Abstract
A new method to assemble time-calibrated supertrees is able to incorporate paleontological and molecular dates. This method, along with new branch length transformations, is implemented in the Stratigraphic Tools for Mesquite. It was used here to analyse a dataset on bone microanatomy, body size and habitat of 46 species of lissamphibians through a variety of methods (Felsenstein independent contrasts, variance partition with phylogenetic eigenvector regression, discriminant analyses and simple regressions). Our analyses showed that the new methods can produce adequate standardization for several characters on a tree whose branch lengths can represent evolutionary time. The analyses confirmed previous conclusions about the presence of an ecological signal in bone microanatomical data.
Keywords: bone compactness, comparative biology, geological age, Lissamphibia, molecular dating, phylogeny, supertree
Introduction
The conquest of land by vertebrates has fascinated generations of paleontologists, morphologists and developmental biologists. Fossilization of mineralized tissues records much biological data in various bony structures. Phylogeny, biomechanical constraints and habitat (e.g. aquatic vs. terrestrial) leave a signature that can be extracted from bone cross-sections (de Buffrénil & Buffetaut, 1981; de Ricqlès et al. 2004; Laurin et al. 2004). The use of microanatomical data offers a unique opportunity to study the processes that document the adaptation of tetrapods to various lifestyles. An extensive database of long-bone cross-sections of extant tetrapods is necessary to identify microanatomical adaptations to habitat (aquatic or terrestrial) and to infer the lifestyle of extinct species on the basis of bone microanatomy.
Histological studies of cross-sections of various extant species (de Ricqlès & de Buffrénil, 2001; Laurin et al. 2004; Kriloff et al. 2008) showed that, in most cases, terrestrial taxa have moderately compact long bones with a large medullary cavity and a compact cortical region, whereas long bones of aquatic vertebrates are generally either more compact with a smaller medullary cavity (e.g. pipids, dugongs, Eocene cetaceans) or of moderate compactness with a medullary spongiosa that obliterates the medullary cavity (e.g. extant cetaceans). Lissamphibia is clearly an excellent group for this kind of investigation because it includes clades displaying three types of lifestyles, i.e. aquatic, amphibious and terrestrial. In addition, most of the numerous species of this group are easily available. In this study, we tried to bring an objective and quantitative point of view on these observations using several statistical methods, most of which take into consideration the effect of phylogenetic relationships, to complement the previous study by Laurin et al. (2004), which suggested that returns to an aquatic lifestyle in lissamphibians are associated with an increase in femoral compactness and body size.
Although the relationship between bone microanatomy and habitat has been studied for nearly a century, most of the published illustrations of bone sections did not include an extensive series of standardized views (here, cross-sections) of a standardized anatomical region (e.g. mid-diaphysis) of a single bone (here, the femur) in several species. Tables reporting quantitative attributes of bone sections are no substitute for images because no model can capture all of the information present in the original data, despite some recent progress in that direction (Girondot & Laurin, 2003; Laurin et al. 2004). Thus, this report includes detailed drawings of femoral cross-sections of 46 species of lissamphibians, which constitute one of the largest comparative sets of long-bone mid-diaphyseal cross-sections ever published. Using these drawings, which were used to extract compactness profile data in Bone Profiler (Girondot & Laurin, 2003), anatomists could perhaps discover new characters of ecological or phylogenetic interest that have so far eluded us.
Most of the earlier analyses (Wall, 1983; Fish & Stein, 1991; de Buffrénil & Rage, 1993; Leclair et al. 1993) were performed before the development of modern comparative methods and therefore their results need to be reassessed using modern comparative techniques, such as phylogenetically independent contrasts [Felsenstein independent contrasts (FIC)] and variance partitioning with phylogenetic eigenvector regression (PVR) (Felsenstein, 1985; Desdevises et al. 2003). Furthermore, the phylogenetic signal in bone microanatomy, which has proven to be a contentious issue (Cubo et al. 2005), needs to be assessed using statistical tests, such as (among other possibilities) PVR (Diniz-Filho et al. 1998) or comparison of the squared length of a character over the reference tree with that of a randomized population of trees produced by reshuffling terminal taxa (Laurin, 2004).
All of the analyses mentioned above, and most comparative analyses in general, require phylogenies with estimated branch lengths (Felsenstein, 1985; Desdevises et al. 2003), which can be compiled from the literature. Indeed, the importance of using adequate branch lengths in comparative analyses has been long established (Díaz-Uriarte & Garland, 1998). This critical step of the analysis is not always easy because, despite the recent proliferation of molecular dating studies, no single published study will include all species represented in a given dataset (in most cases at least). This problem is compounded by the relative paucity of options to quickly generate a set of plausible branch lengths, short of performing molecular dating on a tree, which is not a trivial task (Sanderson, 2003). We propose a way to include both molecular divergence time estimates and minimal dates from the fossil record in a time-calibrated supertree, and we propose new methods of branch length transformations that can be used to adequately standardize the contrasts, and that do not obscure the relationship between branch lengths and evolutionary time. All of these methods were implemented in the Stratigraphic Tools optional package (Josse et al. 2006) of Mesquite (Maddison & Maddison, 2008). These new methods should also be useful for squared-change parsimony optimization (Maddison, 1991), which is a popular method to trace the evolution of continuous characters (e.g. Laurin, 2004; Cubo et al. 2005) and which shares many of the same assumptions as FIC. When the resulting branch lengths are biologically meaningful, evolutionary interpretation of some results (such as evolutionary rates) is facilitated.
