SUMMARY
Terminally misfolded or unassembled secretory proteins are retained in the endoplasmic reticulum (ER) and subsequently cleared by the ER-associated degradation (ERAD) pathway. The degradation of ERAD substrates involves mannose trimming of N-linked glycans, however the mechanisms of substrate recognition and sorting to the ERAD pathway are poorly defined. EDEM1 (ER degradation-enhancing α-mannosidase-like 1 protein) has been proposed to play a role in ERAD substrate signaling or recognition. We show that EDEM1 specifically binds non-native proteins in a glycan-independent manner. Inhibition of mannosidase activity with kifunensine or disruption of the EDEM1 mannosidase-like domain by mutation had no effect on EDEM1 substrate binding, but diminished its association with the ER membrane adapter protein, SEL1L. These results support a model whereby EDEM1 binds non-native proteins and uses its mannosidase-like domain to target aberrant proteins to the ER membrane dislocation and ubiquitination complex containing SEL1L.
INTRODUCTION
Newly synthesized proteins that traverse the secretory pathway enter the endoplasmic reticulum (ER) and interact with a diverse set of chaperones and processing enzymes. These ER proteins assist in the maturation of secretory cargo and administer a quality control test that helps to sort native proteins for exit to the Golgi (Ellgaard and Helenius, 2003; Hebert and Molinari, 2007). Terminally misfolded proteins are sequestered away from the productive folding pathway and are eventually delivered to an ER membrane dislocation and ubiquitination complex for retrotranslocation and subsequent degradation by the cytosolic proteasome in a process termed ER-associated degradation (ERAD)(Vembar and Brodsky, 2008). The partitioning and clearance of aberrant proteins are essential for cellular homeostasis and survival.
N-linked glycans can act as sorting tags during the maturation and quality control processes in the ER (Hebert and Molinari, 2007). As proteins emerge into the ER lumen, glycans comprised of 3 glucoses, 9 mannoses and 2 N-acetylglucosamines (Glc3Man3GlcNAc2) are attached to proteins. Generation of monoglucosylated glycans by successive cleavage of the first two glucoses by glucosidases I and II, allows for the interaction between the immature glycoprotein and the lectin chaperone calnexin and its soluble paralogue, calreticulin (Hebert and Molinari, 2007). Lectin chaperone binding to monoglucosylated glycans facilitates the efficient folding and ER retention of immature glycoproteins. In contrast, the identification of aberrant proteins for destruction appears to involve mannose trimming, as pharmacological or genetic inhibition of mannosidase activity stabilized glycosylated ERAD substrates (Jakob et al., 1998; Liu et al., 1999; Svedine et al., 2004).
The ER contains a number of mannosidase-like or mannose-binding proteins that are involved in ERAD. Degradation appears to involve the extensive mannose trimming of substrates to Man6-5 (Hosokawa et al., 2009; Quan et al., 2008). The glycosylhydrolase family 47 contains two subgroups that reside in the ER including α 1,2-mannosidase I (ER ManI) and the EDEM (ER degradation-enhancing α-mannosidase-like proteins) family. ER ManI removes a single α 1,2-mannose on the B branch and can continue to remove additional mannose residues under higher nonphysiological concentrations of the enzyme (Herscovics et al., 2002). The EDEM family contains three ER proteins EDEM 1–3, and their corresponding genes are targets of transcriptional regulation in response to stress. Overexpression of EDEM1 increased the rate of misfolded glycoprotein trimming and degradation, while the trafficking of correctly folded glycoproteins was unaffected (Molinari et al., 2003; Oda et al., 2003; Olivari et al., 2006). However, the mechanism by which EDEM1 assists glycoprotein quality control remains uncertain.
In the present study, we characterized the binding properties of EDEM1 to investigate how misfolded glycoproteins are selected and targeted for degradation. EDEM1 binding to the model ERAD substrate, α-1-antitrypsin (A1AT) null Hong Kong (NHK), did not require mannose trimming or glycosylation. However, EDEM1 was shown to associate with the downstream ERAD component, SEL1L, in a carbohydrate-dependent manner. Hence, we propose that EDEM1 serves as a quality control receptor that acts as a molecular link between misfolded proteins and the SEL1L-containing ER membrane dislocation and ubiquitination complex.
