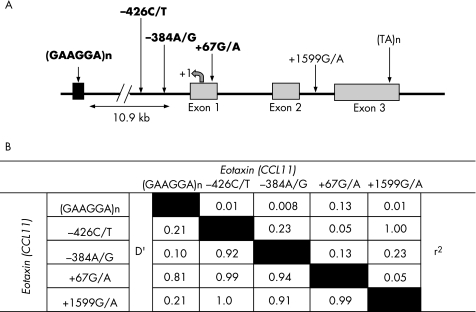
To verify the SNPs and repetitive sequences in and around the eotaxin gene, we sequenced the DNA samples from 40 randomly selected individuals and calculated the LD among the identified polymorphic eotaxin variants (fig 1). The –426C/T and +1599G/A SNPs were found to be in complete LD (D′ = 1, r2 = 1); thus, the +1599 G/A SNP was exempted from further genotyping. Also, the (TA)n repeat was found to be non‐polymorphic, so was not genotyped further. Therefore, a total of three SNPs viz –426C/T, –384A/G and +67G/A, and a (GAAGGA)n repeat were genotyped from patients and controls and from all the recruited family members of the probands with asthma to establish the association of eotaxin gene and asthma. Table 1 summarises the demographic characteristics such as age, sex and other clinical parameters of the study groups.
Figure 1 (A) The schematic representation of eotaxin gene on chromosome 17q21.2 with single‐nucleotide polymorphism (SNP) locations and the flanking microsatellite repeat. The SNPs studied in our population are marked in bold. (B) Pairwise linkage disequilibrium (LD) for all two‐way comparisons among the five polymorphisms investigated in the eotaxin (CCL11) gene as calculated using EMLD.
Association of eotaxin polymorphisms with asthma/atopy
We have identified a polymorphic (GAAGGA)n hexanucleotide repeat, 10.9 kb upstream of the eotaxin gene and have used it for further genotyping in two independent study populations of case–controls and families with asthma. A total of 10 alleles were obtained with a heterozygosity index = 0.814 in our study population (table 2). A highly significant difference was obtained in the allele count in the patients and the controls, when analysed using CLUMP22 with one million simulations (normal χ2 (T1) = 42.36, df = 9, p = 3×10−6 and χ2 from table after collapsing columns with small expected values together (T2) = 40.13, df = 6, p = 10−6). The allele with eight repeats (GAAGGA)8 was found to be most significantly different when compared between patients and controls (χ2 (T3) = 25.6, df = 1, p = 2×10−6). The distribution was significant even after Bonferroni correction (α = 0.008). A significant transmission distortion was also observed for the allele with eight repeats in our FBAT analysis. The association was more significant when atopic individuals, along with patients with asthma, were considered as diseased for the analysis (table 2).
Table 2 Allele frequencies and the results of association analysis of (GAAGGA)n repeat polymorphism in eotaxin gene.
| Repeat size (PCR product size) | Case–control studies | FBAT (p Value) | ||||||
|---|---|---|---|---|---|---|---|---|
| Additive model | Dominant model | |||||||
| Cases | Controls | OR (95% CI) | p Value | Asthma | Atopy* | Asthma | Atopy† | |
| 4 (148) | 0 (0) | 1 (0.21) | – | – | – | – | – | – |
| 5 (154) | 104 (22.22) | 76 (15.97) | 1.5 (1.08 to 2.09) | 0.014 | 0.450 | 0.470 | 0.368 | 0.505 |
| 6 (160) | 0 (0) | 1 (0.21) | – | – | – | – | – | – |
| 7 (166) | 2 (0.43) | 1 (0.21) | – | – | – | – | – | – |
| 8 (172)† | 65 (13.89) | 21 (4.41) | 3.5 (2.1 to 5.81) | 0.000 | 0.016 | 0.005 | 0.006 | 0.002 |
| 9 (178) | 10 (2.14) | 21 (4.41) | – | 0.056 | 0.011 | 0.089 | 0.018 | |
| 10 (184) | 138 (29.49) | 162 (34.03) | 0.6 (0.38 to 0.95) | NS | 0.289 | 0.333 | 0.424 | 0.381 |
| 11 (190) | 32 (6.84) | 52 (10.92) | – | 0.03 | 0.721 | 0.530 | 0.909 | 0.609 |
| 12 (196) | 46 (9.83) | 61 (12.82) | – | NS | 0.117 | 0.170 | 0.055 | 0.091 |
| 13 (202) | 71 (15.17) | 80 (16.81) | – | NS | 0.755 | 1.000 | 0.896 | 0.935 |
| Overall p value | 0.001 | 0.145 | 0.043 | 0.097 | 0.037 | |||
FBAT, family‐based association test; NS, non‐significant; PCR, polymerase chain reaction
*The members without asthma but with atopy of the recruited families were also considered as diseased for the FBAT analysis.
