Abstract
Fatty acid desaturase 1 and 2 (FADS1 and FADS2) code for the key desaturase enzymes involved in the biosynthesis of long chain polyunsaturated fatty acids in mammals. FADS3 shares close sequence homology to FADS1 and FADS2 but the function of its gene product remains unknown. Alternative transcripts (AT) generated by alternative splicing (AS) are increasingly recognized as an important mechanism enabling a single gene to code for multiple gene products. We report the first AT of a FADS gene, FADS3, generated by AS. Aided by ORF Finder, we identified putative coding regions of eight AT for FADS3 with 1.34 kb (classical splicing), 1.14 (AT1), 0.77 (AT2), 1.25 (AT3), 0.51 (AT4), 0.74 (AT6), and 1.11 (AT7). In addition we identified a 0.51 kb length transcript (AT5) that has a termination codon within intron 8–9. The expression of each AT was analyzed in baboon neonate tissues and in differentiated and undifferentiated human SK-N-SH neuroblastoma cells. FADS3 AT are expressed in 12 neonate baboon tissues and showed reciprocal increases and decreases in expression changes in response to human neuronal cell differentiation. FADS3 AT, conserved in primates and under metabolic control in human cells, are a putative mediator of LCPUFA biosynthesis and/or regulation.
Keywords: fatty acid desaturases, alternative splicing, alternative transcripts, LCPUFA biosynthesis
1. Introduction
Long chain polyunsaturated fatty acids (LCPUFA), especially docosahexaenoic acid (DHA; 22:6n-3), eicosapentaenoic acid (EPA, 20:5n-3) and arachidonic acid (ARA; 20:4n-6) are ubiquitous in mammalian tissue and are indispensable for health, development and cognition (Carlson, 2001). They are essential constituents of phospholipid components of the biological membranes, are precursors for signaling eicosanoids and docosanoids that are major drug targets (e.g., COX-1, COX-2 inhibitors, leukotriene receptor antagonists) and are also known to regulate transcription of genes involved in lipid metabolism (Jump, 2002; Schmitz and Ecker, 2008). They can be obtained from the diet or are endogenously synthesized in the liver among many tissues, from dietary essential fatty acids (EFA) precursors, alpha-linolenic acid (ALA, 18:3n-3) and linoleic acid (LA, 18:2n-6) by an alternating series of position-specific desaturation and carbon chain-elongation reactions.
Fatty acid desaturases are enzymes that catalyze the introduction of cis double bonds at specific positions in a fatty acid chain. FADS1, FADS2, and FADS3 are members of fatty acid desaturase gene family that arose evolutionarily from gene duplication. They share high degree of sequence homology, are clustered within the 100 kb region on the long arm of human chromosome 11q12-13.1, have the same exon/intron organization with 12 exons and 11 introns, and are similarly organized on mouse chromosome 19 (Nakamura and Nara, 2004). The gene products of FADS1 and FADS2 have well defined functions as front end PUFA desaturases required for LCPUFA biosynthesis in mammals however no function has emerged for FADS3.
Most known desaturase genes involved in PUFA biosynthesis contain the N-terminal cytochrome b5 domain (HPGG) as electron donor and three histidine motifs “HXXXH, HXXHH and QXXHH”, conserved from human to microalga (Sperling et al., 2003), however, there are exceptions (Sayanova et al., 2006). FADS1 codes for a 5-desaturase, and FADS2 codes for a 6-desaturase with many substrates, and has recently been shown to have 8-desaturase activity (Park et al., 2009). Both genes are considered to yield a protein product(s) thought to operate on both n-3 and n-6 PUFA. The Δ6-desaturase (FADS2) operates on 18:3n-3 and 18:2n-6, resulting in the synthesis of 6,9,12,15–18:4 and 6,9,12–18:3 (gamma-linolenic acid), respectively. This step is rate-limiting, and is followed by elongation to 8,11,14,17–20:4 and 8,11,14–20:3 (dihomo-gamma-linolenic acid). A rapid Δ5-desaturation (FADS1) on these PUFA produces EPA and ARA. EPA can be further elongated and desaturated to yield DHA, which is accepted as the mammalian pathway (Voss et al., 1991). These steps are certainly localized to the endoplasmic reticulum (ER), and a beta-oxidation step is localized to the peroxisomes, but have been suggested over the years to occur in mitochondria (Infante and Huszagh, 1998), or via a Δ4-desaturase as demonstrated in Thraustochytrium (Qiu et al., 2001). Expression of FADS1 and FADS2 were detected in several human tissues with greatest expression in liver for FADS1 ; FADS2 showed highest expression in the brain followed by liver, lung and heart. Only a single transcript has been identified for both genes (Cho et al., 1999a; Cho et al., 1999b).
FADS3 was cloned by Marquardt et al (Marquardt et al., 2000). It spans 17.9 kb of genomic DNA, shares a high degree of homology with FADS1 (52%) and with FADS2 (62%), and encodes a putative protein of 445 amino acids (aa) with a molecular mass of 51.1 kDa. FADS3 is among the six most highly expressed genes at the implantation site of fertilized mouse embryos in the interimplantation region (prior to implantation), suggesting a crucial role in the initiation of pregnancy (Ma et al., 2006). However, the function of the FADS3 gene product remains unknown.