RESULTS
EDEM1 selectively binds misfolded proteins
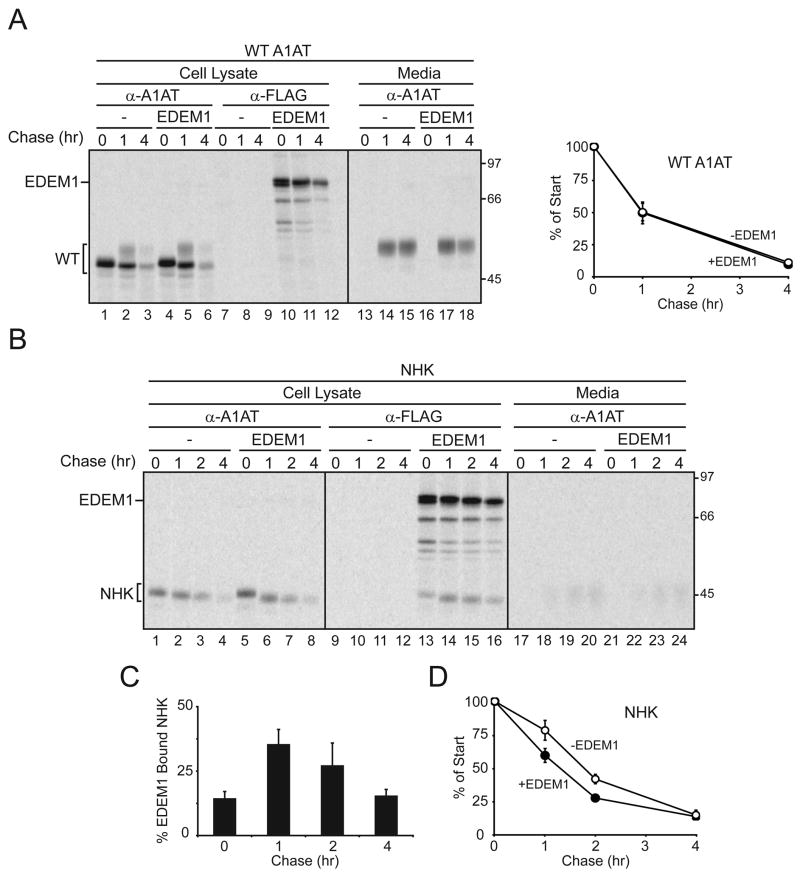
To characterize the binding specificity of EDEM1, a cell-based binding assay was established. Wild type (WT) and mutant forms of A1AT, a model soluble glycoprotein, were separately co-expressed in 293T cells with EDEM1 tagged with a C-terminal FLAG epitope. Cells were radiolabeled with [35S]-Met/Cys for 15 min, chased for various times and solubilized using a non-denaturing detergent prior to immunoprecipitation with A1AT or FLAG antisera. Immune-complexes were washed under harsh conditions to minimize non-specific interactions. These conditions disrupt substrate interactions with calnexin and calreticulin (Hebert et al., 1995).
The ER form of WT A1AT was resolved as a sharp band of ~48 kD (Figure 1A, lanes 1–6). A slower migrating smear appeared after 1 hr of chase that corresponded to protein containing complex sugars received after passage through the Golgi. The complex carbohydrate-containing form of A1AT accumulated in the culture media in a time-dependent manner (Figure 1A, lanes 13–18). The stability of WT A1AT was unaffected by the co-expression of EDEM1 (Figure 1A, plot).
Figure 1. EDEM1 transiently binds to mutant α-1-antitrypsin.
(A) Blank vector (−) or EDEM1-FLAG was co-expressed with (A) WT A1AT or (B) NHK into 293T cells. Cells were radiolabeled for 15 min and chased for the indicated times. Cells were lysed in MNT buffer, A1AT and EDEM1 were isolated using A1AT (α-A1AT) and FLAG (α-FLAG) antisera. Secreted A1AT was also isolated from the cell culture media. The corresponding proteins were resolved on 9% reducing SDS-PAGE. Quantification of (A) percent of cellular WT A1AT, (C) percent EDEM1 bound NHK or (D) percent of cellular NHK were determined.
The NHK mutant variant of A1AT has a frame-shift mutation resulting in a C-terminal truncation of 61 amino acids (Sifers et al., 1988). NHK has served as a model ERAD substrate (Hosokawa et al., 2001; Liu et al., 1999). As expected, the ER glycoform of NHK accumulated and was not found in the culture media (Figure 1B)(Sifers et al., 1988). In contrast to WT A1AT, a significant fraction of NHK co-immunoprecipitated with EDEM1 (Figure 1B, lanes 13–16, and 1C) and its co-expression accelerated the turnover of NHK (Figure 1D), as previously observed (Hosokawa et al., 2001; Oda et al., 2003). EDEM1 binding to NHK reached a maximum level of 35% after 1 hr of chase, indicating that binding was both efficient and transient.