†Significant after Bonferroni correction (α = 0.008 for case–control and α = 0.007 for FBAT after Bonferroni correction for the six and seven tests, respectively).
Numbers in parentheses indicate the frequency (%).
Table 3 depicts the genotype frequencies for the –426C/T, –384A/G and +67G/A polymorphisms. The distributions of genotype frequencies of these three SNPs were consistent with Hardy–Weinberg expectations in patients and controls (p>0.05). Using Armitage's trend test for association, the genotypic distribution of +67G/A SNP was found to be significantly different in patients and controls (χ2 = 6.91, p = 0.009, common OR = 1.96). Also, in our FBAT analysis, significant association was observed when analysed using a dominant model (p = 0.006 for asthma and p = 0.006 for atopy), but not in an additive model (p = 0.75 for asthma and p = 0.77 for atopy; table 4).
Table 3 Genotype/haplotype frequencies, log10 total serum IgE and plasma eotaxin levels with respect to +67G/A, –384A/G and –427C/T polymorphisms and the risk haplotype in eotaxin gene.
| Genotypes | Genotype frequencies of different genotypes in case–control studies | †Log10 TsIgE levels with respect to different genotypes | Plasma eotaxin levels with respect to different genotypes | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patients* | Controls* | Common OR | p Value | Log10 TsIgE levels (±SE) | No of individuals (n) | Overall p Value | Plasma eotaxin levels (±SE) (pg/ml) | No. of individuals (n) | Overall p Value | |
| +67G/A | ||||||||||
| AA | 0 (0.0) | 2 (0.84) | 2.00 (0.41) | 2 | 7 (75) | 1 | ||||
| GA | 47 (20.35) | 70 (29.54) | 1.96 | 0.0086 | 2.55 (0.06) | 104 | 0.42 | 113 (13) | 34 | 0.0057 |
| GG | 184 (79.65) | 165 (69.62) | 2.54 (0.04) | 301 | 153 (7) | 104 | ||||
| −384A/G | ||||||||||
| GG | 55 (23.5) | 57 (24.05) | 2.41 (0.06) | 107 | 124 (13) | 33 | ||||
| GA | 127 (54.27) | 105 (44.3) | 0.843 | 0.176 | 2.58 (0.04) | 202 | 0.016 | 134 (9) | 71 | 0.0072 |
| AA | 52 (22.22) | 75 (31.65) | 2.61 (0.06) | 100 | 177 (13) | 35 | ||||
| −427C/T | ||||||||||
| TT | 14 (5.96) | 16 (6.69) | 2.52 (0.12) | 26 | 128 (29) | 7 | ||||
| CT | 85 (36.17) | 84 (35.15) | 0.975 | 0.936 | 2.51 (0.05) | 138 | 0.58 | 134 (12) | 43 | 0.43 |
| CC | 136 (57.87) | 139 (58.16) | 2.57 (0.04) | 248 | 149 (8) | 88 | ||||
| Haplotype | ||||||||||
| GAC8/GAC8 | 7 (3.13) | 0 (0) | 3.15 (0.24) | 6 | 336 (52) | 2 | ||||
| GAC8/Non GAC8 | 47 (20.98) | 20 (8.93) | 5.01 | <0.0001 | 2.65 (0.08) | 59 | 0.013 | 161 (8) | 80 | <0.0001 |
| Non GAC8/Non GAC8 | 170 (75.89) | 204 (91.07) | 2.52 (0.03) | 324 | 134 (5) | 192 | ||||
TsIgE, total serum IgE.