To date, there are no unambiguous reports of AT for any of the FADS genes. Two isoforms for FADS3 were detected with Northern blotting but an attempt to confirm them using RT-PCR was not successful (Marquardt et al., 2000). In the course of investigation of the role of FADS3 in LCPUFA biosynthesis, we performed RT-PCR analysis using baboon neonate cDNA and detected multiple AT generated by alternative splicing events. We report here characterization of FADS3 AT, their expression in twelve baboon neonate tissues and in human undifferentiated and differentiated neuroblastoma cells.
2. Materials and Methods
2.1 Total RNA extraction and preparation of cDNA
High quality neonate baboon tissues obtained at necropsy, treated with RNAlater, and maintained at −80°C, were used to isolate RNA. Total RNA from 30 mg tissue homogenates was extracted using the RNeasy Mini kit (Qiagen, Valencia, CA). Total RNA yield was assessed by 260 nm UV absorption. RNA quality was analyzed by 260/280 nm ratios of the samples and by agarose gel electrophoresis to verify RNA integrity. One microgram of DNase-treated total RNA was reverse-transcribed into first strand cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). The resulting cDNA was used as template for PCR-reactions.
2.2 RT(Reverse Transcription)-PCR and sequence analysis of FADS3 genes
The protein coding region of baboon FADS3 and FADS3 AT were generated from primers synthesized using human FADS3 (NM_021727) and monkey FADS3 (XR_014740) mRNA sequences. Primer pairs are presented in Supplementary Table 1. PCR primers were ordered from Integrated DNA Technologies (IDT, Coralville, IA) and were amplified by PCR with baboon liver, retina and spleen cDNA as templates and Ampli Taq Gold (Applied Biosystems, USA) in a 30 µl reaction volume containing 1µM of each primer, 0.25 mM dNTPs, 10× PCR buffer, and 1.5 mM MgCl2 using Eppendorf gradient thermal cycler. Touch down PCR was performed for FADS3 amplification reactions, wherein annealing temperature was set between 72°C and 69°C for 1 min for 40 cycles. In addition to the band of expected size ~1.4 kb, some discrete bands were obtained in agarose gels. The products were gel purified, cloned in pGEM T-Easy vector (Promega, USA) and sequenced using T7 forward and SP6 reverse universal primers. DNA sequencing was performed at Cornell University life sciences core laboratories center using the Applied Biosystems automated 3730 DNA analyzer.
2.3 Mammalian cell culture and sample preparation
SK-N-SH neuroblastoma cells (ATCC, USA) were grown at 37°C in a humidified environment with 5% CO2 using DMEM/F-12 with 10% FBS, 2mM L-alanyl-glutamine, and 15 mM HEPES for undifferentiated cells and routine passaging. Cell differentiation was carried out in serum free DMEM/F-12 with 1X N-2 supplement (Invitrogen, USA) containing 100 mg/L human transferrin, 5 mg/L recombinant full chain insulin, .0063 mg/L progesterone, 16.1 mg/L putrescine, and .0052 mg/L sodium selenite (Bottenstein, 1992; Dong and Lim, 1996). Cells were seeded in parallel and grown to 70% confluence in regular growth media with 10% FBS. One flask of undifferentiated cells was then harvested for RNA extraction, while the remaining cells were switched to chemically defined N-2 supplemented media for an additional six days in order to halt growth and induce differentiation before harvesting. RNA was isolated using the QIAshredder and RNeasy Mini kit (Qiagen, Valencia, CA).
2.4 Expression of FADS3AT in baboon tissues and human cells
To analyze the expression levels of each AT, primer pairs specific for each transcript bridging the deleted parts of the exons were designed (Supplementary Table 2). Tissue distribution of each transcript was measured using 12 normal tissues from a baboon neonate (12 weeks old) and undifferentiated (embryonic stage) and differentiated SK-N-SH neuroblastoma cells by RT-PCR. For FADS3 AT PCR amplification reactions, annealing temperature was performed between 60–65°C for 30–40 cycles. The PCR fragments generated from AT were cloned and sequenced. Amplicon size of each AT is presented in Supplementary Table 2.
3. Results and Discussion
3.1 Sequence analysis of baboon FADS3
The sequencing of the expected ~1.4 kb PCR product (Baboon FADS3) revealed the presence of an open reading frame of 1,338 base pairs, encoding a putative protein of 445 aa and a stop codon. It shares a high degree of homology with other FADS genes (51% identity with baboon FADS1 (EF531577) and 62% identity with baboon FADS2 (EU780003)). Like other front-end desaturases, baboon FADS3 protein also has “HPGG” (at aa positions 55–58) characteristic of Cytochrome b5 and three conserved histidine motifs “HXXXH, HXXHH and QXXHH”. The human and baboon FADS3 mRNA sequences align at a level of 97% identity along the entire coding region. Analysis and comparison of aa sequence of baboon FADS3 showed 99% identity and 99% similarity with human FADS3 (NP_068373), and 54% identity and 70% similarity with bifunctional zebrafish desaturase (AAG25710).