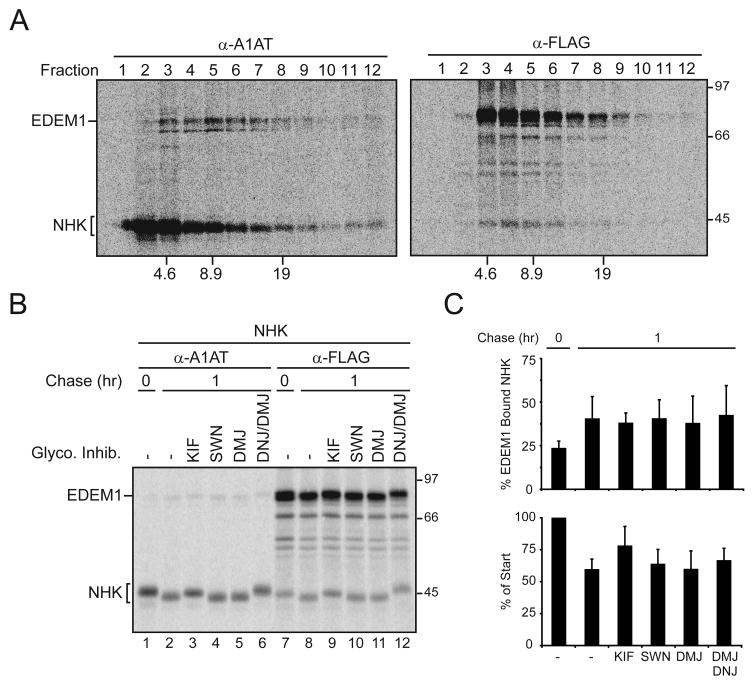
To further characterize the EDEM1-NHK complex, cell lysates were separated by sucrose density gradients ultracentrifugation. Uncomplexed NHK was found at the top of the gradient in fractions 1 and 2 (Figure 2A, α-A1AT). NHK did not co-immunoprecipitate with EDEM1 in these fractions (Figure 2A, α-FLAG). EDEM1-NHK complexes were found most predominately in fraction 4 (~7S, Figure 2A, α-FLAG). The complex size and the presence of only the EDEM1 doublet and NHK in the A1AT immunoprecipitated fraction 4 supports the formation of a complex comprised largely of EDEM1 and NHK (Figure 2A). EDEM1 also efficiently bound and accelerated the turnover of two additional ERAD substrates, the Z-variant of A1AT and a mutant form of the type-I membrane glycoprotein tyrosinase (Figure S1)(Termine et al., 2009)(data not shown). Together, these results indicated that EDEM1 differentiates between native and non-native glycoproteins, and transiently binds aberrant proteins.
Figure 2. Characterization of the EDEM1-NHK complex and its glycan trimming independence.
(A) 293T cells were transfected with EDEM1-FLAG and NHK. After pulse-labeling for 15 min, cells were chased for 1 hr and lysed with MNT buffer. The lysate was layered on a linear sucrose gradient in MNT buffer and centrifuged. Each fraction was subjected to immunoprecipitation using α-A1AT or α-FLAG antibody, and samples were separated by reducing SDS-PAGE. Sedimentation velocity values and molecular weights are denoted at the bottom and right side of the autoradiograms, respectively. (B) EDEM1-FLAG was co-expressed with NHK in 293T cells. Cells were pulse-labeled for 15 min and chased for the indicated times. 150 μM KIF, 50 μM SWN, 1 mM DMJ, and/or 0.5 mM DNJ were present 1 hr prior to radiolabeling, as well as in the radiolabeling and chase medium. A1AT or EDEM1 were isolated from cell lysates using A1AT (α-A1AT, lanes 1–6) and FLAG (α-FLAG, lanes 7–12) antisera, respectively. Complexes were resolved on 9% reducing SDS-PAGE. (C) Quantifications of the percent of cellular NHK or percent of EDEM1 bound NHK are displayed.
EDEM1 binds misfolded glycoproteins in a carbohydrate trimming-independent manner
Mannose trimming of substrate carbohydrate side chains during maturation has been proposed to play a role in marking aberrant proteins for destruction (Cabral et al., 2001; Hebert and Molinari, 2007). To investigate the requirement for carbohydrate trimming as a determinant for EDEM1 binding to misfolded proteins, EDEM1 was co-expressed with NHK in the absence or presence of various glycosidase inhibitors and the binding of EDEM1 to NHK was analyzed by co-immunoprecipitation (Figure 2B and C). Three different mannosidase inhibitors were employed to explore the role of mannose trimming for EDEM1 binding. Kifunensine (KIF) and 1-deoxymannojirimycin (DMJ) inhibit class I mannosidases such as ER ManI (Vallee et al., 2000). In contrast, swainsonine (SWN) inhibits class II mannosidases such as Golgi α-mannosidases (Moremen, 2002).
EDEM1 binding to NHK was unaffected regardless of the mannosidase inhibitor employed (Figure 2B and C). Furthermore, the inclusion of the glucosidase inhibitor, 1-deoxynojirimycin (DNJ), had no affect on EDEM1 binding. As observed previously, only KIF moderately stabilized NHK after a 1 hr chase (Hosokawa et al., 2001). These results demonstrated that EDEM1 bound NHK irrespective of glycan trimming and suggest that EDEM1 recognizes misfolded regions of aberrant proteins.