Materials and methods
The 46 species of lissamphibians listed in Fig. 1(Caudata and Anura) and a total of 127 specimens were studied. These species show different lifestyles (strictly aquatic, amphibious and terrestrial) and different body sizes.
Fig. 1.
Reference phylogeny of sampled taxa. (A) Tree showing the minimal divergence times as established from the fossil record (Marjanović & Laurin, 2007; age of Baurubatrachus and phylogenetic position of Eodiscoglossus corrected) and, when the fossil record is insufficient, from molecular evidence. The extinct terminal taxa were inserted only to provide a time calibration (no microanatomical data are available). A few molecular dates were also inserted and can be identified by the plain font (as opposed to italics used for genuine genus and species names). (B) Tree used in the comparative analyses, produced by enforcing minimal branch lengths of 30 Ma for terminal branches and 20 Ma for internal branches using Stratigraphic Tools (Josse et al. 2006) and subsequently removing all taxa used for the time calibration. The habitat (binary coding) is indicated by shading: aquatic taxa in white; amphibious and terrestrial taxa in black. The presence of both shades on the same branch indicates ambiguity (parsimony optimization was performed in Mesquite). Within each geological period, the succession of stages is indicated by an alternation of gray and white bands. From bottom (oldest) to top (youngest), these are as follows. In the Carboniferous: Visean, Serpukhovian, Bashkirian, Moscovian, Kasimovian and Gzhelian. In the Permian: Asselian, Sakmarian, Artinskian, Kungurian, Roadian, Wordian, Capitanian, Wujiapingian and Changxingian. In the Triassic: Scythian, Anisian, Ladinian, Carnian, Norian and Rhaetian. In the Jurassic: Hettangian, Sinemurian, Pliensbachian, Toarcian, Aalenian, Bajocian, Bathonian, Callovian, Oxfordian, Kimmeridgian and Tithonian. In the Cretaceous: Berriasian, Valanginian, Hauterivian, Barremian, Aptian, Albian, Cenomanian, Turonian, Coniacian, Santonian, Campanian and Maastrichtian. In the Paleogene: Danian, Selandian, Thanetian, Ypresian, Lutetian, Bartonian, Priabonian, Rupelian and Chattian. In the Neogene: Aquitanian, Burdigalian, Langhian, Serravallian, Tortonian, Messinian, Pliocene and Quaternary.
The femur of each specimen (all adults) was dissected and prepared for cross-sections using classic histological methods. Cross-sections of the bone were made at the mid-diaphyseal level to avoid the variation of the compactness profile along the bone and to have the best ecological signal (Laurin et al. 2004). Anatomical drawings of these sections were made with a camera lucida, digitized and analysed with Bone Profiler (Girondot & Laurin, 2003) and Mesquite (Maddison & Maddison, 2008). Most of the analysed data represent bone compactness profiles, which follow a sigmoidal curve and can be represented by four main variables: the size of the medullary region (P), the width of the transition zone between medullary and cortical regions (S), and asymptotic compactness values in the center (Min) and cortex (Max). These values can be computed from whole sections or from small 6°-wide ‘pie-chart’ sections (radial values of P, S, Min and Max; Appendix 1). See Girondot & Laurin (2003) and Laurin et al. (2004) for more details about this model and the meaning of each variable, and Table 1 for an exhaustive list of abbreviations of these variables. The lifestyle was scored mostly from the literature, especially from large compilations by authors familiar with these taxa (e.g. Goin et al. 1978; Duellman & Trueb, 1986; Thorn & Raffaëlli, 2001) and from a few personal communications from herpetologists (J. Castanet, H. Francillon-Vieillot, etc.).
Table 1.