*Numbers in parentheses indicate the frequency (%).
†Analysis with Log10 TsIgE (total serum IgE) levels was done irrespective of the asthma status.
Table 4 Family‐based association test for +67G/A, −384A/G and −427C/T polymorphisms studied in eotaxin gene with asthma and atopy phenotype.
| Allele | Asthma | Atopy* | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Additive model | Dominant model | Additive model | Dominant Model | |||||||||||||||
| Families | S | E (S) | p Value | Families | S | E (S) | p Value | Families | S | E (S) | p Value | Families | S | E (S) | p Value | |||
| +67G/A | ||||||||||||||||||
| A | 86 | 61 | 63 | 0.76 | 89 | 62 | 60.25 | 0.77 | 83 | 65 | 67 | 0.77 | 89 | 73 | 71 | 0.76 | ||
| G | 86 | 177 | 175 | 14 | 19 | 15.25 | 0.005 | 83 | 191 | 189 | 16 | 27 | 23 | 0.006 | ||||
| −384A/G | ||||||||||||||||||
| A | 123 | 180 | 183.5 | 0.74 | 110 | 110 | 105.75 | 0.51 | 130 | 208 | 214 | 0.55 | 114 | 133 | 128.75 | 0.54 | ||
| G | 123 | 188 | 184.5 | 102 | 107 | 99.25 | 0.17 | 130 | 224 | 218 | 107 | 131 | 120.75 | 0.1 | ||||
| −427C/T | ||||||||||||||||||
| C | 108 | 222 | 223.5 | 0.84 | 29 | 32 | 33 | 0.77 | 113 | 254 | 256 | 0.81 | 31 | 42 | 43.75 | 0.63 | ||
| T | 108 | 96 | 94.5 | 110 | 84 | 83.5 | 0.94 | 113 | 116 | 114 | 113 | 101 | 100.75 | 0.97 | ||||
FBAT, family‐based association test.
*The members without asthma but with atopy of the recruited families were also considered as diseased.
The null hypothesis of no association in the presence of linkage was tested using –e option of the FBAT.
Association of eotaxin polymorphisms with total serum IgE levels
TsIgE values were converted to log10 scale for the analysis and were found to follow the normal distribution. The genetic effects of eotaxin polymorphisms were tested on the log10 TsIgE levels in the case–control cohort impertinence to their asthmatic status. No significant association was detected with respect to +67G/A and –426C/T SNPs. Similar results were obtained when families with asthma were analysed for the total evidence of association using QTDT (p>0.05). Nevertheless, a marginally significant association was observed for the –384A/G polymorphism (f = 3.86, df = 2, p = 0.016 using analysis of variance and p = 0.018 using QTDT). A stronger association was observed with (GAAGGA)n hexanucleotide repeat polymorphism, when analysed using analysis of variance (F ratio = 2.9, df = 7, p = 0.006), and was also found to be marginally associated using QTDT (p = 0.05). Allele (GAAGGA)8 was associated with high log10 TsIgE in both the analysis (see supplementary figure 1 available online at http://jmg.bmj.com/supplemental).
Association of eotaxin haplotypes with asthma and TsIgE levels
A total of 38 haplotypes were generated using PHASE and HBAT. However, only eight haplotypes were present at frequencies above 5% in the population (see supplementary figure 2 available online at http://jmg.bmj.com/supplemental). Since the counts per haplotypes were very low, Clump22 was used with 1 000 000 Monte Carlo simulations. On comparing the overall haplotype frequencies, highly significant differences were observed between the cases and controls (normal T1 χ2 = 79.03, df = 37, p = 0.001 and maximum (T4) χ2 = 43.39, p = 0.001). Individually, the haplotype G_A_C_8 showed the highest difference in the frequency in patients versus controls. The odds of patients rather than that of controls having the haplotypes G_A_C_8 was 3.52 with 95% CI 2.08 to 5.92 (Likelihood χ2 = 25.82, df = 1, p = 0.000). This haplotype was also found to be overtransmitted to the offspring with asthma in family studies (p = 0.01). Thus, the haplotype G_A_C_8 is an important risk/susceptibility haplotype, and is positively associated with asthma.