EDEM1 binds misfolded proteins independent of glycans
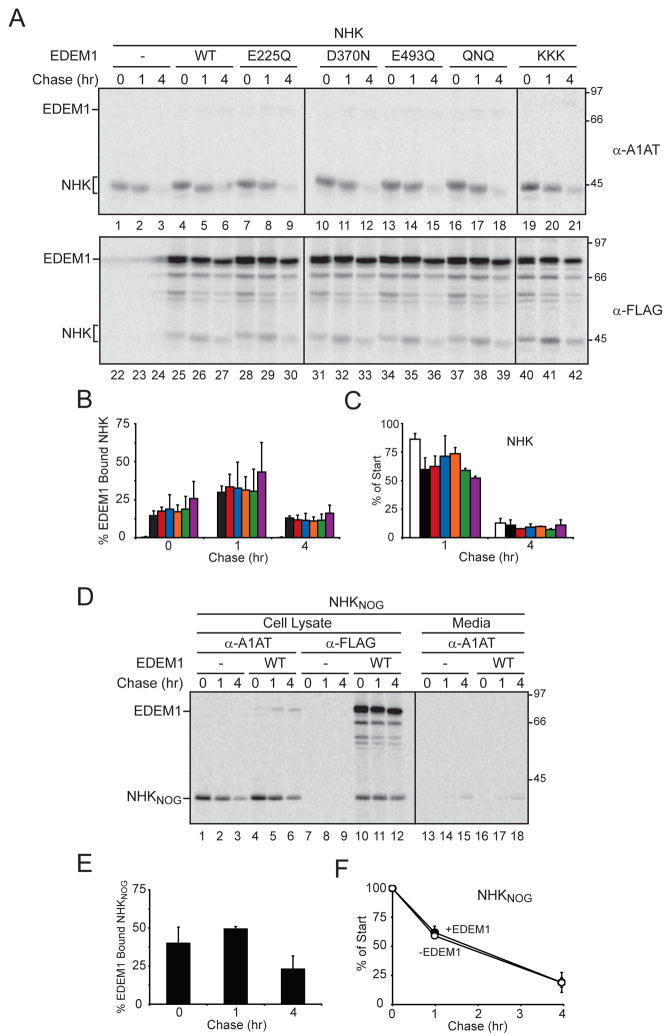
Human ER ManI and EDEM1 share 24% amino acid sequence identity and the three active site residues of ER ManI (Glu330, Asp463, and Glu599) are conserved in EDEM1 (Glu225, Asp370 and Glu493)(Karaveg et al., 2005). Mutation of the acidic residues of ER ManI to neutral amino acids abolished mannosidase activity while either enhancing (Glu330) or diminishing (Asp370 and Glu599) its affinity for Man9 glycans (Karaveg et al., 2005). The similar acidic residues in the mannosidase-like domain of EDEM1 were mutated to neutral amino acids to investigate their role in substrate binding. The EDEM1 mutants possessed half-lives similar to WT-tagged and endogenous EDEM1 (Figure S3B). NHK was co-expressed with either WT EDEM1, EDEM1 single (E225Q, D370N or E493Q) or triple (E225Q/D370N/E493Q; termed QNQ) site mutations and the binding to NHK was monitored.
The binding level to NHK for all three EDEM1 single site mutations was indistinguishable from WT EDEM1 (Figure 3A and B). The peak binding level of ~30% was found after a 1 hr chase (Figure 3B). Similar results were also observed with the triple mutant EDEM1-QNQ. The three acidic residues were also exchanged for bulky and positively charged Lys residues (E225/D370/E493 to K; termed KKK) to determine the effect on substrate binding using a more severe mutation that is expected to abolish any carbohydrate binding activity. Interestingly, EDEM1-KKK binding to NHK increased to 43% after 1 hr of chase indicating that an intact carbohydrate-binding domain was not required for substrate binding.
Figure 3. EDEM1 binding to misfolded glycoproteins does not require carbohydrates.
(A) NHK was co-expressed with blank vector (−), WT, EDEM1-FLAG single (E225Q, D370N, or E493Q) or triple (QNQ or KKK) mutants into 293T cells. Cells were radiolabeled for 15 min and chased for the indicated times. NHK and EDEM1 were isolated from the cell lysates with A1AT (α-A1AT) and FLAG (α-FLAG) antisera. Quantification of (B) % EDEM1 bound NHK and (C) % cellular NHK are displayed. Blank vector (white), WT EDEM1 (black), with the EDEM1 active site mutants as follows: E225Q (red), D370N (blue), E493Q (orange), QNQ (green) and KKK (purple) are designated in the bar graph. (D) NHKNOG was co-expressed with either blank vector (−) or WT EDEM1 in 293T cells. NHKNOG and EDEM1 were analyzed similar to A. Secreted NHKNOG was isolated from the cell culture media using A1AT antiserum. Quantification of (E) % EDEM1 bound NHKNOG and (F) % cellular NHKnog are displayed.
All the EDEM1 single and triple mutants accelerated the turnover of NHK, as the level of NHK observed after 1 hr of chase was decreased (Figure 3A and C). Furthermore, mutation of single or all three putative active site acidic residues combined caused a slight retardation in the mobility of NHK when compared to WT EDEM1 overexpression as observed by SDS-PAGE (Figure 3A, lanes 25–42, and Figure S3A). The difference in mobility was due to glycan trimming, as NHK migrated with identical mobilities after PNGase F treatment (Figure S3A). This observation is in agreement with previous findings using NHK and BACE as ERAD substrates that suggested that EDEM1 possesses mannosidase activity or works as an accessory protein for a mannosidase (Olivari et al., 2006). These results indicate that the putative catalytic residues of the mannosidase-like domain of EDEM1 are not required for binding to misfolded glycoproteins.