Phylogenetic signal in the characters assessed through phylogenetic eigenvector regression (PVR) and variance partitioning
| Character | Explained variance | P-value |
|---|---|---|
| LN(PLg) | 0.0270 | 0.7550 (17) |
| LN(10*MD) | 0.3517 | 0.0004 (2) |
| S0.1 | 0.1624 | 0.0555 (5) |
| P | 0.3805 | 0.0004 (3) |
| Min | 0.0387 | 0.5395 (13) |
| Max | 0.0412 | 0.5807 (14) |
| Srad | 0.0999 | 0.2110 (6) |
| Prad | 0.3723 | 0.0004 (4) |
| Minrad | 0.0289 | 0.6748 (16) |
| Maxrad | 0.0752 | 0.3093 (8) |
| Cc | 0.0449 | 0.4461 (12) |
| Cp | 0.0617 | 0.4099 (10) |
| Cg3 | 0.4578 | 0.0001 (1) |
Each compactness profile and body size parameter was considered as the dependent variable and the lifestyle (binary coding) and some principal coordinates (PCs) (retained by a broken-stick model) were considered as the independent variables (only the portion related exclusively to the phylogeny is shown). The broken-stick model retained only the first three PCs (PC1, PC2, PC3), which represent 60.35% of the phylogenetic variance of the 46 species. Numbers in parentheses next to the probabilities indicate the rank in the family of tests of phylogenetic signal (see Table 5). LN(PLg), natural logarithm of presacral length (from atlas to sacrum) (in cm); LN(10*MD), natural logarithm of 10*maximal diameter (of the cross-section) (in mm); Cg, global compactness of the bone cross-section; P, relative distance from the center to the point of inflection, where the most abrupt change in compactness is observed (P is proportional to the size of the medullary cavity); Prad, radial value of P; LS, lifestyle; S, reciprocal of the slope at the inflection point that generally reflects the width of the transition zone between the cortical compacta and medulla; Min, compactness in the center of the medullary region; Max, compactness in the outermost cortex; Srad, Minrad and Maxrad are respectively the radial values of parameters S, Min and Max; Cp, compactness in the periphery of the cross-section; Cc, compactness in the center of the bone section.
The time-calibrated supertree (Fig. 1) was updated and differs in several respects from the tree used by Laurin et al. (2004). This was required by the publication of comprehensive studies of lissamphibian phylogeny (e.g. Wiens et al. 2005; Frost et al. 2006) and diversification time (Marjanović & Laurin, 2007). We also incorporated data from detailed phylogenies of much smaller clades, such as trees of species of TriturusRafinesque, 1815 (Arntzen et al. 2007), salamandrids (Steinfartz et al. 2007) and Desmognathus Baird, 1850 (Rissler & Taylor, 2003), which suggest, based on molecular data, that the evolutionary radiation of Salamandridae started at least 80 Ma ago, that Triturus is paraphyletic or polyphyletic, and that the radiation between the Desmognathus species sampled here took place between 2 and 16 Ma ago. We resolved the relationships between species of Salamandra Laurenti, 1768 using Weisrock et al. (2006) and obtained approximate divergence time estimates from Steinfartz et al. (2000). Divergence times between Pipa pipa Linnaeus, 1758 and Pipa carvalhoi Miranda-Ribeiro, 1937 are estimated from Evans et al. (2003), who dated the divergence between P. pipa andP. parva Ruthven and Gaige, 1923 at about 55 Ma through molecular dating. However, P. carvalhoi is apparently more closely related to P. pipa than to P. parva, so the divergence betweenP. pipa and P. carvalhoi must be more recent than 55 Ma. In the absence of more specific paleontological and molecular dates, we place this event at about 40 Ma because P. carvalhoi is relatively basal in the taxon Pipa (Cannatella & Trueb, 1988).
The supertree (Fig. 1) was compiled manually using criteria previously proposed (Laurin, 2004, p. 595; Laurin et al. 2004, p. 593) to resolve incompatibilities between various topologies. Trees based on a computer-assisted phylogenetic analysis were preferred over trees based on a manual analysis of a matrix, and the latter were preferred over trees not based on a matrix; phylogenies incorporating a large number of terminal taxa were preferred over trees with few taxa; recent phylogenies were preferred over older ones; trees based on many characters were preferred over those based on few characters; and matrices incorporating both molecular and morphological data were preferred over matrices incorporating only one of these two types of data. We are aware of a variety of methods of using matrix representation parsimony to generate supertrees (Bininda-Emonds & Sanderson, 2001), and these could be used in some cases, but the method that we used has the advantage of being able to incorporate fossils whose systematic position may have been expressed in text without a tree being figured. In any case, the methods that we discuss in this draft concern the calibration of the tree in time using paleontological and molecular data, not how the topology is obtained.