3.2 Sequence analysis of baboon FADS3AT
When PCR products were separated on a 2% agarose gel, discrete bands of various sizes were detected in addition to the cDNA of the expected ~1.4 kb size. All of the PCR products were gel purified, cloned and sequenced. Seven AT for baboon FADS3 were identified in addition to the classical transcript. The generated splice variants correspond to the absence of a portion of exon, complete absence of one or several exons, and retention of an intron. The FADS3 AT sequences are deposited at GenBank and were assigned the following GenBank Accession numbers EU780002, EU780004, FJ641198, FJ641199, FJ641200, FJ641201, FJ641202, and FJ641203.
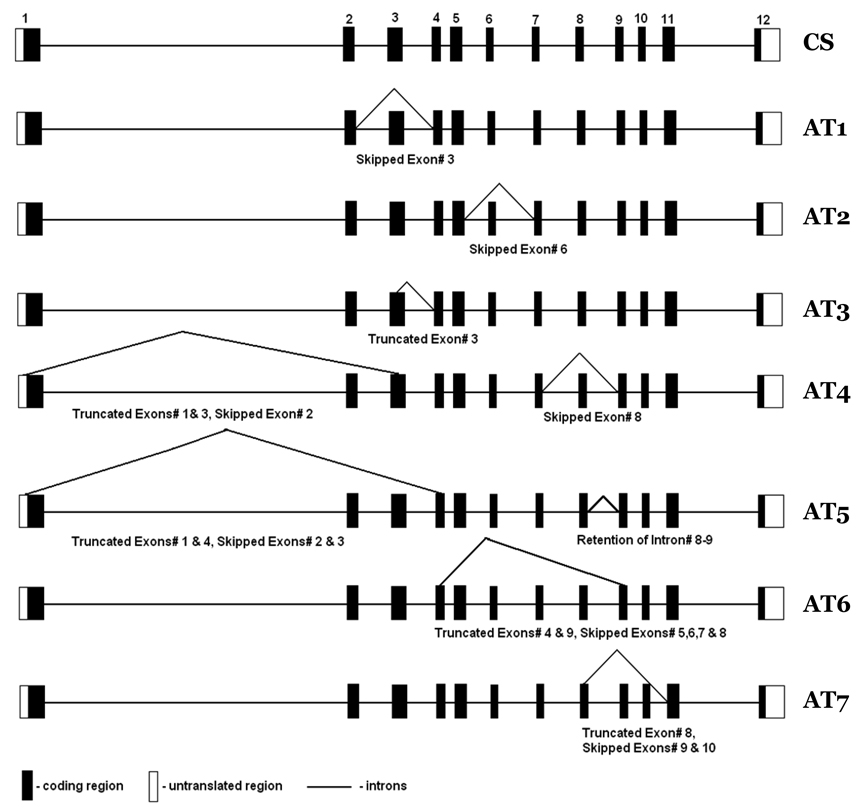
Using sequence analysis data with aid from ORF Finder, we identified the putative coding regions for eight AT for FADS3 with 1.34 kb (classical splicing), 1.14 (AT1), 0.77 (AT2), 1.25 (AT3), 0.51 (AT4), 0.74 (AT6), and 1.11 (AT7). Figure 1 presents the structure of these AT. In addition we identified an AT of 0.51 kb (AT5) length that has a termination codon within intron 8–9. Predicted protein sequences of the FADS3 AT indicate that three variants, AT1, AT3 and AT7, would produce shorter versions of the protein resulting from missing exon(s), whereas AT2, AT4, AT5 and AT6 would produce truncated proteins. AT1 lacks exon 3 due to an in-frame loss of 66 aa and AT3 lacks a portion of exon 3 resulting from an in-frame loss of 28 aa. Both AT1 and AT3 retain all the conserved motifs characteristic of front end desaturases (“HPGG” Cytochrome b5 motif and three histidine repeats “HDLGH, HFQHH, QIEHH). The absence of a portion of exon 8 together with exons 9–10 (AT7) result in an in-frame loss of 76 a.a and also results in the loss of the last histidine repeat “QIEHH”.
Figure 1. Baboon FADS3 and FADS3 AT.
Missing spans within AT are shown. Numbers 1 to 12 are exons; CS, classical splicing; AT1-AT7 denote alternative transcripts.
In AT2, the skip of exon 6 causes a reading frameshift that results in the loss of the last histidine repeat “QIEHH”, and generates a truncated protein of 257 aa. AT4, AT5, and AT6 result from multiple exon deletions. AT4 lost HPGG and QIEHH, AT5 lost HPGG, HDLGH, and QIEHH, whereas, AT6 retains only HPGG while all the histidine repeats are lost from splicing events. Hastings et al. reported that substrate specificity can vary depending upon the differences in the sequence of a desaturase even though the catalytic activities are identical (Hastings et al., 2004). This raises the possibility that FADS3 AT might have different substrate specificities even though some of them share all the conserved catalytic domains.