To further examine the carbohydrate requirement for EDEM1 binding to non-native proteins, the ability of EDEM1 to associate with NHK lacking all three glycosylation sites (NHKNOG) was analyzed. The three Thr in the N-linked glycosylation sites were mutated to Ala. EDEM1 bound 50% of NHKNOG after 1 hr of chase. The binding level of EDEM1 to NHKNOG was greater than that found for NHK containing all of its glycans (Figure 3D, lanes 10–12 and 3E compared to 1C) and it appeared to be found in a similar size complex to glycosylated NHK (Figures S2A). However, the degradation kinetics of the misfolded unglycosylated protein was unaffected by the overexpression of WT EDEM1 (Figure 3D, lanes 1–6, and 3F). Altogether, these results indicate that EDEM1 can recognize and bind misfolded proteins in a glycan-independent manner.
The mannosidase-like domain of EDEM1 is involved in SEL1L binding
Since the mannosidase-like domain of EDEM1 does not appear to be required for ERAD substrate binding, we explored the possibility that it was utilized for binding to downstream ERAD machinery. In S. Cerevisiae, an ER membrane ubiquitin-ligase complex involved in the dislocation and ubiquitination of proteins containing lumenal lesions was identified that contained Hrd 1p, Hrd3p and Der1p (Carvalho et al., 2006; Denic et al., 2006). Hrd1p is an ubiquitin E3 ligase, while Hrd3p is an adapter protein that regulates its function (Vembar and Brodsky, 2008). Hrd3p has a large lumenal domain that supports interactions with lumenal quality control machinery such as Yos9p, a proposed quality control receptor (Denic et al., 2006). The mammalian homologue of yeast Hrd3p is SEL1L (Mueller et al., 2008). As EDEM1 can specifically recognize aberrant proteins, we next determined whether EDEM1 interacts with SEL1L.
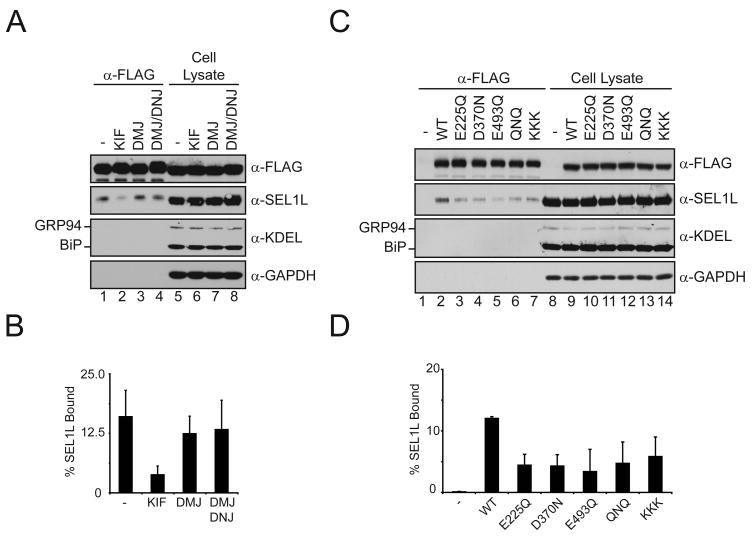
EDEM1 was expressed in 293T cells and immuno-isolated. Immunoblotting of the isolated fractions with antisera to ER and cellular control proteins revealed that EDEM1 bound ~15% of the total SEL1L (Figure 4A and B). EDEM1 binding to GRP94, BiP or GAPDH was not observed under these conditions. The interaction between EDEM1 and SEL1L was investigated using glycosidase inhibitors to characterize the requirement for carbohydrate trimming. EDEM1 binding to SEL1L was only affected by the addition of KIF, which caused a 5-fold decrease in binding. The nature of the EDEM1-SEL1L interaction was further explored by determining if the putative active site residues of the mannosidase-like domain of EDEM1 were utilized for interacting with SEL1L. Mutation of the mannosidase-like domain acidic residues decreased the binding for SEL1L by 2 to 3-fold (Figure 4C and D). Altogether, these results suggest that the putative catalytic residues of the mannosidase-like domain of EDEM1 are involved in binding to SEL1L, and not the binding or recognition of ERAD substrates.
Figure 4. EDEM1 requires its mannosidase-like domain to interact with SEL1L.
(A) WT EDEM1-FLAG was transfected into 293T cells, which were treated in the absence or presence of 150 μM KIF, 1 mM DMJ, or 1 mM DMJ and 0.5 mM DNJ for 5 hr. EDEM1 was immunoprecipitated using FLAG antiserum (α-FLAG). Isolated complexes and 50% of their representative total cell lysate were resolved on a 10% reducing SDS-PAGE, and immunoblotted with antisera for the indicated proteins. (C) Blank vector (−), WT and mannosidase-like domain mutants of EDEM1 were transfected into 293T cells. EDEM1 complexes were analyzed similar to A. Quantification of percent SEL1L bound by EDEM1 from A (B) and C (D) are displayed.