It is not straightforward to combine data from the fossil record, which provide minimal divergence date estimates (Marjanović & Laurin, 2007), and molecular estimates (Arntzen et al. 2007; Steinfartz et al. 2007), which attempt to date the actual divergence. The simple branch length manipulation algorithms implemented in Stratigraphic Tools (Josse et al. 2006) can push hypothetical ancestors back in time, while keeping the geological age of all terminal (known) taxa constant. This procedure is based on the fact that the age of terminal taxa is usually known (with variable precision) but that of hypothetical ancestors is usually only constrained by the fossil record to a given minimum. The method also compensates somewhat for the fact that paleontological data often do not yield reliable estimates of internal branch lengths, such as when several nested clades seem to appear simultaneously in the fossil record. For a series of paleontological dates, two families of branch lengths can be obtained (Marjanović & Laurin, 2007): one assuming that each fossil included for calibration occupied a whole geological stage, which may be appropriate when the exact age of a fossil within the stage is uncertain, as is often the case (using this algorithm, only the minimal length of internal branches can be manipulated), and a second that places each fossil at the top of the geological stage in which it occurs, and in which minimal terminal and internal branch lengths can be specified (these two parameters can be set independently of each other).
Such branch length transformations can be useful in at least two contexts. The first, exemplified by Marjanović & Laurin (2007), is to assess the impact of minimal branch length assumptions on inferred data of appearance of taxa, using the fossil record. The second, exemplified in this draft, is to use paleontological data (in addition to molecular data, if available) to obtain families of branch lengths to perform comparative analyses using methods such as phylogenetic independent contrasts or squared-change parsimony. As suggested by Garland et al. (1992, p. 20), ‘estimates of divergence times would be most appropriate’ to set branch lengths. However, as mentioned in the same paper (p. 22), ‘Regardless of what “starter” branch lengths are employed …, independent contrasts must be adequately standardized so that they will receive equal weighting in subsequent correlation or regression analyses’. By progressively increasing the minimal branch lengths, it is often possible to eliminate statistical artifacts (i.e. to adequately standardize the contrasts) while retaining branch lengths which may reflect evolutionary time (Laurin, 2004; Pouydebat et al. 2008). Thus, as performed in some of our earlier analyses (Laurin, 2004; Marjanović & Laurin, 2007), and following a simple parsimony criterion of not pushing ancestors unnecessarily far back into the past, minimal terminal and internal branch length settings were increased progressively from initially small values (1 Ma for internal and terminal branches). We stopped lengthening branches when no additional characters could be standardized adequately (Fig. 1B). This was assessed using the four tests in the PDAP module for Mesquite.
First, we performed a regression between the absolute value of the standardized contrasts and their expected SD. The latter is estimated from branch lengths; it is the square root of the sum of corrected branch lengths (Felsenstein, 1985). A statistically significant regression would indicate that the selected branch lengths fail to adequately standardize the contrasts. For example, a negative slope would indicate that the evolutionary rate is systematically higher on short branches than on long ones, thus implying that the shortest branches should be lengthened.
Second, we regressed the absolute value of the standardized contrasts against the estimated value of the base node. A statistically significant negative slope would indicate that taxa with a high character value evolve more slowly than taxa with a lower character value, and this could represent a statistical artifact, except in cases in which a trend is expected in a character.
Third, we regressed the absolute value of standardized contrasts against corrected node height (this is equivalent to the geological age of the various hypothetical ancestors). A negative relationship would indicate that evolution slows down upwards in the tree; conversely, a positive slope would indicate acceleration of evolutionary rates. Again, this would be more likely to represent a statistical artifact resulting from a poor choice of branch lengths than a biological phenomenon.
Fourth, we regressed the nodal values against the corrected height of the base nodes. Any significant relationship (with P ≤ 0.05) probably represents a statistical artifact. To be deemed adequate, a tree had to yield non-significant relationships for these four tests and, as we did not perform corrections for multiple tests here, our criteria are rather stringent.
In many cases, some characters will yield artifacts even after various branch length and data transformations; in this case, no FIC or squared-change parsimony was performed because any results would be unreliable. The persistence of such artifacts after various minimal branch length settings have been used probably means that the characters have not evolved according to a Brownian motion model or that the topology (and possibly dates used to set the branch lengths) is wrong. In this case, it may be better to use different branch length transformation procedures, such as Grafen's rho (Grafen, 1989) or the Ornstein-Uhlenbeck (OU) or accelerate or decelerate (ACDC) transformations of Blomberg et al. (2003). However, such methods of branch length transformations will presumably work better if the initial branch lengths are biologically plausible, and will be easier to interpret if the initial branch lengths reflect time. Thus, even in these cases, our methods of branch length manipulations could be useful because they can, to an extent, reduce the adverse impact that the incomplete fossil record may have on time calibration of the tree.
Grafen's rho, OU or ACDC transformations may also be useful if the geological ages implied by the trees are implausible (such as a Devonian age for the first bird). However, all of these transformations imply that the characters did not evolve according to a Brownian motion on the phylogeny. If this reflects the true evolutionary model, such transformations are desirable. But, in other cases, these transformations may be required because the initial branch lengths were too far from the correct phylogeny. Our transformations can be used to check if a plausible set of branch lengths reflecting evolutionary time fits the data under the most simple assumption of all (Brownian motion). It should be possible to devise algorithms to yield the optimal minimal branch lengths directly but this would require software developments beyond the purpose of this study.
