Abstract
Fibroblasts are major cellular components of the tumor microenvironment, regulating tumor cell behavior in part through secretion of extracellular matrix proteins, growth factors and angiogenic factors. In previous studies, conditional deletion of the type II TGF-β receptor in fibroblasts (Tgfbr2FspKO) was shown to promote mammary tumor metastasis in fibroblast: epithelial cell co-transplantation studies in mice, correlating with increased expression of HGF. Here, we advance our findings to show that Tgfbr2FspKO fibroblasts enhance HGF/c-Met and HGF/Ron signaling to promote scattering and invasion of mammary carcinoma cells. Blockade of c-Met and Ron by siRNA silencing and pharmacologic inhibitors significantly reduced mammary carcinoma cell scattering and invasion caused by Tgfbr2FspKO fibroblasts. Moreover, neutralizing antibodies to c-Met and Ron significantly inhibited HGF-induced cell scattering and invasion correlating with reduced Stat3 and p42/44MAPK phosphorylation. Investigation of the Stat3 and MAPK signaling pathways by pharmacologic inhibition and siRNA silencing revealed a cooperative interaction between the two pathways to regulate HGF- induced invasion, scattering and motility of mammary tumor cells. Furthermore, while c-Met was found to regulate both the Stat3 and MAPK signaling pathways, Ron was found to regulate Stat3, but not MAPK signaling in mammary carcinoma cells. These studies demonstrate a tumor suppressive role for TGF-β signaling in fibroblasts, in part by suppressing HGF signaling between mammary fibroblasts and epithelial cells. These studies characterize complex functional roles for HGF and TGF-β signaling in mediating tumor: stromal interactions during mammary tumor cell scattering and invasion, with important implications in the metastatic process.
Keywords: Type II TGF-beta receptor, HGF, Mammary epithelium, Fibroblast, Stat3, MAPK
Introduction
Fibroblasts are a major cellular component of the stroma, influencing tumor cell behavior directly and indirectly through secretion of growth regulators and angiogenic factors, extracellular matrix (ECM) proteins and proteases [1, 2]. Studies have shown that heritable changes in the stroma, including mutations in the TP53 and PTEN genes, correlate with malignancy of breast cancers [3]. Furthermore, co-implantation of lymphoma, bladder or mammary epithelial cells with carcinoma associated, fetal, or irradiated fibroblasts result in enhanced tumor growth and invasion [4–9]. These studies indicate that heritable changes in fibroblasts alter the signaling interactions with epithelial tumor cells to promote tumor progression.
A number of studies have demonstrated an important role for TGF-β signaling in regulating stromal: tumor interactions. TGF-β signaling exerts multiple effects in both stromal and epithelial cells including inhibiting cell proliferation, promoting motility and regulating differentiation. All of these cellular functions are achieved by binding of the TGF-β ligand to its cell surface type II receptor which leads to recruitment and activation of its type I receptor and subsequent downstream signaling in the cytoplasm of multiple pathways including SMAD, MAPK and Rho pathways [10, 11]. In cultured fibroblasts, TGF-β activates fibroblasts, inducing production of growth factors, angiogenic factors, ECM proteins and proteases [12–14]. Studies from our laboratory using a conditional knockout of the TGF-β type II receptor in fibroblasts (Tgfbr2FspKO) [15] have further revealed a significant role for TGF-β signaling in host:tumor interactions. Cre mediated deletion of exon 2 of the Tgfbr2 gene flanked with loxP sites in fibroblasts was shown to abrogate TGF-β signaling in the stroma, resulting in the development of carcinoma of the forestomach and prostatic intraepithelial neoplasias in homozygous Tgfbr2 knockout mice (Tgfbr2FspKO mice) [16]. Interestingly, while mammary development was severely inhibited in Tgfbr2FspKO mice, mammary Tgfbr2FspKO fibroblasts grafted with PyVmT or 4T1 mammary carcinoma cells in mice resulted in enhanced primary tumor growth and metastasis [17] compared to mice grafted with cre negative control fibroblasts (homozygous floxed Tgfbr2 or Tgfbr2Flox/Flox), indicating a unique role for TGF-β signaling in the mammary stromal microenviroment. Tgfbr2 deficiency in fibroblasts was found to result in enhanced HGF secretion and correlate with enhanced phosphorylation c-Met and Ron receptors in the mammary tumor epithelium [16, 17] suggesting that interactions between TGF-β and HGF signaling were important in mediating stromal: epithelial interactions during mammary tumor progression.
Previous studies have established HGF as an important stromal- derived soluble factor in regulating normal and tumorigenic processes. Expressed primarily by fibroblasts, HGF has been shown to act on epithelial cells expressing c-Met receptor tyrosine kinases to regulate a number of cellular processes including cell proliferation, survival, migration and branching, which are necessary for tissue development, including mammary development [11]. These cellular processes have been shown to be regulated by number of signaling pathways including Stat3 and p42/44MAPK pathways [11, 18, 19]. Aberrant expression of HGF or c-Met has been correlated with the development of a number of cancers including colorectal, renal and breast cancers [20], and overexpression or deletion of HGF and c-Met in animal studies confirm the oncogenecity of HGF signaling. Moreover, systemic blockade of c-Met signaling in orthotopic and xenograft mouse models using various inhibitors including soluble receptors and small molecule inhibitors [21–23] have been shown to inhibit primary mammary tumor growth and metastatic spread. While the HGF/c-Met signaling pathways represent a promising therapeutic target, studies have shown that HGF may also regulate other receptor tyrosine kinases including a c-Met related receptor, Ron to regulate tumorigenesis, whose functions in breast cancer still remain unclear [24, 25]. Understanding the multiple signaling pathways activated by HGF has an important implication in cancer therapeutics when predicting possible side effects and therapeutic outcome during patient treatment.
We had recently shown that specific inhibition of c-Met in a tumor xenograft model blocks metastatic spread caused by Tgfbr2FspKO fibroblasts [26]. In the present studies, we advance our previous findings to demonstrate that TGF-β and HGF signaling interact to regulate fibroblast: tumor interactions at the molecular and cellular levels. Using multiple approaches including pharmacologic blockade and siRNA silencing to block HGF signaling, we demonstrate that Tgfbr2FspKO fibroblasts enhance HGF/c-Met and HGF/Ron signaling pathways in mammary carcinoma cells to promote cell scattering and invasion. Furthermore, we demonstrate that HGF/c-Met and HGF/Ron signaling regulates these cellular process by mediating interactions between Stat3 and MAPK signaling. These studies characterize complex functional roles for HGF and TGF-β signaling in mediating tumor: stromal interactions during mammary carcinoma cell scattering and invasion, with important implications in the tumor metastatic process.
Results
Tgfbr2FspKO fibroblasts promote mammary carcinoma cell scattering and invasion associated with enhanced HGF signaling
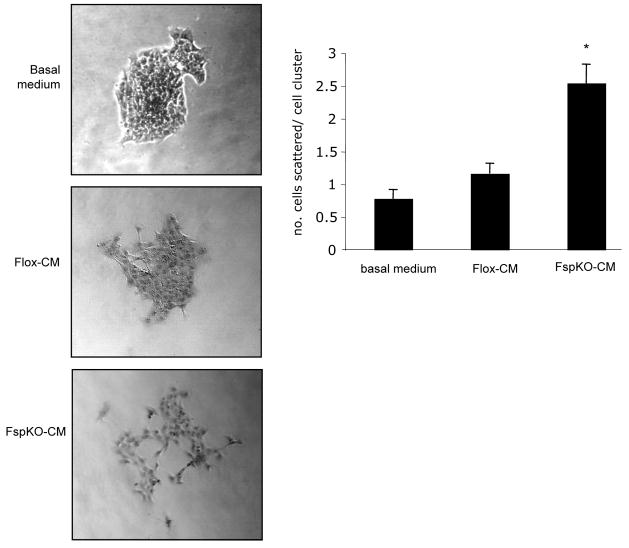
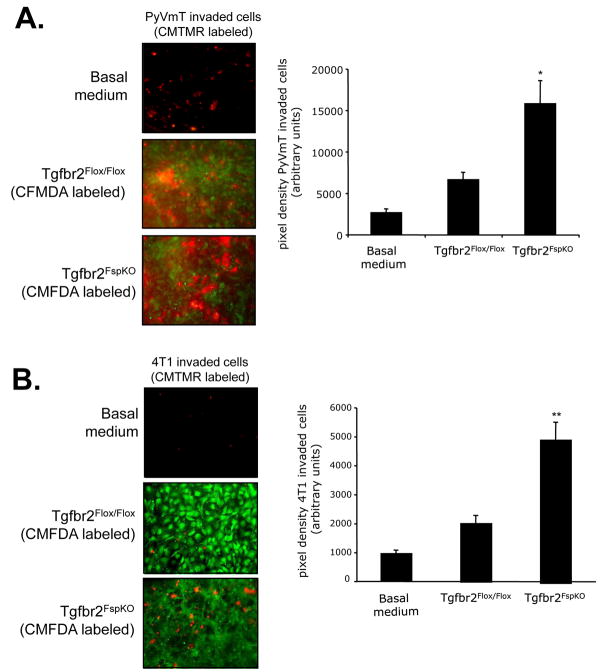
The abilities of epithelial cells to undergo epithelial to mesenchymal transition, disassociate from the primary tumor, migrate and invade through the basement membrane are important to the metastatic process. The effects of Tgfbr2FspKO fibroblasts on cellular scattering and invasion of mammary carcinoma cells were determined through co-culture of carcinoma cells and mammary fibroblasts, using immortalized Tgfbr2flox/flox and Tgfbr2FspKO fibroblasts, which were previously shown to behave in a similar and stable manner to primary mammary fibroblasts [17]. Conditioned medium from Tgfbr2FspKO fibroblasts significantly increased 4T1 cell scattering compared to medium from control Tgfbr2Flox/Flox fibroblasts or basal medium (Figure 1). Furthermore, Tgfbr2FspKO fibroblasts (labeled with CMFDA Celltracker dye) also significantly enhanced invasion of PyVmT and 4T1 mammary carcinoma cells (labeled with CMTMR Celltracker dye) through a Matrigel coated transwell membrane (Figures 2A and B).