DISCUSSION
We characterized the binding properties of EDEM1 and found that EDEM1 is able to discriminate between non-native and native proteins during protein maturation and quality control. EDEM1 binding to ERAD substrates did not require the trimming of substrate glycans or for the substrate to be glycosylated. Instead, EDEM1 appeared to use its mannosidase-like domain to bind to the downstream ERAD machinery target, SEL1L. These results support a role for EDEM1 in the recognition of aberrant proteins and their delivery to an ER membrane ubiquitin-ligase complex by binding to the adapter protein SEL1L.
EDEM1 bound mutant variants of the soluble ERAD substrate A1AT (NHK and Z; Figure 1C and Figure S1A), and the membrane-integrated ERAD substrate tyrosinase (TYR-C71R, Figure S1B). Soluble substrates are solely reliant on lumenal selection for ERAD. We chose to characterize the binding of EDEM1 to NHK in detail since it is arguably the most thoroughly studied soluble mammalian ERAD substrate. Previous studies have demonstrated that pharmacological mannosidase inhibition stabilized NHK, and its turnover was accelerated by the overexpression of mannosidases or EDEM family members (Hosokawa et al., 2003; Hosokawa et al., 2001; Liu et al., 1999). Short hairpin RNA knockdown of HRD1 or SEL1L also decreased the rate of NHK degradation, indicative of their involvement in NHK clearance (Christianson et al., 2008). These results demonstrate that after recognition in the ER lumen as an ERAD substrate, NHK is degraded through the ERAD pathway involving the HRD1/SEL1L complex.
The ERAD process in yeast and mammalian cells appears to utilize a number of protein complexes for recognition, dislocation, ubiquitination and degradation (Carvalho et al., 2006; Christianson et al., 2008; Denic et al., 2006; Mueller et al., 2008). EDEM1 was found recently to reside in a complex with ERdj5 and BiP (Ushioda et al., 2008). ERdj5 possesses reductase activity and a J-domain, which recruits the ER Hsp70 chaperone BiP. This EDEM1-ERdj5-BiP complex is predicted to select and maintain ERAD substrates in a translocation competent form for dislocation to the cytoplasm. Interestingly, we found that the depletion of ERdj5 by siRNA did not affect NHK binding to EDEM1 (Figure S6).
The involvement of protein complexes complicates the interpretation of experiments where the effect on ERAD substrate turnover is monitored after the overexpression of a single subunit from a complex. The creation of potentially orphan subunits might disrupt the function of the complex, producing a dominant-negative effect. For instance, OS-9 and XTP3-B, two putative ERAD receptors observed in complexes with GRP94 and/or BiP, stabilize NHK when they are individually overexpressed (Christianson et al., 2008; Hosokawa et al., 2008). However, overexpression studies can be used effectively to facilitate the probing of the binding properties for the individual components of the ERAD network if the overexpressed factor directly binds the substrate. The ability of EDEM1 to bind NHK in the absence of ERdj5 combined with the size of the EDEM1-NHK complex as determined by ultracentrifugation are consistent with EDEM1 binding directly to NHK in a 1:1 complex.
The current glycoprotein quality control hypothesis predicts that the mannose trimming of ERAD substrate glycans by a mannosidase provides a demannosylated signal on the aberrant secretory cargo for ERAD (Cabral et al., 2001; Hebert and Molinari, 2007). Glycans trimmed to a glycoform ranging from Man8 to Man5 are proposed to create the signal that is recognized by carbohydrate-binding quality control receptors (Jakob et al., 1998). Recent studies suggest that terminal α1,6-linked mannose oligosaccharides provide the signpost for defective glycoproteins (Clerc et al., 2009; Hosokawa et al., 2009; Quan et al., 2008). In this current ER quality control model, the substrate-receptor complex is then targeted to an ER membrane complex for dislocation, ubiquitination and subsequent degradation by the cytosolic proteasome. EDEM1 has been proposed to either act as the mannosidase that creates the demannosylated signal or the quality control receptor that recognizes and sorts mannose-trimmed proteins for ERAD by extracting them from the calnexin binding cycle (Molinari et al., 2003; Oda et al., 2003; Olivari et al., 2006).
Our findings from the analysis of the binding properties of EDEM1 are in conflict with the current models described above. We found that EDEM1 appeared to directly recognize non-native structures, as it associated with NHK irrespective of its glycosylation status. However, EDEM1 did not bind or accelerate the turnover of two naturally nonglycosylated ERAD substrates κ light chain or mutant transthyretin (Figures S4 and S5)(Okuda-Shimizu and Hendershot, 2007; Sekijima et al., 2005). The ability of EDEM1 to recognize aberrant structures appears to be highly substrate-dependent. If EDEM1 can also act as a mannosidase, it must behave in a nontraditional manner as it selectively, efficiently and persistently bound substrate, properties not shared by other glycosidases.