The advantage of this procedure to obtain branch lengths over many other transformations of branch lengths, such as logarithmic, exponential or square-root transformations, is that the branch lengths remain biologically easy to interpret (they represent evolutionary time), as long as the hypothetical ancestors do not need to be pushed unreasonably far back into the past. Other options, which may not be as appealing, consist of discarding initial branch lengths and using unitary branch lengths, or using methods that rely purely on topology or rank to assign lengths and transforming these using a method such as Grafen's rho (Grafen, 1989). However, the method of Grafen (1989) (and unitary branch lengths) assumes either that the species in the comparative sample are representative of the number of extant and extinct lineages of the clade and that the evolutionary model is speciational, if topology (or number of species above each node) is used to assign branch lengths, or that absolute ranks have an objective basis, if ranks are used to determine node height. None of these assumptions seem to be justified in most cases, and Linnean ranks are known to be arbitrary constructs (Laurin, 2008). Thus, our procedure (assigning minimal ages to each node and gradually lengthening minimal branch lengths until adequate contrast standardization is achieved) should provide sets of biologically meaningful branch lengths and thus facilitate interpretation of evolutionary rates.
In order to be able to use both molecular and paleontological dates in constraining the minimal ages of the clades in the supertree, we have inserted relevant extinct taxa, mostly from the supertree of Marjanović & Laurin (2007). We have also inserted molecular dates, which constrain various clades whose age was estimated using molecular data, mostly from Steinfartz et al. (2007), as terminal taxa in the tree, to mimic paleontological data that also provide minimal age estimates. However, as Stratigraphic Tools cannot make hypothetical ancestors younger than any of their known descendants (Josse et al. 2006; Marjanović & Laurin, 2007), we used the upper (younger) bound of the 95% confidence interval in these cases, rather than the point estimate, whenever a confidence interval was reported. We are aware that several factors influence molecular dating and that some molecular dates are poorly constrained (Graur & Martin, 2004; Britton, 2005). However, we are not concerned here with how these dates were computed but rather with what can be done with a set of paleontological and molecular dates. In our experience, the best results can be obtained by the simultaneous use of both types of data; we recently confirmed previous findings (Brochu, 2004) that the most important source of variance in molecular dates is the choice of calibration dates (Marjanović & Laurin, 2007).
We could not accurately date much of the evolutionary radiation within Ranidae because the phylogeny within this group remains highly contentious (Che et al. 2007). Thus, in this case, we simply used Stratigraphic Tools (Josse et al. 2006) to set minimum branch lengths between these species, and empirically chose a global minimum branch length setting that ensured adequate standardization of phylogenetically independent contrasts. This procedure was applied to obtain crude estimates of (mostly terminal) branch lengths for divergences betweenRana iberica Boulenger, 1879 andRana temporaria Linnaeus, 1758, and betweenLithobates catesbeianus Shaw, 1802 andLithobates vaillanti Brocchi, 1877. Similarly, we could not date the divergence between the species of Salamandra, which must be very recent (as argued by Shaffer & McKnight, 1996); it has been set as occurring in the Pleistocene. The study by Zhang et al. (2008) was published too late to be incorporated into our starting tree (Fig. 1A) but the tree that we used for our analyses (Fig. 1B) implies divergence dates between Pleurodeles poireti and Pleurodeles waltl of 30 Ma, only slightly older than suggested by Zhang et al. (2008). Given the increasing number of studies that estimate molecular divergence dates, the length of most of these branches will probably be much better known in a few years.
We tested the presence of a phylogenetic signal in microanatomical and body-size characters by a variance-partitioning method with PVR (Desdevises et al. 2003). Thus, a phylogenetic-distance matrix was produced using Stratigraphic Tools for Mesquite (Josse et al. 2006) and then converted into a matrix of principal coordinates (PCs) using the R Package version 4.0 for the Macintosh (Casgrain et al. 2004). The PC analyses generated n – 1 PCs for n species (45 PCs in this analysis). However, only a subset of the resulting 45 axes can be used because otherwise no degree of freedom would be left. These axes were selected using a broken-stick model (Diniz-Filho et al. 1998). Vectors of character data were exported using the StratAdds module (Faure et al. 2006). A succession of multiple linear regressions (with 9999 permutations) was then performed using Permute (Casgrain, 2005). To detect the phylogenetic signal, each compactness profile and body size parameter was considered a dependent variable and the lifestyle and retained PCs were considered the independent variables.