FIGURE 1. Tgfbr2FspKO fibroblasts promote mammary carcinoma cell scattering.
4T1 cells were cultured in clusters of 50–75 cells and treated with basal medium (DMEM/F12/0.5% ABS) or conditioned medium fromTgfbr2Flox/Flox (Flox-CM) or Tgfbr2FspKO fibroblasts (FspKO-CM) for 24 hours. The number of cells scattered per cell cluster were counted. Statistical significance was determined by Two Tailed T-test; * vs. Flox-CM, p<0.05. Data are represented as mean±standard error of the mean (SEM).
FIGURE 2.
Tgfbr2FspKO fibroblasts promote mammary carcinoma cell invasion. Fibroblasts were labeled with CMFDA (green) fluorochrome and plated on the underside of a Matrigel coated transwell membrane. A. PyVmT tumor cells or B. 4T1 cells were labeled with CMTMR (red) fluorochrome and plated on topside of the transwell filter in the presence or absence of Tgfbr2Flox/Flox control fibroblasts or Tgfbr2FspKO fibroblasts for 8 h. As a control, PyVmT and 4T1 tumor cells were seeded onto Matrigel coated transwell filters in basal medium containing DMEM/F12/0.5% ABS, in the absence of fibroblasts. Invaded tumor cells were visualized by fluorescence microscopy and micrographed at 20× magnification; an overlay of invaded carcinoma cells and fibroblasts is shown. Statistical significance was determined by Two Tailed T- test; *vs. Tgfbr2□lox/Flox fibroblasts, p<0.05, ** vs. Tgfbr2Flox/Flox fibroblasts, p<0.001. Data are represented as mean± SEM.
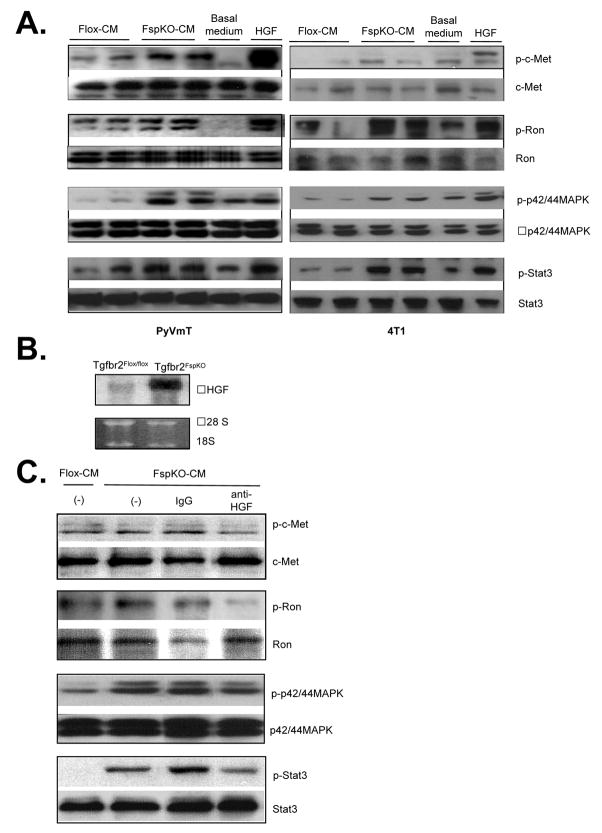
In previous studies, primary tumors grafted with Tgfbr2FspKO fibroblasts in nude mice showed increased levels of phosphorylated c-Met and Ron receptor proteins [17, 27]. To determine if this enhanced phosphorylation was specifically due to altered paracrine signaling interactions between the Tgfbr2FspKO fibroblasts and mammary epithelial cells, 4T1 and PyVmT cells were treated with conditioned medium from Tgfbr2Flox/flox control fibroblasts or Tgfbr2FspKO fibroblasts and analyzed by western blot. Tgfbr2FspKO fibroblast conditioned medium significantly enhanced phosphorylation of the c-Met and Ron receptors and the downstream signaling proteins Stat3 and p42/44MAPK (Figure 3A). These effects correlated with increased levels of HGF transcripts in Tgfbr2FspKO fibroblasts, according to northern analysis (Figure 3B). Furthermore, neutralizing antibodies to HGF reduced phosphorylation of c-Met, Ron, Stat3 and p42/44MAPK proteins in 4T1 cells enhanced by Tgfbr2FspKO fibroblast conditioned medium (Figure 3C). These results indicate that increased HGF expression from Tgfbr2 deficient fibroblasts enhance c-Met and Ron signaling in mammary carcinoma cells.
FIGURE 3.
Conditioned medium from Tgfbr2FspKO fibroblasts enhances HGF signaling in mammary carcinoma cells. A. PyVmT (left panel) or 4T1 (right panel) carcinoma cells were treated with basal medium containing DMEM/F12/0.5% ABS, HGF (60 ng/ml) or fibroblast conditioned medium from Tgfbr2Flox/Flox control or Tgfbr2FspKO fibroblasts, and analyzed by western blot analysis for phosphorylation status of indicated proteins. Experiments were performed in quaduplicate with duplicate samples. Representative results are shown. B. Northern blot analysis of Tgfbr2Flox/Flox control and Tgfbr2FspKO fibroblasts. C. 4T1 cells were treated with fibroblast conditioned medium in the presence or absence of anti-HGF or control IgG and analyzed by western blot for the phosphorylation status of indicated proteins.