Furthermore, when we analysed haplotypes with respect to log10 TsIgE, this risk haplotype (G_A_C_8) was found to be associated with TsIgE levels (F ratio = 4.4, df = 2, p = 0.013). The individuals homozygous for this haplotype had higher log10 TsIgE levels as compared with the heterozygous individuals (table 3).
Functional correlation of eotaxin polymorphisms with its plasma levels
To find any functional correlation of eotaxin gene polymorphisms with its level of expression, the levels of eotaxin in plasma samples of 140 unrelated patients were estimated. The +67G/A polymorphism was found to affect the plasma eotaxin levels. The presence of G allele is associated with higher concentrations of eotaxin (F ratio = 8.3, df = 1, p = 0.004), and this effect seemed to be dose dependent as the GA heterozygotes had higher than AA but lower mean levels of eotaxin then the GG homozygotes (F ratio = 5.4, df = 2, p = 0.006) (table 3). The –384A/G also showed a significant association with plasma eotaxin levels (F ratio = 5.1, df = 2, p = 0.007). The eotaxin levels were found to be significantly different with different alleles of the (GAAGGA)n repeat (F ratio = 3.3, df = 6, p = 0.004). Particularly, alleles (GAAGGA)11 and 12 were associated with low eotaxin levels, whereas alleles with repeat sizes (GAAGGA)8 and 10 had higher levels (see supplementary figure 1 available online at http://jmg.bmj.com/supplemental).
Further, when haplotypes (counts ⩾5) were analysed for eotaxin levels, a significant difference was observed (F ratio = 3.6, df = 6, p = 0.001). Individuals homozygous for the risk haplotype G_A_C_8 had the higher eotaxin levels when compared with individuals heterozygous and homozygous for other haplotypes (table 3).
Discussion
To elucidate the role of eotaxin in asthma, we evaluated various known eotaxin SNPs along with a hexanucleotide repeat, 10.9 kb upstream of eotaxin gene, for their association with asthma/atopy, TsIgE and eotaxin levels. As there are evidences for transcriptional regulation of eotaxin expression, we focused our study on various promoter polymorphisms and a non‐synonymous variant (+67G/A), which have already been shown to be linked to eotaxin levels or to have some functional role in the regulation of gene expression.20,22 We have found a significant association of +67 G/A SNP, which causes a single amino acid change from alanine (G) to threonine (A) at the signal peptide cleavage site, with asthma in our case–control studies. The p value for the null hypothesis of no association in the presence of linkage was also significant in the dominant model of FBAT analysis for asthma and atopy phenotypes. It is important to mention here that the association between eotaxin variants and atopy in our analysis might be biased by asthma, as our sample collection was focused on probands with asthma. Nevertheless, these results present an important piece of information as individuals with atopy could be predisposed to asthma after getting appropriate environmental stimuli. Our results are supported by the findings of Nakamura et al20 and Tsunemi et al,21 who also showed a significant association of +67 G/A with lung‐function test in a disperse and ethnically divergent US population, and with atopic dermatitis in a Japanese population, respectively. We further demonstrated the protective role of +67 G/A‐Thr variant, as the individuals with A allele were found to have lower mean eotaxin levels in their plasma. Although associated with eotaxin levels, this particular SNP was not found to be associated with log10 TsIgE levels. Nevertheless, the promoter –384C/T SNP was associated with log10 TsIgE as well as eotaxin levels. Similar results were obtained in a study on Japanese patients with atopic dermatitis, but was not reproduced in other studies.21 Probable reasons for the observed inconsistencies in the results include different ethnic background (genetic make‐up), differential age‐grouped patients, differential selection criteria for the control population and different study designs. For example, we have excluded patients and controls who were smoking in the past 3 years because we have also done the genetic analysis with TsIgE levels, a trait affected by the smoking status of an individual. This selection also helped us to exempt the cases of intrinsic asthma. Similarly, we have recruited the patients and controls on the basis of their family history of asthma/atopy (presence/absence) to exclude any misdiagnosis of the disease (http://www.nhlbi.nih.gov/guidelines/asthma/asthgdln.pdf). Further, most of the asthma association studies on eotaxin gene conducted to date are case–control studies, except for the recently published study by Raby et al.16 In general, case–control studies could provide better directions on promising loci because of their ample power, but at times might be affected by cryptic population substructuring/stratification, resulting in spurious positive/negative results.27 On the other hand, asthma being a complex genetic disorder, due to small effects of individual genes, a family‐based study may fail to detect significant association even when it exists.28 Thus, herein we have undertaken both the designs simultaneously. We have tried to rule out the possibility of population stratification in our case–control studies by recruiting the age, sex and ethnicity‐matched patients, and controls by taking the comprehensive information about their geographical area, ethnic background and migration status. Replication of our results in a more robust family‐based study design adds further confidence to our results. Thus, it is very unlikely that our results are due to stratification or an inherent statistical bias.