To detect the ecological signal in the variation of lifestyle, we performed an additional analysis of variance partition with PVR. The dependent variable was the lifestyle and the independent variables were the compactness profile parameters, body size parameters and PCs representing the phylogeny. We used backward-elimination and forward-selection procedures (in Permute) to determine which variables show the most important ecological signal. The lifestyle had either a ternary coding (aquatic, amphibious, terrestrial) or a binary coding (aquatic vs. amphibious to terrestrial) because most previous studies on this topic showed that it is difficult to distinguish between amphibious and terrestrial taxa on the basis of bone microanatomy (Laurin et al. 2004; Germain & Laurin, 2005; Canoville & Laurin, 2009). We also performed a FIC analysis of characters whose contrasts were adequately standardized on our tree using the PDAP module of Mesquite (Midford et al. 2003; Maddison & Maddison, 2008). Finally, we performed a few simple, non-phylogenetic T-tests in Microsoft Excel to compare with the results of the independent contrast analyses, to determine if a significant amount of ecological data is included in our microanatomical and body size data. This was done because independent contrast analyses have little power when the independent character is discrete, especially when the number of transitions between states is relatively small, as is the case here.
As our study involves testing the presence of a phylogenetic and of an ecological signal in several characters and using various techniques, we have used the false discovery rate procedure to account for multiple comparisons (Benjamini & Hochberg, 1995; Curran-Everett, 2000). We have divided our tests into two families, namely those concerning a phylogenetic signal and those concerning the ecological signal in the data.
Finally, a linear-discriminant analysis was carried out using Statistica 6 (StatSoft France, 2003). This method, which does not require linearity between categories of discrete variables (the lifestyle) and does not consider phylogenetic relationships, gives the probability of the modeled lifestyle. The dependent variable was once again the lifestyle and the independent variables were the bone compactness and body size parameters. This analysis indicates the proportion of correct inferences that can be made from the models, which could be used on fossil femora to provide paleoecological interpretations.
Results
Qualitative examination of the mid-diaphyseal cross-sections of femora
Urodeles
The diaphysis of the long bones of urodeles generally presents a simple histological structure (de Ricqlès, 1977) but the microstructure can show more interspecific variability.
Urodeles of small and average body size
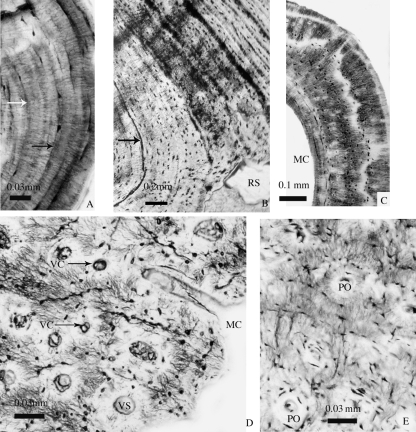
As is common in lissamphibians (de Ricqlès, 1995), the avascular parallel-fibered bone tissue shows extensive lines of arrested growth in most individuals (Fig. 2A,B). Most sections lack vascularization, resorption cavities or a spongiosa (Figs 3, 4) but endosteal resorption is observed in various species, such as Pleurodeles waltl Michahelles, 1830 (Fig. 3F), Triturus cristatus Laurenti, 1768 (Fig. 3I), Triturus helveticus Razoumovsky, 1789 (Fig. 3K) and Desmognathus quadramaculatus Holbrook, 1840 (Fig. 3M). Unlike the other species in the present dataset, Ambystoma andersoni Krebs and Brandon, 1984 shows numerous resorption cavities (Fig. 3D). Vascularization, represented by one or a few canals, was observed in only a few terrestrial species, such as Salamandrella keyserlingii Dybowski, 1870 (Fig. 3N),Ambystoma opacum Gravenhorst, 1870 (Fig. 3O) andDesmognathus monticola Dunn, 1916 (Fig. 3T).
Fig. 2.
Histological mid-diaphyseal cross-sections of femora of urodeles (A, B) and anurans (C–E). (A) Desmognathus monticola; (B) Andrias japonicus; (C) Rana ridibunda; (D) Lithobates catesbeianus; (E) Rhinella marina. In D and E, cortical pseudolamellar bone shows numerous primary osteons, which (in E) are linked by radial anastomoses. White and black arrows in A and B show the lines of arrested growth. MC, medullary cavity; PO, primary osteon; RS, resorption space; VC, vascular canals; VS, vascular spaces.
Fig. 3.