c-Met and Ron signaling cooperate to mediate mammary carcinoma cell scattering and invasion enhanced by Tgfbr2FspKO fibroblasts
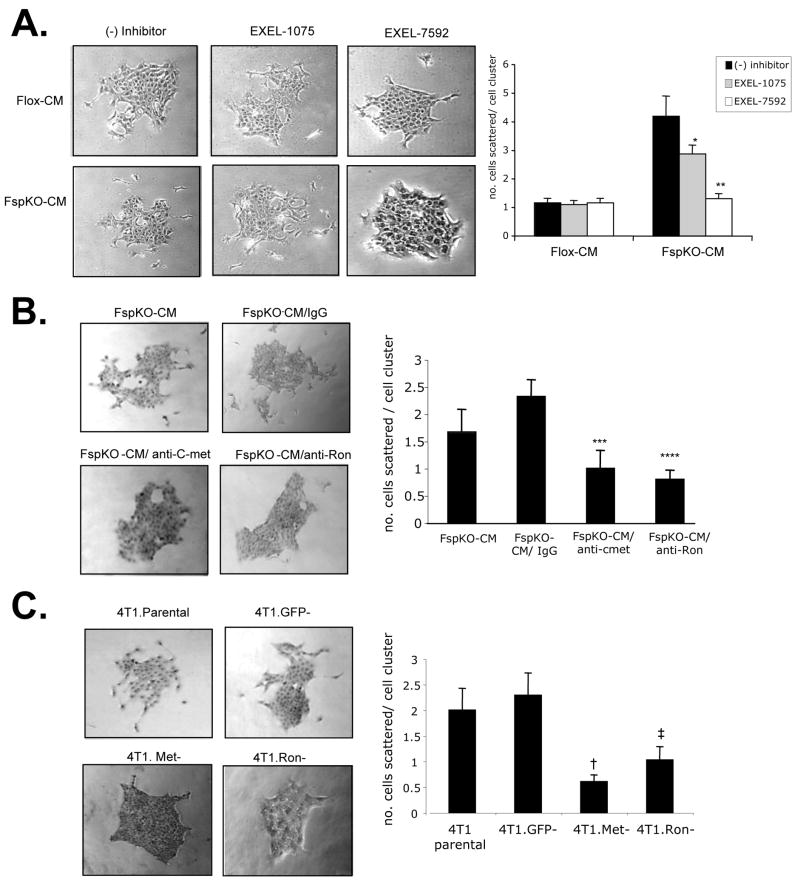
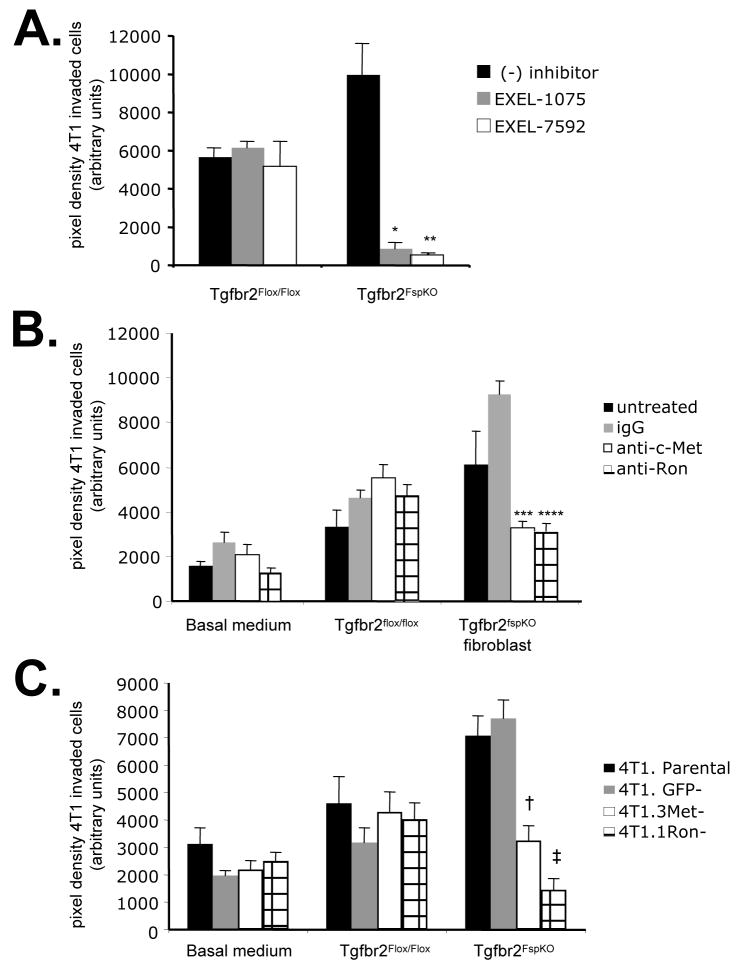
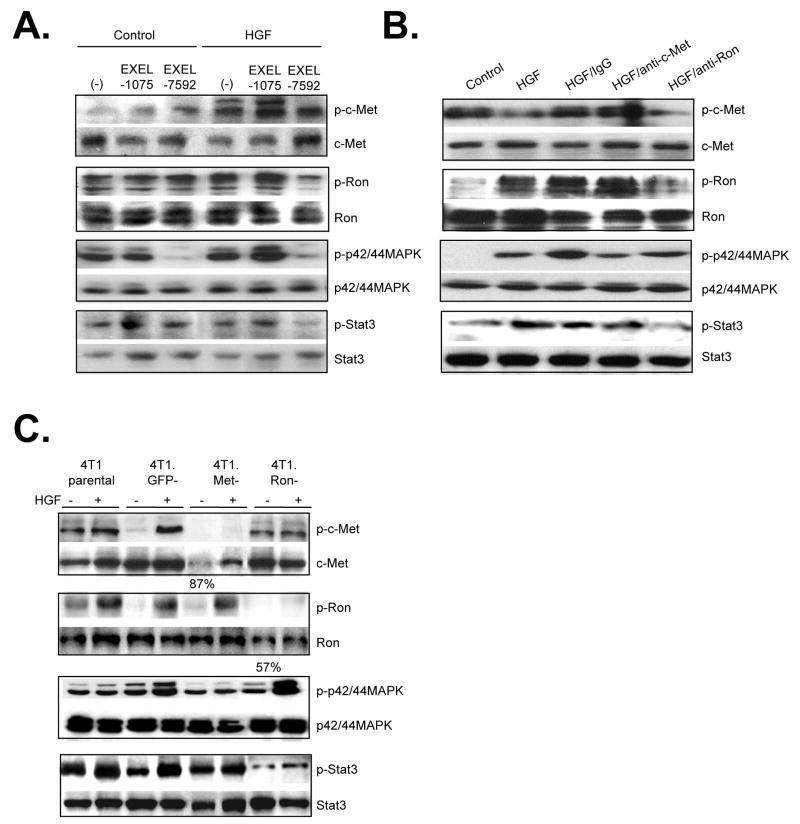
To determine the functional contribution of HGF derived from Tgfbr2FspKO fibroblasts in carcinoma cell scattering and invasion, HGF signaling was inhibited using multiple approaches. In previous studies, tumor bearing mice treated with a VEGFR/c-Met inhibitor, EXEL-7592, potently inhibited tumor growth and metastases, and significantly decreased c-Met phosphorylation in primary mammary tumor tissues, compared to inhibition of VEGFR alone with EXEL-1075 [26]. These studies indicated a potential clinical significance for targeting the c-Met pathway in breast cancer with EXEL-7592, with a need to further determine the molecular and cellular mechanisms of this inhibitor. In the present studies, treatment of 4T1 cells with 250 nM EXEL-7592 significantly reduced cell scattering induced by Tgfbr2FspKO fibroblast conditioned medium, with increased numbers of rounded cell clusters (Figure 4A). EXEL-1075 treatment also reduced cell scattering induced by Tgfbr2FspKO fibroblasts, but was not found to be statistically significant. The inhibitory effect of EXEL-1075 was consistent with the low level VEGFR2 expression detected in 4T1 tumor cells by western analysis (data not shown). Neither EXEL-7592 nor EXEL-1075 significantly affected scattering of 4T1 cells treated with control Tgfbr2Flox/Flox fibroblast conditioned medium, further indicating that Tgfbr2FspKO fibroblasts specifically enhanced HGF signaling of 4T1 carcinoma cells.
FIGURE 4.
Inhibition of c-Met and Ron receptors block 4T1 cell scattering enhanced by Tgfbr2FspKO fibroblast conditioned medium. 4T1 cells were treated with basal medium (DMEM/F12/0.5% ABS), or conditioned medium from Tgfbr2Flox/flox (Flox-CM) or Tgfbr2FspKO fibroblasts (FspKO-CM) in the presence or absence of the following c-Met or Ron inhibitors for 24 hours: A. Treatment of cells with 250 nM EXEL-7592 (VEGFR/c-Met inhibitor) or EXEL-1075 (VEGFR control inhibitor). B. 4T1 cells were treated with 1 μg/ml anti-c-Met, anti-Ron or control IgG. C. siRNA silencing of c-Met, Ron or control GFP, treatment with FspKO-CM. Quantitation of the expression of c-Met and Ron by densitometry analysis of western blots indicated an 87% reduction in c-Met and 57% reduction in Ron expression relative to expression in 4T1 Parental cells. Statistical significance was determined by two tailed T-test: * vs FspKO-CM; p=0.12, ** vs. FspKO-CM/EXEL1075; p<0.01, *** vs. FspKO-CM/IgG; p<0.01, **** vs FspKO-CM/IgG; p<0.01, † vs. 4T1.GFP-; p<0.01, ‡ vs. 4T1.GFP-; p<0.01. Data are represented as mean± SEM.
As Tgfbr2FspKO fibroblast conditioned medium enhanced the levels of phosphorylated c-Met and Ron receptor proteins, we determined the specific contribution of these receptors in cell scattering. Neutralizing antibodies to c-Met and Ron both reduced 4T1 cell scattering induced by Tgfbr2FspKO fibroblasts (Figure 4B), compared to control IgG treatment. The inhibitory effects of anti-c-Met and anti-Ron were also observed with MCF-7 and MDCK cell scattering enhanced by Tgfbr2FspKO fibroblast conditioned medium (Supplemental Figure 1A and 1B). In further studies, stable expression small interfering RNAs (siRNAs) to c-Met and Ron in 4T1 cells resulted in significant reductions cell scattering in response to Tgfbr2FspKO fibroblast conditioned medium, compared to 4T1 parental cells or control cells expressing siRNAs to GFP (4T1.GFP-) (Figure 4C). The results indicate that HGF derived from Tgfbr2FspKO fibroblasts significantly contribute 4T1 carcinoma cell scattering through both c-Met and Ron receptor activation.
We next determined the functional contribution of HGF signaling enhanced by Tgfbr2FspKO fibroblasts in 4T1 carcinoma cell invasion. EXEL-7592 treatment inhibited carcinoma cell invasion more significantly than EXEL-1075. EXEL-1075 also inhibited 4T1 invasion enhanced by Tgfbr2FspKO fibroblasts, consistent with low level VEGFR2 expression in 4T1 cells detected by western analysis (data not shown). Neither EXEL inhibitor significantly affected cellular invasion stimulated by control fibroblasts (Figure 5A). In addition, antibody neutralization of c-Met and Ron significantly inhibited 4T1 cell invasion enhanced by Tgfbr2 deficient fibroblasts, compared to control IgG treatment (Figure 5B). Similar effects of anti-c-Met and anti-Ron were also observed with PyVmT carcinoma cell invasion (Supplemental Figure 1C). Consistently, c-Met deficient and Ron deficient 4T1 cells (4T1.Met- and 4T1.Ron-) showed significantly decreased invasion through Matrigel matrix, compared to 4T1 parental or 4T1.GFP- cells (Figure 5C). These data indicate that Ron and c-Met receptor signaling in 4T1 enhanced by Tgfbr2 deficient fibroblasts is required for 4T1 carcinoma cell invasion.