3.3 FADS3 conservation and putative subcellular localization of baboon FADS genes
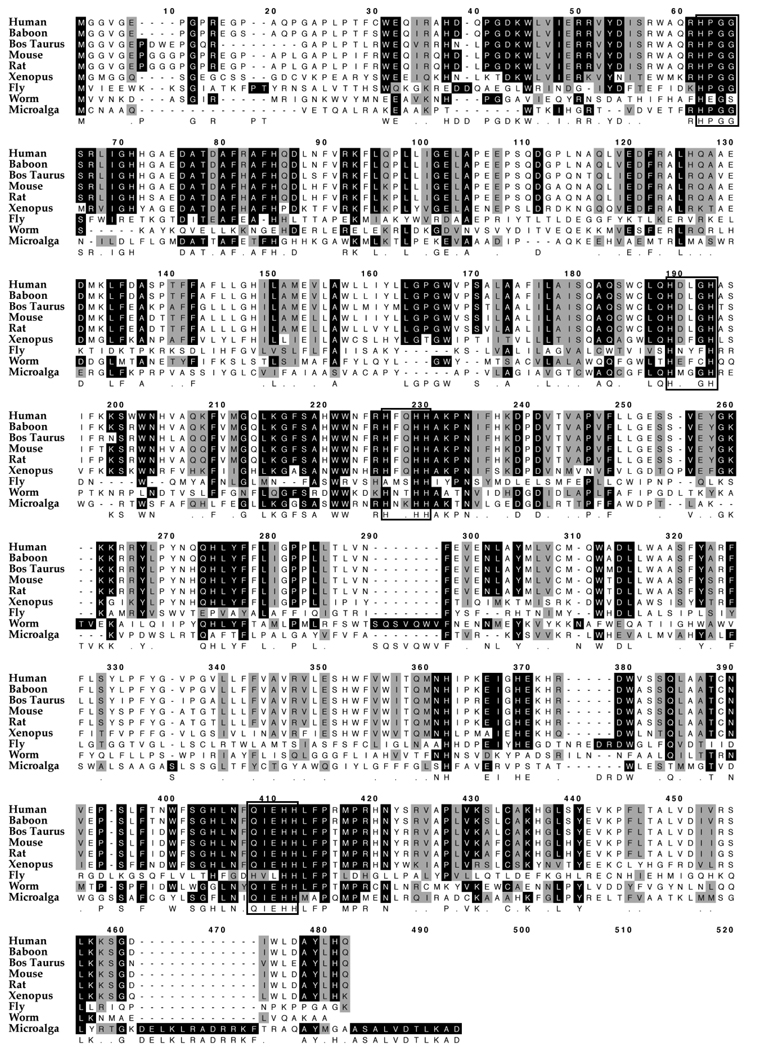
Figure 2 shows the alignment of FADS3 and its putative homologs from various species. It can be seen that the “HPGG” characteristic of Cytochrome b5 and three conserved histidine motifs “HXXXH, HXXHH and QXXHH” are well conserved from human to microalga.
Figure 2. Alignment of amino acid sequences of FADS3.
Using MacVector software and ClustalW alignment, FADS3 aa sequences were aligned from various species. Well conserved motifs common for desaturases are depicted in boxes. The “HPGG” characteristic of a cytochrome b5 domain and three conserved histidine motifs “HXXXH, HXXHH and QXXHH” are shown in boxes.
We used the Proteome analyst computational tool to query the subcellular localization of the FADS genes (Lu et al., 2004). Amino acid sequence information is entered and the software provides an estimate of the probability that the protein is localized to a specific organelle. FADS1, FADS2, and FADS3 were all found to be localized to ER with a probability score of 1.000; FADS1 and FADS2 are known to code for ER proteins. Similarly an equivalent test for the mitochondria shows that FADS1 and FADS2 had probability scores of 0.529 and 0.562, respectively, whereas FADS3 was found to have a probability score of 0.999 for mitochondrial localization. This result suggests that FADS3 contains domains that are more commonly found in the mitochondria compared to FADS1 and FADS2, and may point to a mitochondrial role for one or more proteins for which the FADS3 codes.
3.4 Expression of FADS3AT in baboon tissues and human cells
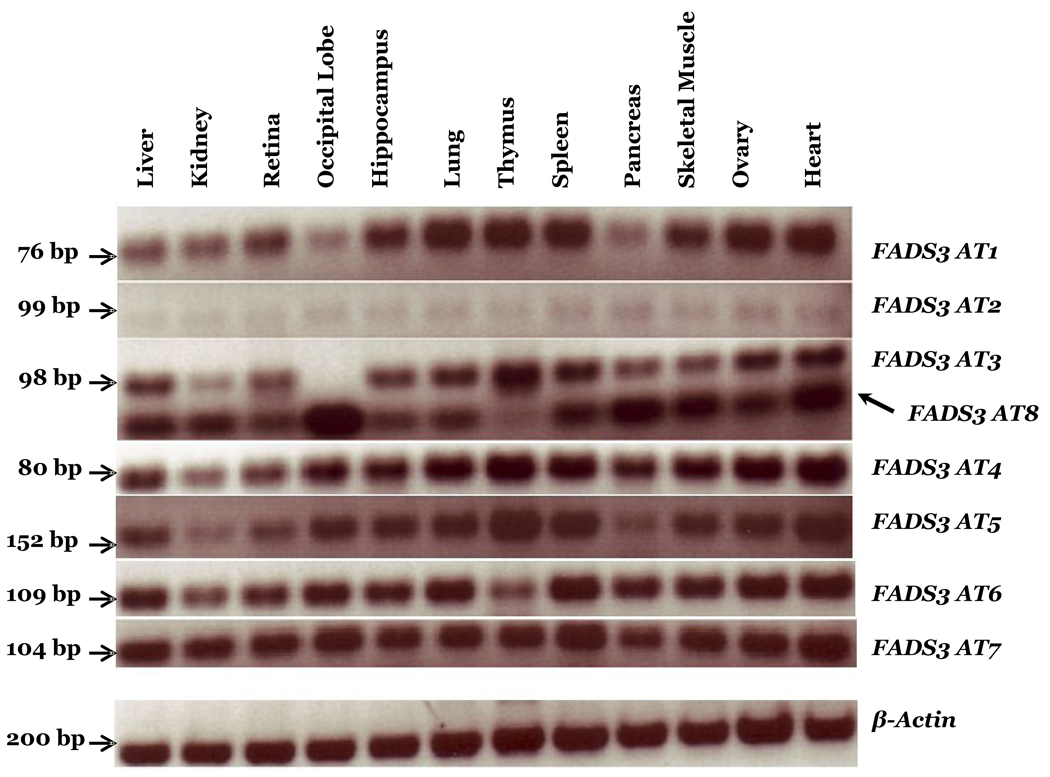
Although FADS3 was cloned in 2000 (Marquardt et al., 2000), and was shown to be expressed in human heart, liver, lung, uterus and brain tissues, nothing is known about its function in LCPUFA biosynthesis. We determined the tissue distribution and expression patterns of FADS3 AT in a baboon neonate and also determined the regulation patterns in human NB cells. To study the tissue distribution of FADS3 AT we performed semiquantitative RT-PCR using primer pairs bridging the deleted parts of the exons specific for each transcript, as shown in Supplementary Table 2. Figure 3 shows that all the variants are expressed in the 12 baboon neonate tissues