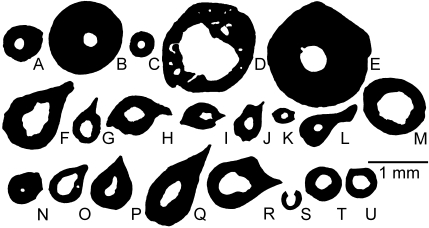
Mid-diaphyseal cross-sections of femora of urodeles of small body size. (A) Onychodactylus fischeri(Hynobiidae); (B) Amphiuma means (Amphiumidae); (C) Proteus anguineus (Proteidae); (D)Ambystoma andersoni (Ambystomatidae); (E)Necturus maculosus(Proteidae); (F) Pleurodeles waltl (Salamandridae); (G)Pleurodeles poireti(Salamandridae); (H) Triturus marmoratus (Salamandridae); (I) Triturus cristatus (Salamandridae); (J) Mesotriton alpestris (Salamandridae); (K) Triturus helveticus(Salamandridae); (L) Triturus montandoni (Salamandridae); (M)Desmognathus quadramaculatus (Plethodontidae); (N)Salamandrella keyserlingii(Hynobiidae); (O)Ambystoma opacum (Ambystomatidae); (P)Salamandra atra(Salamandridae); (Q)Salamandra lanzai(Salamandridae); (R)Salamandra salamandra(Salamandridae); (S) Desmognathus ochrophaeus(Plethodontidae); (T) Desmognathus monticola (Plethodontidae); (U)Plethodon glutinosus (Plethodontidae). A–E are aquatic, F–M are amphibious and N–U are terrestrial. Amphiuma means is included here even though its body size is large because its limbs are minute. The section of Desmognathus ochrophaeus(S) is damaged; only the intact portion, which was modeled using Bone Profiler (Girondot & Laurin, 2003), is shown here.
Fig. 4.
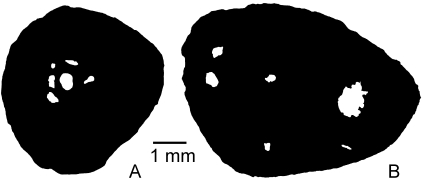
Mid-diaphyseal cross-sections of femora of urodeles of large body size. (A) Cryptobranchus alleganiensis (Cryptobranchidae); (B) Andrias japonicus(Cryptobranchidae). Both species are aquatic.
Urodeles of large body size
Cryptobranchids are the largest extant urodeles. Andrias japonicus Temminck, 1836, for example, can exceed 1.3 m and 25 kg. Cryptobranchids are strictly aquatic and no terrestrial or amphibious urodeles showing this length and weight are known. The cortex of the femur is composed of simple periosteal lamellar bone, and the medullary region is small (Fig. 4A) and retains bone trabeculae in Cryptobranchus alleganiensis (Fig. 4B). Resorption cavities are present in both species but secondary osteons are lacking, despite the presumably relatively great age of the sectioned specimen of A. japonicus (judging by its presacral length of 34 cm from atlas to sacrum).
Anurans
The bony tissues of adult anurans are generally of two types (Enlow & Brown, 1956): lamellar, avascular bone in some small species or vascular bone, mostly in species of intermediate to large body size.
Anurans of small body size
Resorption cavities are numerous in the aquatic Bombina orientalis Boulenger, 1890 (Fig. 5A), whereas they are lacking in almost all of the other species, as in Rana esculenta Linnaeus, 1758 (Fig. 5D) or Rana ridibunda Pallas, 1771 (Fig. 5E). In most small amphibious and terrestrial anurans, the cortical bone is lamellar and avascular, as first described by Foote (1916). However, ‘Bufo’pentoni Anderson, 1893 shows a few vascular and resorption spaces (Fig. 5I). No spongiosa is found in the medullary cavity in small anurans.
Fig. 5.
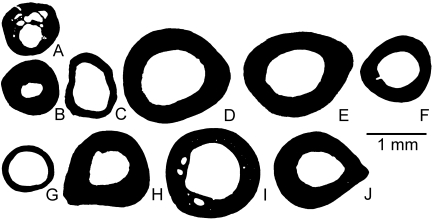
Mid-diaphyseal cross-sections of femora of anurans of small body size. (A) Bombina orientalis; (B) Ascaphus truei; (C) Discoglossus sp.; (D) Rana esculenta (a hybrid of R. ridibunda and R. lessonae); (E) Rana ridibunda; (F) Rana iberica; (G) Pelobates fuscus; (H) Rana dalmatina; (I) ‘Bufo’pentoni; (J) Pachymedusa dacnicolor. A is aquatic, B–F are amphibious and G–J are terrestrial.