FIGURE 5.
Inhibition of c-Met and Ron receptors block 4T1 carcinoma cell invasion induced by Tgfbr2Fspko fibroblasts. 4T1 cells labeled with CMTMR CellTracker dye were seeded onto Matrigel coated transwell membrane coated with Tgfbr2Flox/Flox or Tgfbr2FspKO fibroblasts labeled with CMFDA CellTracker dye and assessed for invasion after 8 h in the presence or absence of the following c-Met and Ron inhibitors: A. 250 nM EXEL-7592 or EXEL-1075. B. 1 μg/ml anti-c-Met, anti-Ron or control goat IgG. C. siRNA silencing of c-Met, Ron or control GFP. Experiments were performed in replicates of four and repeated in triplicate. Quantitation of carcinoma cells was determined by measuring pixel density of CMTMR labeled cells using Image J software (arbitrary units). Statistical significance was determined by Two Tailed T-test: * vs. Tgfbr2FspKO fibroblasts/(−) inhibitor, p<0.001; **vs. Tgfbr2FspKO fibroblasts/(−) inhibitor, p<0.001; ***vs Tgfbr2FspKO fibroblasts/IgG, p<0.01; ****vs Tgfbr2FspKO fibroblasts/IgG, p<0.01; † vs. 4T1.GFP-/Tgfbr2FspKO fibroblasts, p<0.05; ‡ vs. 4T1.GFP-/Tgfbr2FspKO fibroblasts, p<0.05. Data are represented as mean±SEM.
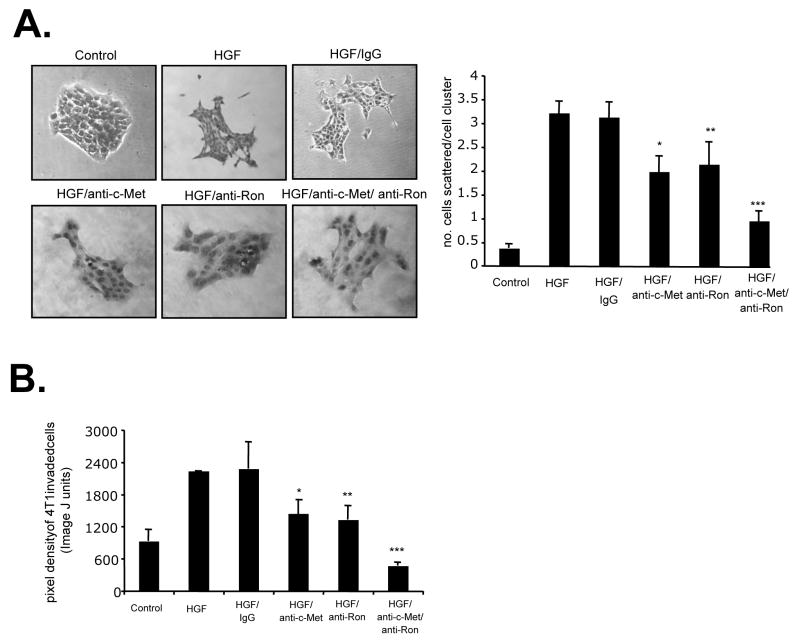
As both c-Met and Ron receptors were shown to regulate cellular scattering and invasion enhanced by Tgfbr2FspKO fibroblasts, we determined if these receptors cooperated in 4T1 cell scattering and invasion using neutralizing antibodies to c-Met and Ron. Treatment of 4T1 cells with both anti-c-Met and anti-Ron resulted in a more significant inhibition of scattering and invasion, compared to treatment with anti-c-Met or anti-Ron alone (Figures 6A and B). These data indicated an additive interaction between c-Met and Ron in regulating HGF- induced cell scattering and invasion in mammary carcinoma cells.
FIGURE 6.
Ron and c-Met receptors cooperate to mediate HGF- induced mammary tumor cell scattering, motility and invasion. 4T1 tumor cells were treated with Control (serum free medium) or HGF in the presence or absence of control goat IgG (1μg/ml) or neutralizing antibodies (1μg/ml) to c- Met and Ron, and assessed for the ability to A. scatter B. invade across a Matrigel coated membrane. Pixel density of invaded cells was measured by Image J software. Statistical significance was determined by Two Tailed T-test: *vs. HGF/IgG; p<0.01, **vs. HGF/IgG; p<0.01, and Two Way ANOVA Test: ***vs. HGF/anti-c-Met and HGF/anti-Ron treatments; p<0.05. Data are represented as mean±SEM.
Stat3 and p42/44MAPK signaling pathways cooperate to mediate 4T1 tumor cell scattering and invasion
We further determined the contribution of c-Met and Ron receptors in HGF- induced Stat3 and MAPK signaling in 4T1 cells. EXEL-7592 was found to significantly inhibit HGF-induced phosphorylation of Stat3 and p42/44MAPK proteins, associated with decreased phosphorylation of both c-Met and Ron (Figure 7A). These inhibitory effects of c-Met and Ron in 4T1 cells were consistent with in vitro kinase assays, in which EXEL-7592 inhibited Ron kinase activity with an IC50 of 8 nM and c-Met kinase activity with an IC50 of 0.4 nM. In addition, EXEL-7592 also inhibited several VEGFR isoforms with IC50 nM profiles similar to EXEL-1075 (Supplemental Tables 1 and 2). EXEL-1075 treatment did not significantly affect the phosphorylation status of c-Met, Ron, Stat3 or p424/44MAPK proteins (Figure 7A). Treatment of 4T1 tumor cells with neutralizing antibodies to c-Met inhibited HGF-induced phosphorylation of c-Met, p42/44MAPK and Stat3 proteins, but did not significantly alter Ron receptor phosphorylation. Neutralizing antibodies to Ron reduced the phosphorylation of Ron and Stat3 proteins, but did not significantly affect c-Met phosphorylation or p42/44MAPK phosphorylation (Figure 7B). Consistent with antibody neutralization experiments, 4T1.Met- cells showed decreased phosphorylation of c-Met, Stat3 and p42/44MAPK proteins while 4T1.Ron- cells exhibited reduced phosphorylation of Ron and Stat3 proteins (Figure 7C). These data suggest that c-Met and Ron receptors may function independently of each other, and that in response to HGF, c-Met activates both Stat3 and p42/44MAPK signaling while Ron receptors activate the Stat3 signaling pathway, with possible interactions at the Stat3 pathway.
FIGURE 7.
Ron and c-Met receptors signal independently to activate MAPK and Stat3 pathways in 4T1 carcinoma cells. 4T1 cells were treated with control (serum free medium) or HGF in the presence of absence of the following c-Met and Ron inhibitors and analyzed by western blot analysis for the phosphorylated status of indicated proteins: A. Treatment of cells with 250 nM EXEL-7592 or control KDR inhibitor EXEL-1075 B. Treatment of cells with neutralizing antibodies to c-Met and Ron. C. siRNA silencing of c-Met and Ron. Expression of c-Met and Ron was quantified by densitometry analysis of western blots and is shown as a percentage of expression relative to c-Met and Ron expression in 4T1 Parental cells. Representative results of triplicate experiments are shown.