studied. The AT2 variant was present in reproducibly smaller amounts in all 12 tissues. The primer pair designed to amplify AT3 also revealed the presence of a shorter amplicon (AT8?). AT8 did not initially appear in our cloning screens, but is also expressed in all tissues. The sequencing of the shorter amplicon (~50 base pairs) showed deletion within exon 4. Notably, the AT3 primer pair amplifies only AT8 in the occipital lobe region of the brain, whereas the remaining 11 tissues express both AT3 and AT8 (Figure 3). However, AT3 is highly expressed in the thymus, compared to other tissues. The highest levels of AT8 were observed in occipital cortex lobe, pancreas, skeletal muscle and heart, with much lower levels in thymus. Our RT-PCR analysis using primers specifically designed to amplify AT5 also confirms that intron 8–9 remains in the transcript, and it is strongly expressed in immune tissues (thymus and spleen) and the heart. These data support the hypothesis that FADS3 AT encode functionally important proteins.
Figure 3. FADS3 AT expression in 12 Baboon Neonate Tissues.
RT-PCR analysis of FADS3 AT shows expression in 12 baboon neonate tissues. The amplified products are resolved on a 2% agarose gel, visualized with ethidium bromide and a negative image is shown. Legend: bp, base pair.
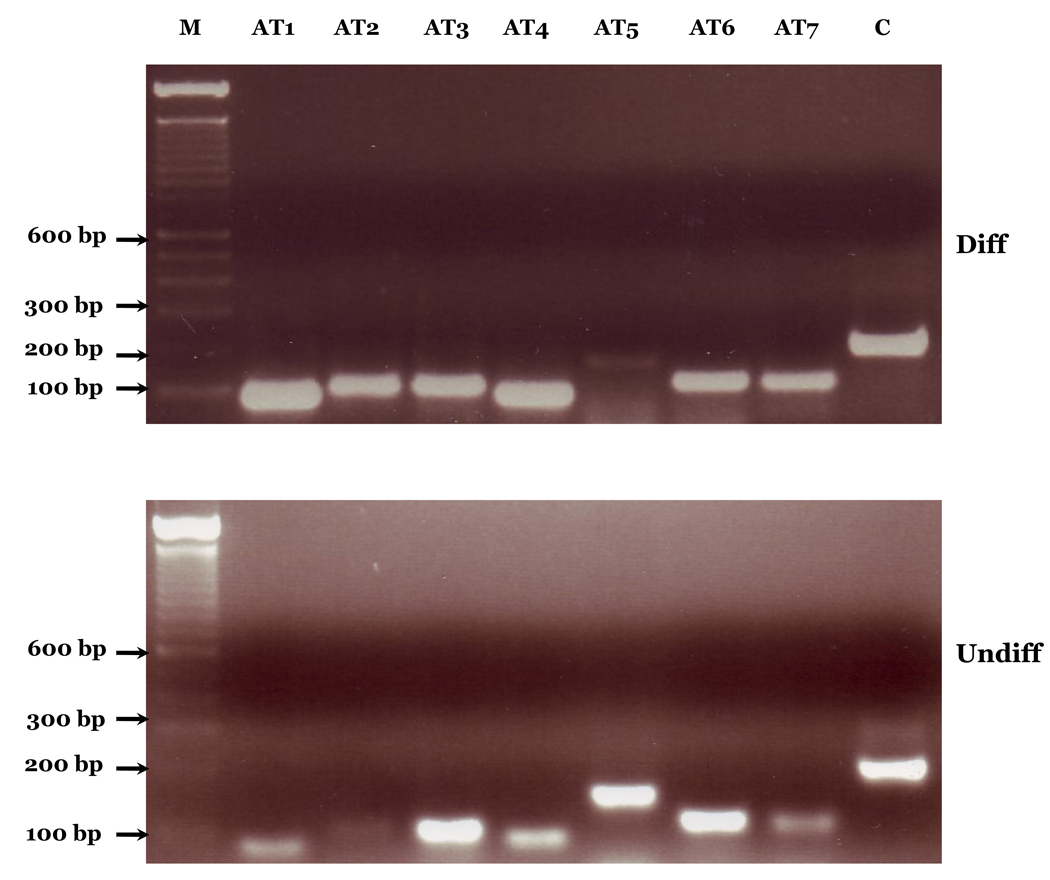
Expression of FADS3 AT in response to cell differentiation was also investigated using human SK-N-SH neuroblastoma (NB) cells. Figure 4 shows that all the variants are found to be expressed in these cells. However, they showed reciprocal increases and decreases in expression pattern in response to cell differentiation (Figure 4). One of the most striking observations is the significant downregulation of AT5 expression in differentiated and significant upregulation in undifferentiated NB cells. Different patterns of expression of AT show that these transcripts are under specific control during the NB cell differentiation. Our data demonstrate that the FADS3 AS events widely detected in baboon tissues are also conserved in human cells. A recent estimate shows splicing level differences of 6–8% between orthologous human and primate (chimpanzee) transcripts and 4% in genes having one or more cassette alternative exons, compared to 80% differences between human and mouse genes (Calarco et al., 2007).