Our results with respect to the (GAAGGA)n repeat are interesting as we have demonstrated the association of (GAAGGA)8 allele with asthma, atopy, TsIgE and plasma eotaxin levels for the first time. Although a strong association between plasma eotaxin levels and hexanucleotide repeat has been observed, its causal relationship with asthma remains to be elucidated. Some of the earlier studies have provided evidence for the involvement of (TCCTTC)n and (CTTCCT)n hexanucleotide repeats in chromosomal loop formation.29 Thus, we speculate that the (GAAGGA)n repeat could be involved in the transcriptional regulation of eotaxin gene by altering the DNA secondary structure. Nevertheless, the possibility of the presence of any other functional polymorphisms present on the 17q21 region in linkage disequilibrium with the (GAAGGA)8 allele could not be overlooked.
To further understand the contributions of these genetic variants to asthma, we constructed four locus haplotypes using PHASE and HBAT in case–control and family studies, respectively. A novel haplotype, G_A_C_8, was found to be associated with asthma/atopy and IgE in both the analyses, and was also associated with plasma eotaxin levels. However, to gain an insight into tissue‐specific expression of eotaxin, these results need to be further validated using bronchoalveolar lavage fluid/lung biopsy/sputum samples.
In conclusion, for the first time, we have identified a hexanucleotide repeat and a novel risk haplotype associated with asthma, atopy, serum IgE and plasma eotaxin levels. As three of the four polymorphisms in our study are associated with plasma eotaxin levels and are also present in LD with each other, it is difficult to state which one of them is directly regulating the eotaxin gene expression. Some confounding in vitro experiments are required to confirm the functional role of the associated variant, specifically (GAAGGA)n hexanucleotide repeat, as no studies so far have been undertaken to elucidate its functional role. The results of our haplotype analysis also reinforce future studies towards the scanning of other chemokine genes, viz, CCL1, CCL2, CCL7, CCL8 and CCL13, present on the same chromosomal locus for their association with asthma and related phenotypes.
Key points
We investigated the association of eotaxin gene variants with the risk of asthma, total serum IgE (TsIgE) and plasma eotaxin levels using case–control and family‐based association studies.
We established a significant association of a newly studied (GAAGGA)n hexanucleotide repeat and the +67G/A single‐nucleotide polymorphism with asthma. Also, the (GAAGGA)n repeat and the –384C/T single‐nucleotide polymorphism were found to be significantly associated with TsIgE levels. We demonstrated further the functional significance of these variants by correlating them with plasma eotaxin levels. Moreover, we identified an important risk haplotype, G_A_C_8, associated with high TsIgE and plasma eotaxin levels.
We concluded that eotaxin polymorphisms could be associated with the development of asthma as well as TsIgE levels by regulating eotaxin levels.
Supplementary table 1 and supplementary figures 1 and 2 are available at http://jmg.bmj.com/supplemental
Acknowledgements
We acknowledge the Department of Biotechnology (DBT project, GAP‐0014) and Council of Scientific and Industrial Research (Task Force project‐SMM0006), Government of India for financial assistance. JB acknowledges CSIR for her fellowship. We thank all participating clinicians and volunteers for helping in this study. We also thank Mr A Kumar, Dr U Mabalirajan, Mr Rajshekhar Chatterjee, Dr Naresh Singh and Ms Deepti Maan for their assistance.