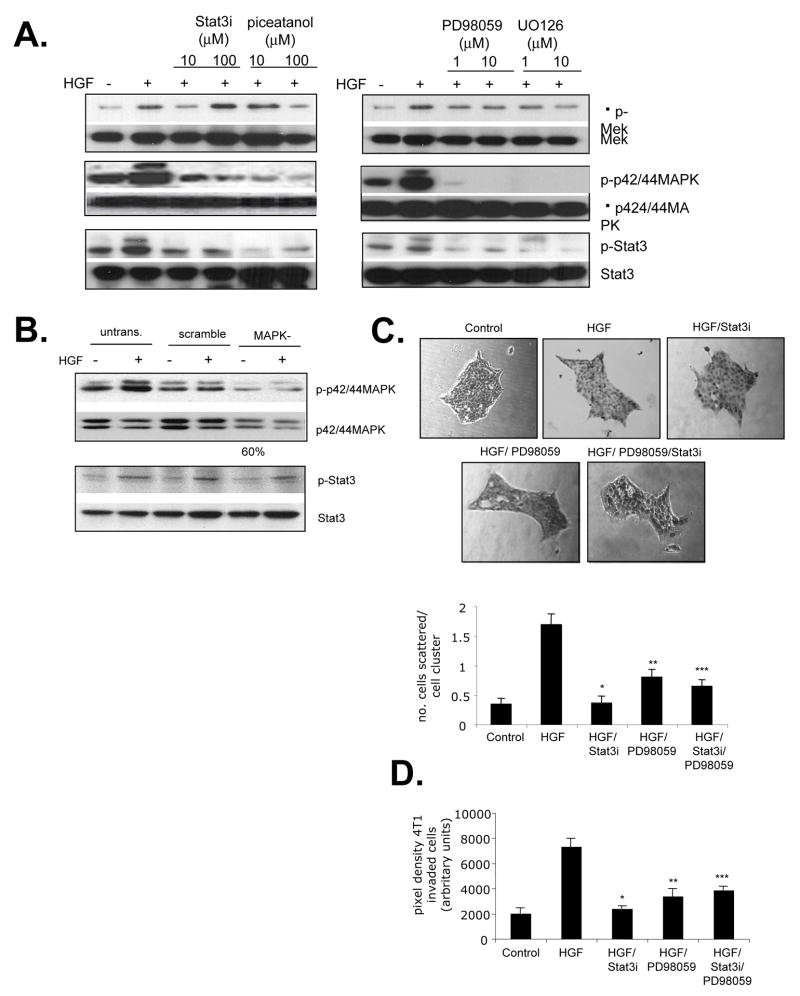
We further analyzed the functional contribution of Stat3 and p44/42MAPK in HGF- induced scattering and invasion of 4T1 mammary carcinoma cells. Interestingly, the Stat3 peptide inhibitor [28] and the small molecule Stat3 inhibitor, piceatannol [29] significantly blocked HGF- induced phosphorylation of MEK and its downstream effector, p42/44MAPK, while the MEK inhibitors U0126 and PD98059 inhibited Stat3 phosphorylation, in both 4T1 cells (Figure 8A) and PyVmT cells (Supplemental Figure 2A). Because U0126 and PD98059 inhibited phosphorylation of both MEK and p42/44MAPK, we determined if p42/44MAPK was specifically required for Stat3 signaling. 4T1 cells were transfected with siRNA duplexes to silence expression of p42 and p44 MAPK isoforms, resulting in a 60% decrease in p42/44MAPK expression, and a significant decrease in HGF-induced phosphorylation. 4T1 cells deficient in p42/44MAPK expression did not show significant changes in Stat3 phosphorylation (Figure 8B), suggesting that MEK, but not p42/44MAPK was required for Stat3 signaling. While treatment of 4T1 cells with PD98059 and Stat3 peptide inhibitors individually reduced HGF-induced scattering and invasion (Figures 8C and D), simultaneous inhibition of Stat3 and p42/44MAPK signaling pathways did not result in either additive or synergistic blockade (Figures 8C,D). Similar results on cellular scattering were observed with MCF-7 and MDCK cells while similar effects on cellular invasion were observed in PyVmT carcinoma cells (Supplemental Figure 2B an 2C). These results suggest that Stat3 and MEK/MAPK signaling pathways interact to mediate HGF- induced cell scattering and invasion of 4T1 mammary tumor cells.
FIGURE 8.
Stat3 and p42/44 MAPK pathways cooperate to mediate HGF induced invasion and scattering A. 4T1 tumor cells were treated with indicated doses of the Stat3 inhibitors: Stat3 inhibitor peptide or piceatanol and indicated doses of the MAPK inhibitors PD98059 or U0126 and analyzed by western blot for the phosphorylation status of the indicated proteins. B. p42/44MAPK expression was inhibited in 4T1 cells by siRNA silencing. 4T1 cells were treated with HGF and analyzed by western blot analysis for the phosprhoylatin status of indicated proteins. C and D: 4T1 cells were treated with Control (serum free medium), HGF (60 ng/ml) in the presence or absence of 10 μM Stat3 peptide inhibitor (Stat3i) or 1 μM PD98059, and analyzed for the ability to C. scatter and D. invade through a Matrigel coated membrane. Statistical significance was determined by Two T-tailed T-test: * vs. HGF; p<0.01, ** vs. HGF; p<0.01 and Two Way ANOVA: *** vs. HGF/PD98059 and HGF/Stat3i treated cells; p>0.05. Data are represented as mean±SEM.
Discussion
In previous studies using a tumor xenograft model, we demonstrated an important role for TGF-β signaling in fibroblasts in regulating HGF/c-Met signaling between the stroma and mammary tumor epithelium during tumor progression [26]. As c-Met has been shown to interact with additional receptor tyrosine kinases including Ron [25], these studies suggest that enhancement of HGF signaling may have broader and more significant effects in altering fibroblast: carcinoma cell interactions, than is currently known. In the present studies, we demonstrate that increased HGF expression by Tgfbr2FspKO fibroblasts enhances mammary carcinoma cell scattering and invasion, through Ron and c-Met receptor signaling, which in part regulated through cooperative interactions between the Stat3 and MAPK signaling pathways (Figure 9). Given that multiple cancer cell lines showed significant responses to Tgfbr2FspKO fibroblasts, which were inhibited in the presence of c-Met and Ron inhibitors, these data indicate that the effects of Tgfbr2FspKO fibroblasts are not restricted to one cell line, but represent more global mechanisms of action.
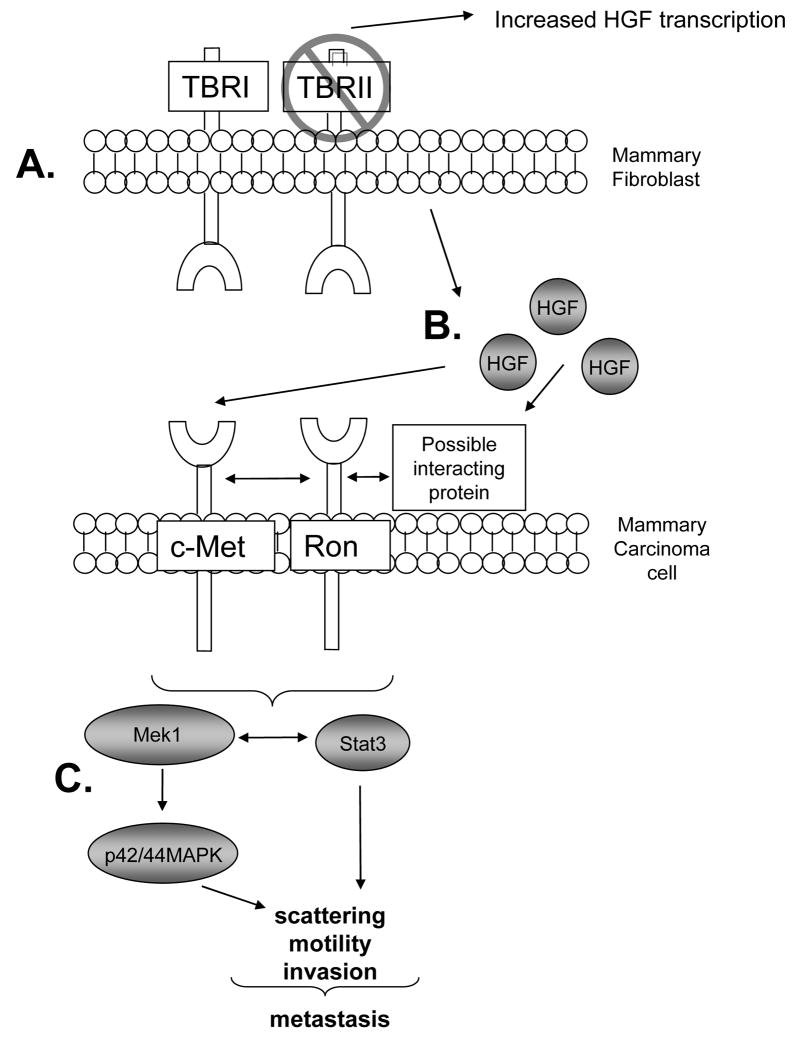
FIGURE 9.
Model of TGF-β and HGF signaling in fibroblast: tumor interactions. A. Cre mediated deletion of the TGF-β type II receptor in mammary fibroblasts inhibits TGF-β signaling, resulting in upregulated HGF expression at the RNA level as well as protein secretion [17,26] B. HGF binds to c-Met directly as may interact with Ron indirectly to enhance c-Met and Ron mediated downstream signaling in mammary epithelial cells. C. Increased cooperative interactions between Stat3 and p42/44MAPK signaling results in mammary carcinoma cell scattering, motility and invasion, important processes involved in metastatic spread.