Figure 4. FADS3 AT expression in human SK-N-SH Neuroblastoma (NB) Cells.
RT-PCR analysis of FADS3 AT shows expression in human SK-N-SH neuroblastoma (NB) cells. M represents a 100 base pair molecular size marker; AT1-AT7 represent alternatively spliced variants; C-Beta-actin Control; Diff, differentiated cells, Undiff, undifferentiated cells.
3.5 FADS-coded PUFA desaturases and putative isoforms
AS, first proposed by Gilbert (Gilbert, 1978), is rapidly gaining importance as an essential molecular mechanism whereby a single gene transcript can give rise to a number of spliced mRNAs to generate remarkable protein diversity. Blencowe suggested that AS may be the most extensively used posttranscriptional modification of genes (Blencowe, 2006). Modrek et al. performed genome-wide analysis to test AS events using a human EST database analysis and found that the majority of splicing events (74%) occur in coding regions, followed by 5’-UTR (22%) and 3’-UTR (4%) (Modrek et al., 2001).
Modrek et al. (Modrek et al., 2001) assessed the systemic functions affected by AS events in human genes and found that many were specific to immune and nervous systems. LCPUFA are highly concentrated in membranes of the nervous system and serve as substrates for signaling molecules. Diets rich in n-6 PUFA induce various physiological and metabolic changes leading towards proinflammatory status with production of series 2 prostaglandins and series 4 leukotrienes, whereas n-3 PUFA have opposite effects (Benatti et al., 2004). The well known, pioneering 1963 work of Mohrhauser and Holman showed that rat liver PUFA composition is related to the ratio of 18:3n-3 and 18:2n-6 in the diet (Mohrhauer and Holman, 1963a; Mohrhauer and Holman, 1963b). Vertebrate desaturase and elongation enzymes operate on both the n-3 and n-6 PUFA families. However, alternative hypotheses involving n-3 or n-6 specific desaturases have been proposed from time to time which suggest that the competition may, at least in part, be “apparent”. A compelling example is the reciprocal changes in concentration of 22:6n-3 and 22:5n-6 that are found associated with specific metabolic conditions, for instance in tissues of Zellweger’s syndrome (Martinez, 1992) or adrenoleukodystrophy (ALD) patients(Martinez, 1990), acyl CoA knockout (Acox ko) mice (Infante et al., 2002), and in vitamin A deficient mice (Zhou et al., 2004). In Zellweger’s, ALD, and Acox ko, 22:6n-3 drops while 22:5n-6 rises; however, in vitamin A deficient mice, the opposite is found: 22:6n-3 rises and 22:5n-6 drops. The latter paper (Zhou et al., 2004) specifically suggests, as we have (Infante et al., 2002), that common enzyme pathways for 22:6n-3 and 22:5n-6 cannot explain these results. Plausible hypotheses are that isoforms based on FADS AT exist that operate with high selectivity on specific PUFA and are differently regulated, and/or that the unknown function(s) of FADS3 are PUFA-specific. A vertebrate 5-desaturase with all the characteristics of microsomal desaturases, including 63% identity with the human 5-desaturase (FADS1), and with significant activity only towards n-3 PUFA (20:4n-3) has been reported in Atlantic salmon (Hastings et al., 2004). An early study of PUFA synthesis in mammalian cells concluded that separate enzymes may exist for the n-3 and n-6 PUFA (Maeda et al., 1978). A mitochondrial pathway involving separate PUFA desaturases for n-6 and n-3 fatty acids was postulated to explain observed features of LCPUFA biosynthesis (Infante and Huszagh, 1998).
Many isoforms of stearoyl CoA desaturase (SCD) are known in many species, expressed in a tissue specific manner (Dridi et al., 2007), but thusfar no AT of any of the PUFA-specific desaturases are known, other than the ones that we present in this study. The data of figure 3 and figure 4 show that AT of PUFA-specific desaturases exist in primates, and support the hypothesis that AT have different roles because of their differential expression. Molecular regulation of PUFA desaturases produced by classical splicing of FADS1 and FADS2, respectively, are well studied, but nothing is known about FADS3 regulation. Both FADS1 and FADS2 are regulated by macro and micronutrients, hormones and multiple transcription factors (Brenner, 1981; Nakamura and Nara, 2004; Zolfaghari and Ross, 2003). Identifying the substrate specificities of these novel FADS3 transcripts and functions associated with them will help in better understanding the cellular processes mediated by PUFA and their metabolites.