Abbreviations
FBAT - family‐based association test
HBAT - haplotype‐based association testing
LD - linkage disequilibrium
QTDT - quantitative trait disequilibrium test
SNP - single‐nucleotide polymorphism
SPT - skin prick test
TsIgE - total serum IgE
Footnotes
Competing interests: None.
Supplementary table 1 and supplementary figures 1 and 2 are available at http://jmg.bmj.com/supplemental
References
- 1.Rothenberg M E, Hogan S P. The eosinophil. Annu Rev Immunol 200624147–174. [DOI] [PubMed] [Google Scholar]
- 2.Broide D, Sriramarao P. Eosinophil trafficking to sites of allergic inflammation. Immunol Rev 2001179163–172. [DOI] [PubMed] [Google Scholar]
- 3.Rothenberg M E. Eotaxin. An essential mediator of eosinophil trafficking into mucosal tissues. Am J Respir Cell Mol Biol 199921291–295. [DOI] [PubMed] [Google Scholar]
- 4.Pease J E, Williams T J. Eotaxin and asthma. Curr Opin Pharmacol 20011248–253. [DOI] [PubMed] [Google Scholar]
- 5.Uguccioni M, Mackay C R, Ochensberger B, Loetscher P, Rhis S, LaRosa G J, Rao P, Ponath P D, Baggiolini M, Dahinden C A. High expression of the chemokine receptor CCR3 in human blood basophils: role in activation by eotaxin, MCP‐4, and other chemokines. J Clin Invest 19971001137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sallusto F, Mackay C R, Lanzavecchia A. Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science 19972772005–2007. [DOI] [PubMed] [Google Scholar]
- 7.Kudlacz E, Whitney C, Andresenl C, Conklyn M. Functional effects of eotaxin are selectively upregulated on IL‐5 transgenic mouse eosinophils. Inflammation 200226111–119. [DOI] [PubMed] [Google Scholar]
- 8.Hadjicharalambous C, Dent G, May R D, Handy R L, Anderson I K, Davies D E, Djukanovic R. Measurement of eotaxin (CCL11) in induced sputum supernatants: validation and detection in asthma. J Allergy Clin Immunol 2004113657–662. [DOI] [PubMed] [Google Scholar]
- 9.Miotto D, Christodoulopoulos P, Olivenstein R, Taha R, Cameron L, Tsicopoulos A, Tonnel A B, Fahy O, Lafitte J J, Luster A D, Wallaert B, Mapp C E, Hamid Q. Expression of IFN‐gamma‐inducible protein; monocyte chemotactic proteins 1, 3, and 4; and eotaxin in TH1‐ and TH2‐mediated lung diseases. J Allergy Clin Immunol 2001107664–670. [DOI] [PubMed] [Google Scholar]
- 10.Lamkhioued B, Renzi P M, Abi‐Younes S, Garcia‐Zepada E A, Allakhverdi Z, Ghaffar O, Rothenberg M D, Luster A D, Hamid Q. Increased expression of eotaxin in bronchoalveolar lavage and airways of asthmatics contributes to the chemotaxis of eosinophils to the site of inflammation. J Immunol 19971594593–4601. [PubMed] [Google Scholar]
- 11.Brown J R, Kleimberg J, Marini M, Sun G, Bellini A, Mattoli S. Kinetics of eotaxin expression and its relationship to eosinophil accumulation and activation in bronchial biopsies and bronchoalveolar lavage (BAL) of asthmatic patients after allergen inhalation. Clin Exp Immunol 1998114137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura H, Weiss S T, Israel E, Luster A D, Drazen J M, Lilly C M. Eotaxin and impaired lung function in asthma. Am J Respir Crit Care Med 19991601952–1956. [DOI] [PubMed] [Google Scholar]
- 13.Tateno H, Nakamura H, Minematsu N, Nakajima T, Takahashi S, Nakamura M, Fukunaga K, Asano K, Lilly C M, Yamaguchi K. Plasma eotaxin level and severity of asthma treated with corticosteroid. Respir Med 200498782–790. [DOI] [PubMed] [Google Scholar]
- 14.Bu L M, Bradley M, Soderhall C, Wahlgren C F, Kockum I, Nordenskjold M. Susceptibility loci for atopic dermatitis on chromosome 21 in a Swedish population. Allergy 200661617–621. [DOI] [PubMed] [Google Scholar]