Using multiple approaches, we analyzed the functional contribution of enhanced HGF expression in Tgfbr2 deficient fibroblasts on mammary carcinoma cell scattering and invasion. . A partial reduction of c-Met and Ron phosphorylation with anti-HGF treatment suggests that additional factors secreted by Tgfbr2FspKO fibroblasts may contribute to c-Met and Ron signaling in 4T1 mammary carcinoma cells, including TGF-α [17]. Signaling interactions between EGFR and c-Met receptors have been reported in human epidermoid carcinoma cells, PyVmT and 4T1 mammary carcinoma cells [31, 32]. The use of the small molecular inhibitor, EXEL-7592 most potently inhibited 4T1 cell scattering and invasion enhanced by Tgfbr2FspKO fibroblasts. Interestingly, EXEL-1075 also slightly inhibited 4T1 cell scattering and potently inhibited invasion induced by Tgfbr2FspKO fibroblasts. The low level VEGFR2 expression in 4T1 cells is consistent with previous studies which have correlated expression of endothelial markers in metastatic breast cancer cells [36]. It would of interest in the future to further investigate a potential role for VEGFR signaling in breast cancer cellular invasiveness. Specific inhibition of c-Met and Ron by receptor neutralizing antibodies and siRNA silencing also resulted in significant reductions in cell scattering and invasion induced by Tgfbr2FspKO fibroblasts. As neutralizing antibodies to HGF inhibited phosphorylation of c-Met, Ron, p42/44MAPK and Stat3 proteins, these data provide strong evidence that HGF derived from Tgfbr2 deficient fibroblasts enhance c-Met and Ron signaling in mammary carcinoma cells to promote cell scattering and invasion.
In addition to c-Met, our data suggest HGF may also signal through Ron receptor tyrosine kinases to regulate mammary carcinoma cell motility and invasion. Studies have associated high level expression of Ron with breast cancer malignancy in histological and mouse tumor xenograft studies [30, 31]; yet, little is currently known regarding the mechanisms of Ron signaling in fibroblast: mammary carcinoma interactions. Our studies indicate that Tgfbr2FspKO fibroblasts enhance epithelial cellular scattering and invasion partly through Ron signaling in multiple cell lines including 4T1 PyVmT, MCF-7 and MDCK cell lines. While studies have shown that Ron is activated through direct ligand binding of MSP [25, 32] or through heterodimerization of Met and Ron [25], we observed that Ron phosphorylation was not significantly affected by c-Met inhibitors, suggesting that c-Met is not required for Ron phosphorylation. In recent studies, c-Met was not required for HGF binding of nucleolin and activation of downstream signaling in prostate cancer cells [33] indicating that HGF has other binding partners. However, it is unlikely that HGF would bind Ron directly as studies have shown that HGF has no significant effect on Ron phosphorylation and does not interfere with Ron binding to MSP in ligand binding studies [34, 35]. Evidence to support a possible indirect association between HGF and Ron comes from studies in which HGF binds to vitronectin and fibronectin in endothelial cells promotes an integrin –c-Met downstream signaling [36]. In addition, HGF binding to heparin sulfate proteoglycans, in particular CD44 in colorectal cancer cells resulting in CD44 interactions with c-Met to promote downstream signaling [37]. It is possible that similar mechanisms may exist involving HGF and Ron, in which HGF may bind to non-c-Met proteins to promote Ron signaling.
In our present studies, a cooperative interaction was also observed between Stat3 and MAPK signaling pathways in mammary carcinoma cells. Cooperative roles between the MEK/MAPK and Stat3 signaling pathways have been reported in renal epithelial cells [38] and endothelial cells [39] in mediating cellular proliferation and survival, while antagonistic roles between Stat3 and MEK/MAPK signaling pathways have been reported in keratinocytes and liver progenitor cells [40, 41]. Such studies indicate that the interactions between Stat3 and MEK/MAPK signaling pathways are cell type specific, with important implications in targeting therapeutics. Studies have implicated both the MEK/MAPK and Stat3 signaling pathways in cytoskeletal reorganization through regulation of rac and rho GTPases, respectively [42, 43]. In addition, p42/44MAPK has been shown to regulate expression of extracellular matrix proteinases including MMP9 [42, 44]; Stat3 regulation of MMP-1 and MMP2 has been implicated in tumor invasiveness [45]. Together, these studies suggest that Stat3 and MEK/MAPK signaling pathways function cooperatively in mammary carcinoma cells to regulate cell scattering and invasion, possibly directly through regulation of the actin cytoskeleton, and indirectly, through expression of MMPs to alter the stromal microenvironment.
In summary, our studies demonstrate important functional roles of TGF-β and HGF signaling in mediating fibroblast: tumor interactions, by demonstrating that HGF derived from Tgfbr2FspKO fibroblasts regulates mammary tumor cell invasion, mediated through Ron and c-Met receptor signaling networks. Carcinoma cell invasiveness through the extracellular matrix is an important step in tumor metastasis, a process which remains a leading cause of death among cancer patients [46]. The challenges are due in part to limited understanding of the metastatic process, which result in difficulties in clinically detecting and treating micro-metastases. It is therefore important to identify and understand the interplay of molecules involved in the metastatic process that would aid the design in more effective cancer therapeutics.
Materials and Methods
Chemicals
RNase free buffer was purchased from Ambion (Austin, TX). Prime-IT RmT kit was purchased from Stratagene (La Jolla, CA). Growth Factor Reduced Matrigel was purchased from BD Biosciences (Franklin Lakes, NJ). Rabbit IgG antibodies conjugated to horseradish peroxidase (anti-rabbit-hrp) and ECL Plus were purchased from Amersham/GE (Piscatway, NJ). Stat3 peptide inhibitor and piceattannol were purchased from Calbiochem (San Diego, CA). PD98059, U0126, and antibodies to phospho-c-Met (Y1234/1235), c-Met, phospho-Stat3, Stat3, phospho-p42/44MAPK, p424/44MAPK were purchased from Cell signaling Technologies (Boston, MA). EXEL-7592 and EXEL-1075 were synthesized and purified in the Medicinal Chemistry Department at Exelixis, Inc [47] (South San Franciso, CA). A 10 mM stock solution was prepared in DMSO and diluted in optimal assay buffers or culture medium. Trizol Reagent, Hybond Nylon Membrane, and CMTMR and CMFDA CellTracker Dyes were purchased from Invitrogen/Molecular Probes (Carlsbad, CA). Recombinant mouse HGF and neutralizing antibodies to c-Met and Ron were purchased from R&D systems (Minneapolis, MN). siRNAs to p42/44MAPK, scramble control siRNAs, siRNA transfection reagent, siRNA transfection medium and antibodies to phospho-Ron (Y1238) and Ron were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). Goat IgG, protease inhibitor cocktail, sodium orthovanadate, gelatin and fibronectin were purchased from Sigma (St. Louis, MO). 10% neutral formalin buffer was purchased from vWR (West Chester, PA).
Cell lines
Mammary fibroblasts from control Tgfbr2Flox/Flox and Tgfbr2FspKO mice were spontaneously immortalized and previously characterized [17]. Fibroblast cell lines were cultured on plastic in DMEM/F12 5% Adult Bovine Serum (ABS). PyVmT mammary carcinoma cells were isolated from MMTV-PyVmT transgenic mice as previously described [17] and cultured on plastic in DMEM media containing 10% FBS with antibiotics. 4T1 mammary carcinoma cells, (generously provided by the laboratory of Fred Miller (University of Michegan, Ann Arbor, MI) and the following 4T1 sublines: 4T1.GFP-, 4T1.3Met-, 4T1.1Ron- were cultured on plastic in DMEM media containing 10% FBS with antibiotics. Generation of c-Met deficient (4T1.3MET) and siRNA specificity control (4T1.GFP-) were generated and characterized as previously described [26]. Generation of a Ron deficient 4T1 cell line was generated through stable expression of pGabe retroviral vector carrying siRNAs to Ron according to the methods previously described [26].
Generation of fibroblast-conditioned medium
Tgfbr2Flox/Flox and Tgfbr2FspKO fibroblasts were plated in 10 cm dishes in DMEM/F12/5% ABS for 24 h. The complete media was aspirated and cells were then incubated in 4 ml of DMEM/F12 containing 0.1% ABS for 24 h. The conditioned medium was then removed, filtered through 0.4 micron filtration unit and used for the experiments indicated.
Northern Blot analysis
Tgfbr2Flox/flox and Tgfbr2Fspko fibroblasts were plated in 10 cm dishes (300,000 per plate) in complete medium for 24 h. The medium was aspirated and the cells were lysed in 1 ml Trizol Reagent (Invitrogen), vortexed with 500 μl phenol/choloroform, and centrifuged for 5 min at 12000 rpm. The aqueous layer was extracted and the RNA was precipitated with 500 μl isopropanol at −20°C overnight. Samples were pelleted at 12000 rpm for 10 min, washed with 75 % ethanol, and resuspended in RNase free buffer (RNAse Secure). 10 μg of total RNA was resolved in 1% agarose gel containing: 2.2 M formaldehyde, 1X 3-[N-Morpholino] proponesulfonic acid (MOPS), DEPC and ethidium bromide. The RNA was transferred to a Hybond nylon membrane, and probed with an HGF cDNA fragment labeled with 32P-dCTP using a Prime-IT RmT kit according to manufacturer’s protocol. Membranes were washed with 2×SSC at room temperature, followed by two stringent washes (1×SSC at 65 °C for 10 min and 0.5×SSC at 65 °C for 10 min), before exposure to film.
siRNA silencing of p42/44MAPK
p42/44MAPK expression was silenced by transient transfection of siRNAs in 4T1 cells, according to manufacterer’s protocol. Briefly, 4T1 cells (200,000) were plated in a 6 well dish and incubated in serum free medium with a complex of 500 ng of p42/44MAPK siRNA or control scramble siRNA and 16 μl siRNA transfection reagent for 8 h. The transfected cells were incubated with siRNA/transfection reagent complexes in complete medium for an additional 24 h. The medium containing siENA/transfection reagent complexes was aspirated and replaced with fresh complete medium.