4. Conclusion
This is the first finding of AT of FADS genes. We present in vivo and in vitro evidence that at least 9 novel FADS3 AT, including AT8, are generated by splicing events. The FADS3 AT are expressed in many tissues and showed changes in abundance in response to human neuronal cell differentiation. FADS3 AT function is as yet unknown but their structure, and high sequence homology to functional FADS1 and FADS2, strongly implies that they are involved in LCPUFA biosynthesis. LCPUFA are ubiquitous components of cells and tissues that can be, but are not always, obtained in the diet. Intense interest in LCPUFA metabolism is related to the wide range of human intakes compared to requirements for optimal health. The most recent frontiers include diseases of aging (e.g., Alzheimer’s disease) and psychiatric/affective disorders. Determining the function of FADS3 AT, conserved among species, provides a putative mechanism for understanding of LCPUFA biosynthetic regulation, and how it differs among individuals.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM071534 and a Cornell University Center for Vertebrate Genomics Seed grant. The authors thank Alexander Philip Limjuco, Sara Huh, and Satya Prasanthi Kothapalli for technical assistance. We thank Dr.Ling Qi for his help with MacVector software and valuable suggestions.
Abbreviations
- LCPUFA
long chain polyunsaturated fatty acids
- DHA
docosahexaenoic acid
- EPA
eicosapentaenoic acid
- ARA
arachidonic acid
- AT
alternative transcripts
- FADS1
fatty acid desaturase 1
- FADS2
fatty acid desaturase 2
- FADS3
fatty acid desaturase 3
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Woo Jung Park, Email: wp48@cornell.edu.
Kumar SD Kothapalli, Email: ksk25@cornell.edu.
Holly T Reardon, Email: htr2@cornell.edu.
Luke Y. Kim, Email: lyk4@cornell.edu.
References
- Benatti P, Peluso G, Nicolai R, Calvani M. Polyunsaturated fatty acids: Biochemical, nutritional and epigenetic properties. J. Am. Coll. Nutr. 2004;23:281–302. doi: 10.1080/07315724.2004.10719371. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Bottenstein JE. Environmental influences on cells in culture. In: Boulton A, Baker G, Walz W, editors. Neuromethods. The Humane Press Inc; 1992. pp. 63–85. [Google Scholar]
- Brenner RR. Nutritional and hormonal factors influencing desaturation of essential fatty acids. Prog. Lipid Res. 1981;20:41–47. doi: 10.1016/0163-7827(81)90012-6. [DOI] [PubMed] [Google Scholar]
- Calarco JA, Xing Y, Caceres M, Calarco JP, Xiao X, Pan Q, Lee C, Preuss TM, Blencowe BJ. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson SE. Docosahexaenoic acid and arachidonic acid in infant development. Semin. Neonatol. 2001;6:437–449. doi: 10.1053/siny.2001.0093. [DOI] [PubMed] [Google Scholar]
- Cho HP, Nakamura M, Clarke SD. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J. Biol. Chem. 1999a;274:37335–37339. doi: 10.1074/jbc.274.52.37335. [DOI] [PubMed] [Google Scholar]
- Cho HP, Nakamura MT, Clarke SD. Cloning, expression, and nutritional regulation of the mammalian delta-6 desaturase. J. Biol. Chem. 1999b;274:471–477. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- Dong JM, Lim L. Selective up-regulation of alpha 1-chimaerin mrna in sk-n-sh neuroblastoma cells by k+/−induced depolarisation. Eur. J. Biochem. 1996;236:820–826. doi: 10.1111/j.1432-1033.1996.00820.x. [DOI] [PubMed] [Google Scholar]
- Dridi S, Taouis M, Gertler A, Decuypere E, Buyse J. The regulation of stearoyl-coa desaturase gene expression is tissue specific in chickens. J. Endocrinol. 2007;192:229–236. doi: 10.1677/JOE-06-0070. [DOI] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Hastings N, Agaba MK, Tocher DR, Zheng X, Dickson CA, Dick JR, Teale AJ. Molecular cloning and functional characterization of fatty acyl desaturase and elongase cdnas involved in the production of eicosapentaenoic and docosahexaenoic acids from alpha-linolenic acid in atlantic salmon (salmo salar) Mar. Biotechnol. (NY) 2004;6:463–474. doi: 10.1007/s10126-004-3002-8. [DOI] [PubMed] [Google Scholar]
- Infante JP, Huszagh VA. Analysis of the putative role of 24-carbon polyunsaturated fatty acids in the biosynthesis of docosapentaenoic (22:5n-6) and docosahexaenoic (22:6n-3) acids. FEBS Lett. 1998;431:1–6. doi: 10.1016/s0014-5793(98)00720-0. [DOI] [PubMed] [Google Scholar]