- 15.Koppelman G H, Stine O C, Xu J, Howard T D, Zheng S L, Kauffman H F, Bleecker E R, Meyers D A, Postma D S. Genome‐wide search for atopy susceptibility genes in Dutch families with asthma. J Allergy Clin Immunol 2002109498–506. [DOI] [PubMed] [Google Scholar]
- 16.Raby B A, Van Steen K, Lazarus R, Celedon J C, Silverman E K, Weiss S T. Eotaxin polymorphisms and serum total IgE levels in children with asthma. J Allergy Clin Immunol 2006117298–305. [DOI] [PubMed] [Google Scholar]
- 17.Chae S C, Lee Y C, Park Y R, Shin J S, Song J H, Oh G J, Hong S T, Pae H O, Choi B M, Chung H T. Analysis of the polymorphisms in eotaxin gene family and their association with asthma, IgE, and eosinophil. Biochem Biophys Res Commun 2004320131–137. [DOI] [PubMed] [Google Scholar]
- 18.Shin H D, Kim L H, Park B L, Jung J H, Kim J Y, Chung I Y, Kim J S, Lee J H, Chung S H, Kim Y H, Park H S, Choi J H, Lee Y M, Park S W, Choi B W, Hong S J, Park C S. Association of eotaxin gene family with asthma and serum total IgE. Hum Mol Genet 2003121279–1285. [DOI] [PubMed] [Google Scholar]
- 19.Miyamasu M, Sekiya T, Ohta K, Ra C, Yoshie O, Yamamoto K, Tsuchiya N, Tokunaga K, Hirai K. Variations in the human CC chemokine eotaxin gene. Genes Immun 20012461–463. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura H, Luster A D, Nakamura T, In K H, Sonna L A, Deykin A, Israel E, Drazen J M, Lilly C M. Variant eotaxin: its effects on the asthma phenotype. J Allergy Clin Immunol 2001108946–953. [DOI] [PubMed] [Google Scholar]
- 21.Tsunemi Y, Saeki H, Nakamura K, Sekiya T, Hirai K, Fujita H, Asano N, Tanida Y, Kakinuma T, Wakugawa M, Torii H, Tamaki K. Eotaxin gene single nucleotide polymorphisms in the promoter and exon regions are not associated with susceptibility to atopic dermatitis, but two of them in the promoter region are associated with serum IgE levels in patients with atopic dermatitis. J Dermatol Sci 200229222–228. [DOI] [PubMed] [Google Scholar]
- 22.Chang H S, Kim J S, Lee J H, Cho J I, Rhim T Y, Uh S T, Park B L, Chung I Y, Park C S, Shin H D. A single nucleotide polymorphism on the promoter of eotaxin1 associates with its mRNA expression and asthma phenotypes. J Immunol 20051741525–1531. [DOI] [PubMed] [Google Scholar]
- 23.Batra J, Singh T P, Mabalirajan U, Sinha A, Prasad R, Ghosh B. Association of inducible nitric oxide synthase (iNOS) with asthma severity, total serum IgE and blood eosinophil levels. Thorax 20076216–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasieni P D. From genotypes to genes: doubling the sample size. Biometrics 1997531253–1261. [PubMed] [Google Scholar]
- 25.Sham P C, Curtis D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet 19955997–105. [DOI] [PubMed] [Google Scholar]
- 26.Stephens M, Smith N J, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 200168978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hintsanen P, Sevon P, Onkamo P, Eronen L, Toivonen H. An empirical comparison of case‐control and trio based study designs in high throughput association mapping. J Med Genet 200643617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Gastwirth J L, Gail M H. Power and related statistical properties of conditional likelihood score tests for association studies in nuclear families with parental genotypes. Ann Hum Genet 200569296–314. [DOI] [PubMed] [Google Scholar]
- 29.Hampel K J, Ashley C, Lee J S. Kilobase‐range communication between polypurine polypyrimidine tracts in linear plasmids mediated by triplex formation: a braided knot between two linear duplexes, Biochemistry 1994335674–5681. [DOI] [PubMed] [Google Scholar]