Western blot analyses of cultured 4T1 tumor cells
4T1 parental and sublines were cultured to 80 % confluency, starved in serum free media for 24 h, and then treated with 60 ng/ml HGF in the presence or absence of inhibitors in serum free media for 30 min. The tumor cells were lysed in RIPA buffer containing 10 mM Tris-HCl, pH 8.0, 0.1 mM EDTA, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl supplemented with protease inhibitor cocktail containing aprotinin, leupeptin, bestatain and pepstatin A and 10 mM sodium orthovanadate. 50 μg of protein were fractionated on 8–12 % SDS PAGE gels. The proteins were transferred to nitrocellulose membranes and then probed with antibodies (1:1000) to the phosphorylated and unphosphorylated forms of c-Met, Ron, p42/44MAPK or Stat3. Specific immunoreaction was detected using anti-rabbit-hrp and ECL Plus chemiluminesence.
Scatter assay
Scatter assays were performed as described [48]. Briefly, 500 4T1 cells per well were plated in a 24 well plate and cultured to clusters of 20–30 cells. After 24 hours of starvation in serum free media, cells were stimulated in 60 ng/ml HGF in the presence or absence of inhibitors for 24 hours, fixed in neutral formalin buffer and stained in Mayer’s hematoxylin for 30 min at RT. Cells were visualized by phase contrast microscopy, micrographed at 20× magnification with 10 fields per sample. Quantitation of cell scattering was determined by calculating the number of cell scattered per cell cluster. Each experimental group was plated in triplicate wells, with a minimum count of 10 samples per well. Experiments were performed in triplicate. The data are represented as mean values with error bars reflected as standard error of the mean.
Invasion assay
Tumor cells were labeled with 1.5 μM CMTMR (red) flourochome while mammary fibroblasts were labeled with 1.5 μM CMFDA (green) fluorochrome in serum free medium for 45 min at 37°C. The medium containing dye was aspirated and cells were allowed to recover for 24 h. For HGF stimulation experiments, tumor cells were serum starved for 24 h and then seeded on top of the transwell (75,000) in the presence or absence of 60 ng/ml HGF, and inhibitors and incubated at 37 °C for 8 h. For fibroblast:tumor co-culture experiments, 100,000 fibroblasts were seeded on the underside of transwell 8 μm pore filter (BD Biosciences) coated with Growth Factor Reduced Matrigel. CMTMR labeled tumor cells (75,000) were then seeded in DMEM/F12 0.5% ABS on top of the transwell filter in the presence or absence of fibroblasts and inhibitors and incubated for 8 h. The cells were fixed in 10 % neutral formalin buffer for 10 min. Tumor cells on the topside of the filter were removed by cotton swab. The transwell filter was mounted on glass slides in aqua-based mounting medium. Invaded tumor cells were visualized by fluorescence microscopy and micrographed at 20x magnification, three fields per sample. Experiments were performed in replicates of four and repeated in triplicate. Quantitation of tumor cells was determined by measuring pixel density of red labeled cells using Image J (NIH) software (arbitrary units). The data are represented as mean values with error bars reflected as standard error of the mean.
Statistical Analysis
Analyses of study results focused on estimating the association between the study groups, e.g., Tgfbr2FspKO fibroblasts vs. Tgfbr2Flox/Flox fibroblasts, and study endpoints. The tests of hypotheses concerning the single measurement of outcome data, e.g., number of invaded cells, were carried out using either through the Two tailed Student’s t-test or ANOVA. All tests of significance were two-sided, and differences were considered statistically significant when p-value < 0.05.
Supplementary Material
Acknowledgments
This work was supported by grants number CA102162 and CA85492 (to HLM) from the National Cancer Institute DHHS, and by the TJ Martell Foundation.
Nonstandard abbreviations uses
- HGF
hepatocyte growth factor/scatter factor
- VEGF
vascular endothelial growth factor
- Tgfbr2
type II TGF-β receptor gene
- Tgfbr1Flox/Flox
Tgfbr2 with loxP sites flanking exon 2
- Tgfbr2FspKO
cells in which conditional knockout of Tgfbr2 was accomplished by expression of Fsp1-Cre
- MDCK
Madin Darbey Canine Kidney
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Bhowmick NA, Moses HL. Tumor-stroma interactions. Curr Opin Genet Dev. 2005;15:97–101. doi: 10.1016/j.gde.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 3.Kurose K, Gilley K, Matsumoto S, Watson PH, Zhou XP, Eng C. Frequent somatic mutations in PTEN and TP53 are mutually exclusive in the stroma of breast carcinomas. Nat Genet. 2002;32:355–357. doi: 10.1038/ng1013. [DOI] [PubMed] [Google Scholar]
- 4.Barcellos-Hoff MH, Ravani SA. Irradiated mammary gland stroma promotes the expression of tumorigenic potential by unirradiated epithelial cells. Cancer Res. 2000;60:1254–1260. [PubMed] [Google Scholar]
- 5.Camps J, Chang S, Hsu T, Freeman M, hong S, Zhau H, von Eschenbach A, Chung L. Fibroblast-mediated acceleration of human epithelial tumor growth in vivo. Proc Natl Acad Sci U S A. 1990;87:75–79. doi: 10.1073/pnas.87.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuxhorn J, Ayala G, Smith M, Smith V, Dang T, Rowley D. Reactive Stroma in Human Prostate cancer: Induction of Myofibroblast Phenotype and Extracellular Matrix Remodeling. Clinical Cancer Research. 2002a;8:2912–2923. [PubMed] [Google Scholar]
- 7.Maffini MV, Soto AM, Calabro JM, Ucci AA, Sonnenschein C. The stroma as a crucial target in rat mammary gland carcinogenesis. J Cell Sci. 2004;117:1495–1502. doi: 10.1242/jcs.01000. [DOI] [PubMed] [Google Scholar]
- 8.Sakakura T, Sakagami Y, Nishizuka Y. Accelerated mammary cancer development by fetal salivary mesenchyma isografted to adult mouse mammary epithelium. J Natl Cancer Inst. 1981;66:953–959. [PubMed] [Google Scholar]
- 9.Shekhar MP, Werdell J, Santner SJ, Pauley RJ, Tait L. Breast stroma plays a dominant regulatory role in breast epithelial growth and differentiation: implications for tumor development and progression. Cancer Research. 2001;61:1320–1326. [PubMed] [Google Scholar]
- 10.Daniel CW, Robinson S, Silberstein GB. Bioactive Components of Human Milk. New York: Plenum Publishers; 2001. The transforming growth factors beta in development and functional differentiation of the mouse mammary gland; pp. 61–69. [DOI] [PubMed] [Google Scholar]
- 11.Pollard J. Tumour-stromal interactions. Transforming growth factor-beta isoforms and hepatocyte growth factor/scatter factor in mammary gland ductal morphogenesis. Breast Cancer res. 2001;3:230–237. doi: 10.1186/bcr301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abraham S, Sawaya BE, Safak M, Batuman O, Khalili K, Amini S. Regulation of MCP-1 gene transcription by Smads and HIV-1 Tat in human glial cells. Virology. 2003;309:196–202. doi: 10.1016/s0042-6822(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 13.Akhurst RJ, Derynck R. TGF-B signaling in cancer- a double edged sword. Trends in Cell Biology. 2001;11:S44–S50. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- 14.Ihn H. Pathogenesis of fibrosis: role of TGF-β and CTGF. Curr Op Rheum. 2002;14:681–685. doi: 10.1097/00002281-200211000-00009. [DOI] [PubMed] [Google Scholar]
- 15.Chytil A, Magnuson MA, Wright CVE, Moses HL. Conditional inactivation of the TGF-B type II receptor using Cre-Lox. Genesis. 2002;32:73–75. doi: 10.1002/gene.10046. [DOI] [PubMed] [Google Scholar]
- 16.Bhowmick N, Chytiil A, Plieth D, Gorska A, Dumont N, Shappel S, Washington M, Neilson E, Moses H. TGF-β signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:847–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 17.Cheng N, Bhowmick NA, Chytil A, Gorksa AE, Brown KA, Muraoka R, Arteaga CL, Neilson EG, Hayward SW, Moses HL. Loss of TGF-beta type II receptor in fibroblasts promotes mammary carcinoma growth and invasion through upregulation of TGF-alpha-, MSP- and HGF-mediated signaling networks. Oncogene. 2005;24:5053–5068. doi: 10.1038/sj.onc.1208685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosario M, Birchmeier W. How to make tubes: signaling by the Met receptor tyrosine kinase. Trends Cell Biol. 2003;13:328–335. doi: 10.1016/s0962-8924(03)00104-1. [DOI] [PubMed] [Google Scholar]