An intact mannosidase-like domain of EDEM1, expected to be required for substrate recognition or signaling, was instead required for binding to the downstream ERAD machinery. EDEM1 binding to SEL1L was disrupted by mutation of the acidic amino acids that are putative mannose interacting residues in EDEM1 or mannosidase inhibition with KIF. KIF and DMJ treatment both inhibit mannose trimming resulting in the accumulation of Man9 glycans (Avezov et al., 2008). However, only KIF treatment inhibited EDEM1 binding to SEL1L. As KIF is a mannose analogue, binding inhibition may involve its direct association with the EDEM1 mannosidase-like domain rather than the accumulation of untrimmed mannose side chains.
Kopito and colleagues recently demonstrated that OS-9 and XTP3-B interacted with SEL1L (Christianson et al., 2008). Mutations to their mannose-6-phosphate receptor homology (MRH) domains perturbed associations with SEL1L indicating that the MRH domains of OS-9 and XTP3-B are used for binding to SEL1L, likely through the glycans of SEL1L. Human SEL1L possesses five N-linked glycosylation sites and the bulk of its lumenal domain is comprised of 11 tetratricopeptide repeats (TPR). TPR domains mediate protein-protein interactions supporting the recruitment of a variety of proteins including chaperones from the Hsp70 and Hsp90 families (D’Andrea and Regan, 2003). Therefore, EDEM1 binding to SEL1L may involve bipartite interactions including its mannosidase-like domain binding the large and flexible glycans of SEL1L, and protein-protein association with the TPR domains. Interactions with the TPR domains of SEL1L maybe mediated directly through EDEM1 or a member of the EDEM1 complex such as BiP or ERdj5. Together, these results support a modified model whereby the mannosidase-like domain or the MRH domains of these quality control receptors mediate interactions with the downstream ERAD apparatus providing an alternative glycan-dependent mechanism for targeting and delivery of aberrant secretory cargo through the ERAD pathway.
A detailed understanding of the recognition and delivery process for the ERAD pathway will require the reconstitution of these processes using purified components. In vitro activity studies are needed to understand whether the additional trimming observed after EDEM1 overexpression was due to EDEM1 directly possessing mannosidase activity or the potential association of EDEM1 with a mannosidase. In support of the later possibility, EDEM1 was recently found to bind and stabilize ER ManI (Termine et al., 2009). As most of the proteins involved in the ER quality control selection and delivery process are themselves glycosylated, the interpretation of results using chemical glycosidase inhibitors or the modulation of glycosidase levels through overexpression or RNAi approaches is complicated. It has been widely assumed that perturbations to the glycan status are mediated through substrate glycans. However, results presented here suggest that the glycan status of the ERAD machinery itself also appears to play an important role in shepherding ERAD substrates and factors through the ERAD pathway.
EXPERIMENTAL PROCEDURES
Cell culture and transfections
Human embryonic kidney (HEK) 293T cells were maintained in DMEM supplemented with 10% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin and incubated at 37°C in 5% CO2. Single and co-transfections of plasmids into cells were accomplished using Lipofectamine 2000, following the manufacturer’s instructions. For all co-transfections, EDEM1 and its mutants were transfected at a plasmid molar ratio of 2:1 to the plasmid encoding the substrate protein. Transfections were incubated 16 hr before pulse labeling or harvesting.
Starvation free pulse-chase labeling, immunoprecipitation, and immunoblotting
Pulse chase labeling, immunoprecipitation and immunoblotting was performed as previously described, with a modified exclusion of the starvation period from the pulse-chase experiments (Svedine et al., 2004). Isolation of post-nuclear supernatant (PNS) and all subsequent immunoprecipitation (IP) steps were conducted at 4 °C. Cells were lysed in 0.5% Triton X-100 in MNT buffer [20 mM MES, 100 mM NaCl, 20 mM Tris-HCl (pH 7.5)] containing protease inhibitors. The PNS was cleared with 10% Zysorbin for 1 hr. The clarified supernatant was incubated with the indicated antiserum complexed to protein A-sepharose and rotated for 16 hr. Immune-complexes were washed twice with 0.05% Triton X-100, 0.1% SDS, 300 mM NaCl, 10 mM Tris-HCl (pH 8.6), resuspended in reducing sample buffer, and resolved on SDS-PAGE.
Statistical analysis
Quantification of % of start = ([35S-protein at an indicated chase time]/[35S-protein at the 0 hr chase time])* 100. % EDEM1 bound A1AT = ([A1AT at the indicated chase time after α-Flag IP]/[A1AT at the 0 hr chase time after α-A1AT IP])* 100. % SEL1L bound = ([SEL1L protein after α-FLAG IP]/[SEL1L cell lysates*2])*100. Error bars represent the standard deviation for at least three independent experiments.