- Infante JP, Tschanz CL, Shaw N, Michaud AL, Lawrence P, Brenna JT. Straight-chain acyl-coa oxidase knockout mouse accumulates extremely long chain fatty acids from alpha-linolenic acid: Evidence for runaway carousel-type enzyme kinetics in peroxisomal beta-oxidation diseases. Mol. Genet. Metab. 2002;75:108–119. doi: 10.1006/mgme.2001.3279. [DOI] [PubMed] [Google Scholar]
- Jump DB. Dietary polyunsaturated fatty acids and regulation of gene transcription. Curr. Opin. Lipidol. 2002;13:155–164. doi: 10.1097/00041433-200204000-00007. [DOI] [PubMed] [Google Scholar]
- Lu Z, Szafron D, Greiner R, Lu P, Wishart DS, Poulin B, Anvik J, Macdonell C, Eisner R. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics. 2004;20:547–556. doi: 10.1093/bioinformatics/btg447. [DOI] [PubMed] [Google Scholar]
- Ma XH, Hu SJ, Ni H, Zhao YC, Tian Z, Liu JL, Ren G, Liang XH, Yu H, Wan P, Yang ZM. Serial analysis of gene expression in mouse uterus at the implantation site. J. Biol. Chem. 2006;281:9351–9360. doi: 10.1074/jbc.M511512200. [DOI] [PubMed] [Google Scholar]
- Maeda M, Doi O, Akamatsu Y. Metabolic conversion of polyunsaturated fatty acids in mammalian cultured cells. Biochim. Biophys. Acta. 1978;530:153–164. [PubMed] [Google Scholar]
- Marquardt A, Stohr H, White K, Weber BH. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- Martinez M. Severe deficiency of docosahexaenoic acid in peroxisomal disorders: A defect of delta 4 desaturation? Neurology. 1990;40:1292–1298. doi: 10.1212/wnl.40.8.1292. [DOI] [PubMed] [Google Scholar]
- Martinez M. Abnormal profiles of polyunsaturated fatty acids in the brain, liver, kidney and retina of patients with peroxisomal disorders. Brain Res. 1992;583:171–182. doi: 10.1016/s0006-8993(10)80021-6. [DOI] [PubMed] [Google Scholar]
- Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrhauer H, Holman RT. The effect of dose level of essential fatty acids upon fatty acid composition of the rat liver. J. Lipid Res. 1963a;4:151–159. [PubMed] [Google Scholar]
- Mohrhauer H, Holman RT. Effect of linolenic acid upon the metabolism of linoleic acid. J. Nutr. 1963b;81:67–74. doi: 10.1093/jn/81.1.67. [DOI] [PubMed] [Google Scholar]
- Nakamura MT, Nara TY. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Ann. Rev. Nutr. 2004;24:345–376. doi: 10.1146/annurev.nutr.24.121803.063211. [DOI] [PubMed] [Google Scholar]
- Park WJ, Kothapalli KS, Lawrence P, Tyburczy C, Brenna JT. An alternate pathway to long chain polyunsaturates: The FADS2 gene product 8-desaturates 20:2n-6 and 20:3n-3. J Lipid Res. 2009 doi: 10.1194/jlr.M800630-JLR200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hong H, MacKenzie SL. Identification of a delta 4 fatty acid desaturase from thraustochytrium sp. Involved in the biosynthesis of docosahexanoic acid by heterologous expression in saccharomyces cerevisiae and brassica juncea. J. Biol. Chem. 2001;276:31561–31566. doi: 10.1074/jbc.M102971200. [DOI] [PubMed] [Google Scholar]
- Sayanova O, Haslam R, Qi B, Lazarus CM, Napier JA. The alternative pathway C20 delta8-desaturase from the non-photosynthetic organism acanthamoeba castellanii is an atypical cytochrome b5-fusion desaturase. FEBS Lett. 2006;580:1946–1952. doi: 10.1016/j.febslet.2006.02.062. [DOI] [PubMed] [Google Scholar]
- Schmitz G, Ecker J. The opposing effects of n-3 and n-6 fatty acids. Prog. Lipid Res. 2008;47:147–155. doi: 10.1016/j.plipres.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Sperling P, Ternes P, Zank TK, Heinz E. The evolution of desaturases. Prostaglandins Leukot. Essent. Fatty Acids. 2003;68:73–95. doi: 10.1016/s0952-3278(02)00258-2. [DOI] [PubMed] [Google Scholar]
- Voss A, Reinhart M, Sankarappa S, Sprecher H. The metabolism of 7,10,13,16,19-docosapentaenoic acid to 4,7,10,13,16,19-docosahexaenoic acid in rat liver is independent of a 4-desaturase. J. Biol. Chem. 1991;266:19995–20000. [PubMed] [Google Scholar]
- Zhou D, Zaiger G, Ghebremeskel K, Crawford MA, Reifen R. Vitamin A deficiency reduces liver and colon docosahexaenoic acid levels in rats fed high linoleic and low alpha-linolenic acid diet. Prostaglandins Leukot. Essent. Fatty Acids. 2004;71:383–389. doi: 10.1016/j.plefa.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Zolfaghari R, Ross AC. Recent advances in molecular cloning of fatty acid desaturase genes and the regulation of their expression by dietary vitamin a and retinoic acid. Prostaglandins Leukot. Essent. Fatty Acids. 2003;68:171–179. doi: 10.1016/s0952-3278(02)00267-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.