- 19.Soriano JV, Pepper MS, Orci L, Montesano R. Roles of hepatocyte growth factor/scatter factor and transforming growth factor-beta1 in mammary gland ductal morphogenesis. J Mammary Gland Biol Neoplasia. 1998;3:133–150. doi: 10.1023/a:1018790705727. [DOI] [PubMed] [Google Scholar]
- 20.Christensen JG, Burrows J, Salgia R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett. 2005;225:1–26. doi: 10.1016/j.canlet.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 21.Date K, Matsumoto K, Kuba K, Shimura H, Tanaka M, Nakamura T. Inhibition of tumor growth and invasion by a four-kringle antagonist (HGF/NK4) for hepatocyte growth factor. Oncogene. 1998;17:3045–3054. doi: 10.1038/sj.onc.1202231. [DOI] [PubMed] [Google Scholar]
- 22.Wang M, Wang D, Chen Y. Oncogenic and invasive potentials of human macrophage-stimulating protein receptor, the Ron receptor tyrosine kinase. Carcinogenesis. 2003;23:1291–1300. doi: 10.1093/carcin/bgg089. [DOI] [PubMed] [Google Scholar]
- 23.Mazzone M, Basilico C, Cavassa S, Pennacchietti S, Risio M, Naldini L, Comoglio PM, Michieli P. An uncleavable form of pro-scatter factor suppresses tumor growth and dissemination in mice. J Clin Invest. 2004;114:1418–1432. doi: 10.1172/JCI22235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maggiora P, Lorenzato A, Fracchioli S, Costa B, Castagnaro M, Arisio R, Katsaros D, Massobrio M, Comoglio P, Flavia Di Renzo M. The RON and MET oncogenes are co-expressed in human ovarian carcinomas and cooperate in activating invasiveness. Exp Cell Res. 2003;288:382–389. doi: 10.1016/s0014-4827(03)00250-7. [DOI] [PubMed] [Google Scholar]
- 25.Follenzi A, Bakovic S, Gual P, Stella MC, Longati P, Comoglio PM. Cross-talk between the proto-oncogenes Met and Ron. Oncogene. 2000;19:3041–3049. doi: 10.1038/sj.onc.1203620. [DOI] [PubMed] [Google Scholar]
- 26.Cheng N, Chytil A, Shyr Y, Joly A, Moses HL. Enhanced Hepatocyte Growth Factor Signaling by Type II Transforming Growth Factor-{beta} Receptor Knockout Fibroblasts Promotes Mammary Tumorigenesis. Cancer Res. 2007;67:4869–4877. doi: 10.1158/0008-5472.CAN-06-3381. [DOI] [PubMed] [Google Scholar]
- 27.Joseph H, Gorska AE, Sohn P, Moses HL, Serra R. Overexpression of a kinase-deficient transforming growth factor-B type II receptor in mouse mammary stroma results in increased epithelial branching. Molecular Biology of the Cell. 1999;10:1221–1234. doi: 10.1091/mbc.10.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Catalano RD, Johnson MH, Campbell EA, Charnock-Jones DS, Smith SK, Sharkey AM. Inhibition of Stat3 activation in the endometrium prevents implantation: a nonsteroidal approach to contraception. Proc Natl Acad Sci U S A. 2005;102:8585–8590. doi: 10.1073/pnas.0502343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alas S, Bonavida B. Inhibition of constitutive STAT3 activity sensitizes resistant non-Hodgkin’s lymphoma and multiple myeloma to chemotherapeutic drug-mediated apoptosis. Clin Cancer Res. 2003;9:316–326. [PubMed] [Google Scholar]
- 30.Camp ER, Liu W, Fan F, Yang A, Somcio R, Ellis LM. RON, a tyrosine kinase receptor involved in tumor progression and metastasis. Ann Surg Oncol. 2005;12:273–281. doi: 10.1245/ASO.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Welm AL, Sneddon JB, Taylor C, Nuyten DS, van de Vijver MJ, Hasegawa BH, Bishop JM. The macrophage-stimulating protein pathway promotes metastasis in a mouse model for breast cancer and predicts poor prognosis in humans. Proc Natl Acad Sci U S A. 2007;104:7570–7575. doi: 10.1073/pnas.0702095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang MH, Zhou YQ, Chen YQ. Macrophage-stimulating protein and RON receptor tyrosine kinase: potential regulators of macrophage inflammatory activities. Scand J Immunol. 2002;56:545–553. doi: 10.1046/j.1365-3083.2002.01177.x. [DOI] [PubMed] [Google Scholar]
- 33.Tate A, Isotani S, Bradley MJ, Sikes RA, Davis R, Chung LW, Edlund M. Met-Independent Hepatocyte Growth Factor-mediated regulation of cell adhesion in human prostate cancer cells. BMC Cancer. 2006;6:197. doi: 10.1186/1471-2407-6-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaudino G, Follenzi A, Naldini L, Collesi C, Santoro M, Gallo KA, Godowski PJ, Comoglio PM. RON is a heterodimeric tyrosine kinase receptor activated by the HGF homologue MSP. Embo J. 1994;13:3524–3532. doi: 10.1002/j.1460-2075.1994.tb06659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang MH, Ronsin C, Gesnel MC, Coupey L, Skeel A, Leonard EJ, Breathnach R. Identification of the ron gene product as the receptor for the human macrophage stimulating protein. Science. 1994;266:117–119. doi: 10.1126/science.7939629. [DOI] [PubMed] [Google Scholar]
- 36.Rahman S, Patel Y, Murray J, Patel KV, Sumathipala R, Sobel M, Wijelath ES. Novel hepatocyte growth factor (HGF) binding domains on fibronectin and vitronectin coordinate a distinct and amplified Met-integrin induced signalling pathway in endothelial cells. BMC Cell Biol. 2005;6:8. doi: 10.1186/1471-2121-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wielenga VJ, van der Voort R, Taher TE, Smit L, Beuling EA, van Krimpen C, Spaargaren M, Pals ST. Expression of c-Met and heparan-sulfate proteoglycan forms of CD44 in colorectal cancer. Am J Pathol. 2000;157:1563–1573. doi: 10.1016/S0002-9440(10)64793-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arany I, Megyesi JK, Nelkin BD, Safirstein RL. STAT3 attenuates EGFR-mediated ERK activation and cell survival during oxidant stress in mouse proximal tubular cells. Kidney Int. 2006;70:669–674. doi: 10.1038/sj.ki.5001604. [DOI] [PubMed] [Google Scholar]
- 39.Nakagami H, Morishita R, Yamamoto K, Taniyama Y, Aoki M, Matsumoto K, Nakamura T, Kaneda Y, Horiuchi M, Ogihara T. Mitogenic and antiapoptotic actions of hepatocyte growth factor through ERK, STAT3, and AKT in endothelial cells. Hypertension. 2001;37:581–586. doi: 10.1161/01.hyp.37.2.581. [DOI] [PubMed] [Google Scholar]
- 40.Quadros MR, Peruzzi F, Kari C, Rodeck U. Complex regulation of signal transducers and activators of transcription 3 activation in normal and malignant keratinocytes. Cancer Res. 2004;64:3934–3939. doi: 10.1158/0008-5472.CAN-04-0214. [DOI] [PubMed] [Google Scholar]
- 41.Yeoh GC, Ernst M, Rose-John S, Akhurst B, Payne C, Long S, Alexander W, Croker B, Grail D, Matthews VB. Opposing roles of gp130-mediated STAT-3 and ERK-1/2 signaling in liver progenitor cell migration and proliferation. Hepatology. 2007;45:486–494. doi: 10.1002/hep.21535. [DOI] [PubMed] [Google Scholar]
- 42.Ray RM, Vaidya RJ, Johnson LR. MEK/ERK regulates adherens junctions and migration through Rac1. Cell Motil Cytoskeleton. 2006 doi: 10.1002/cm.20172. [DOI] [PubMed] [Google Scholar]
- 43.Debidda M, Wang L, Zang H, Poli V, Zheng Y. A role of STAT3 in Rho GTPase-regulated cell migration and proliferation. J Biol Chem. 2005;280:17275–17285. doi: 10.1074/jbc.M413187200. [DOI] [PubMed] [Google Scholar]
- 44.Lakka SS, Jasti SL, Gondi C, Boyd D, Chandrasekar N, Dinh DH, Olivero WC, Gujrati M, Rao JS. Downregulation of MMP-9 in ERK-mutated stable transfectants inhibits glioma invasion in vitro. Oncogene. 2002;21:5601–5608. doi: 10.1038/sj.onc.1205646. [DOI] [PubMed] [Google Scholar]
- 45.Xie TX, Wei D, Liu M, Gao AC, Ali-Osman F, Sawaya R, Huang S. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23:3550–3560. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- 46.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 47.Bannen LCDS, Chen J, Dalrymple L. c-Met Modulators and Methods of Use. US Patent WO2005030140. [Google Scholar]
- 48.Santos OF, Moura LA, Rosen EM, Nigam SK. Modulation of HGF-induced tubulogenesis and branching by multiple phosphorylation mechanisms. Dev Biol. 1993;159:535–548. doi: 10.1006/dbio.1993.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.