Supplementary Material
Acknowledgments
We would like to thank Brad Pearse and Kristina Moody for helpful comments, and Dr. M. Ziak, J. Roth (Zurich), L. Hendershot (Memphis, TN) and J. Kelly (La Jolla, CA) for providing constructs. This work was supported by US Public Health grant CA79864 (to D. N. H.) and the Uehara Memorial Foundation (to T. T.).
Footnotes
The Supplemental Data includes Experimental Procedures and six figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avezov E, Frenkel Z, Ehrlich M, Herscovics A, Lederkremer GZ. Endoplasmic Reticulum (ER) Mannosidase I Is Compartmentalized and Required for N-Glycan Trimming to Man5 6GlcNAc2 in Glycoprotein ER-associated Degradation. Mol Biol Cell. 2008;19:216–225. doi: 10.1091/mbc.E07-05-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral CM, Liu Y, Sifers RN. Dissecting glycoprotein quality control in the secretory pathway. Tren Biochem Sci. 2001;26:619–624. doi: 10.1016/s0968-0004(01)01942-9. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1/SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nature Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Foellmer B, Helenius A. Glucose trimming and reglucosylation determines glycoprotein association with calnexin. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Molinari M. In and Out of the ER: Protein Folding, Quality Control, Degradation, and Related Human Diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- Herscovics A, Romero PA, Tremblay LO. The specificity of the yeast and human class I ER alpha1,2-mannosidase involved in ER quality control is not as strict as previously reported. Glycobiology. 2002;12:14G–15G. [PubMed] [Google Scholar]
- Hosokawa N, Kamiya Y, Kamiya D, Kato K, Nagata K. Human OS-9, a lectin required for glycoprotein ERAD, recognizes mannose-trimmed N-glycans. J Biol Chem. 2009 doi: 10.1074/jbc.M809725200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Tremblay LO, You Z, Herscovics A, Wada I, Nagata K. Enhancement of endoplasmic reticulum (ER) degradation of misfolded null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J Biol Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, Nagata K. A novel ER a-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001;2:415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Nagasawa K, Moriyama T, Okawa K, Nagata K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J Biol Chem. 2008;283:20914–20924. doi: 10.1074/jbc.M709336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob CA, Burda P, Roth J, Aebi M. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J Cell Biol. 1998;142:1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaveg K, Siriwardena A, Tempel W, Liu ZJ, Glushka J, Wang BC, Moremen KW. Mechanism of class 1 (glycosylhydrolase family 47) {alpha}-mannosidases involved in N-glycan processing and endoplasmic reticulum quality control. J Biol Chem. 2005;280:16197–16207. doi: 10.1074/jbc.M500119200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Choudhury P, Cabral CM, Sifers RN. Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J Biol Chem. 1999;274:5861–5867. doi: 10.1074/jbc.274.9.5861. [DOI] [PubMed] [Google Scholar]
- Molinari M, Calanca V, Galli C, Lucca P, Pagnetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- Moremen KW. Golgi alpha-mannosidase II deficiency in vertebrate systems: implications for asparagine-linked oligosaccharide processing in mammals. Biochim Biophys Acta. 2002;1573:225–235. doi: 10.1016/s0304-4165(02)00388-4. [DOI] [PubMed] [Google Scholar]
- Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci U S A. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Hosokawa N, Wada I, Nagata K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299:1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari S, Cali T, Salo KE, Paganetti P, Ruddock LW, Molinari M. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem Biophys Res Commun. 2006;349:1278–1284. doi: 10.1016/j.bbrc.2006.08.186. [DOI] [PubMed] [Google Scholar]
- Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, Weissman JS. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekijima Y, Wiseman RL, Matteson J, Hammarstrom P, Miller SR, Sawkar AR, Balch WE, Kelly JW. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Sifers RN, Brashears-Macatee S, Kidd VJ, Muensch H, Woo SL. A frameshift mutation results in a truncated alpha 1-antitrypsin that is retained within the rough endoplasmic reticulum. J Biol Chem. 1988;263:7330–7335. [PubMed] [Google Scholar]
- Svedine S, Wang T, Halaban R, Hebert DN. Carbohydrates act as sorting determinants in ER-associated degradation of tyrosinase. J Cell Sci. 2004;117:2937–2949. doi: 10.1242/jcs.01154. [DOI] [PubMed] [Google Scholar]
- Termine DJ, Moremen KW, Sifers RN. The mammalian UPR boosts glycoprotein ERAD by suppressing the proteolytic downregulation of ER mannosidase I. J Cell Sci . 2009;122:976–984. doi: 10.1242/jcs.037291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- Vallee F, Karaveg K, Herscovics A, Moremen KW, Howell PL. Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases. J Biol Chem. 2000;275:41287–41298. doi: 10.1074/jbc.M006927200. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber C, Cormier JH, Guhl B, Santimaria R, Hebert DN, Roth J. EDEM1 reveals a quality control vesicular transport pathway out of the endoplasmic reticulum not involving the COPII exit sites. Proc Natl Acad Sci U S A. 2007;104:4407–4412. doi: 10.1073/pnas.0700154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.