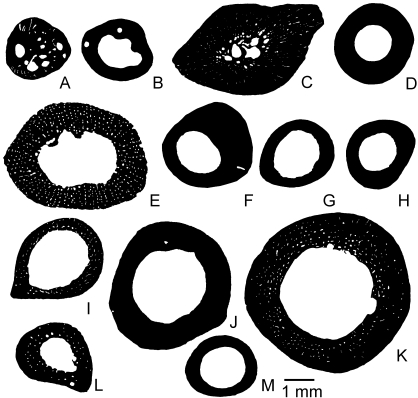
Anurans of large body size
Most of the sections of aquatic species show little if any spongiosa (Fig. 6). However, the presence of resorption cavities or vascular spaces is variable in the lamellar bone. The very thick cortex of Pipa pipa Linnaeus, 1758 displays primary osteons and numerous vascular and resorption spaces (Fig. 6C), whereas Pipa carvalhoi Miranda-Ribeiro, 1937 (Fig. 6B) and Telmatobius culeusGarman, 1875 (Fig. 6D) show few or no cavities. The presence of vascular spaces in amphibious species is variable: Lithobates catesbeianus (Figs 2D, 6E) shows a network of vascular spaces in the paralleled-fibered bone (Fig. 2D), whereas Rana temporaria Linnaeus, 1758 (Fig. 6F), Lithobates vaillanti (Fig. 6G) and Pantherana forreri Boulenger, 1883 (Fig. 6H) lack vascularization. In terrestrial species, vascularization is almost always present, as in Ceratophrys aurita Raddi, 1823 (Fig. 6I), Leptodactylus pentadactylus Laurenti, 1768 (Fig. 6J) and Bufo bufo Linnaeus, 1758 (Fig. 6L). Hypsiboas boans Linnaeus, 1758 (Fig. 6M), which presents avascular lamellar bone, seems to be an exception. In the cortex of Rhinella marina (Figs 2E, 6K), numerous longitudinal primary osteons are visible, mostly near the medullary cavity, where resorption zones occur.
Fig. 6.
Mid-diaphyseal cross-sections of femora of anurans of large body size. (A) Xenopus laevis; (B) Pipa carvalhoi; (C) Pipa pipa; (D) Telmatobius culeus; (E) Lithobates catesbeianus; (F) Rana temporaria; (G) Lithobates vaillanti; (H) Pantherana forreri; (I) Ceratophrys aurita; (J) Leptodactylus pentadactylus; (K) Rhinella marina; (L) Bufo bufo; (M) Hypsiboas boans. A–D are aquatic, E–H are amphibious and I–M are terrestrial.
Even though a single bone of each species has been illustrated for most taxa, the authors have seen other sections of several of these species. In most cases, the amount of intraspecific variation is moderate and studying more specimens would presumably not alter our conclusions substantially (Fig. 7).
Fig. 7.
Mid-diaphyseal cross-sections of femora of the terrestrial urodele Salamandra salamandra(Salamandridae) to show the amount of intraspecific variation. The individual already shown in another plate (Fig. 3R) is not represented again.
Statistical analyses
Phylogenetic and ecological signals in the data
We succeeded in eliminating statistical artifacts of six characters (out of 13) for FIC analyses and phylogenetic signal detection using the reference tree for which the minimum length of terminal branches was set to 30 Ma and that of internal branches to 20 Ma, and the following parameters (some of which were transformed as shown): natural logarithm of presacral length (LN[PLg]), natural logarithm of 10*maximal section diameter (in mm; LN[10*MD]), cube of global compactness (Cg3), P based on global values (P), P based on radial values (Prad), lifestyle (ternary coding) and S0.1. We initially tried standard data transformations with obvious biological justification, such as a logarithmic transformation, but when this failed to standardize contrasts, we tried various alternatives.
The broken-stick model retained only the first three PCs (PC1, PC2, PC3), which represent 60.35% of the phylogenetic variance of the 50 species. The variance partitioning method performed using Permute (Casgrain, 2005) indicated that LN(10*MD), P, Prad and Cg3 contain phylogenetic information (Table 1).
In the analyses of ecological signal, the backward elimination procedure in Permute retained the parameters LN(PLg), Max and Cp as the most significant variables (Table 2) and the variance partitioning analyses showed that these parameters exhibit an ecological signal (P = 0.0003), and that they explained nearly 35% of the lifestyle variance (44% when the covariance with the phylogeny is added), when lifestyle is considered a binary variable. The part of the lifestyle variance explained by the phylogeny alone was not statistically significant (P = 0.2711). An important fraction (47.73%) remained unexplained by the variables studied (Table 2). A forward-selection procedure retained different parameters (PLg, Minrad, Min) as the most significant variables that exhibit an ecological signal (P = 0.0016; Table 3) and they explain 28.42% of the lifestyle variance (40% when the covariance with the phylogeny is added), when lifestyle is considered a binary variable. Again, the part of lifestyle variance explained by the phylogeny alone is not statistically significant and an important fraction remained unexplained (Table 2). Thus, aquatic taxa seem to have a lower compactness in the outermost cortex (lower values of the parameters Max and Cp, Table 3) than their amphibious and terrestrial relatives. Moreover, P has lower values (the medullary cavity is smaller) in aquatic taxa than in terrestrial and amphibious species (Table 3).
Table 2.
Variance of habitat explained by phenotypic characters (body size and bone microanatomical characters) and phylogenetic position (represented by phylogenetic principal coordinates)
| Parameters that reflect the lifestyle, selected by a backward -elimination procedure: LN(PLg), Max, Cp |
Parameters that reflect the lifestyle, selected by a forward-selection procedure: PLg, Minrad, Min |
|||||||
|---|---|---|---|---|---|---|---|---|
| LS 3 states | P-value | LS 2 states | P-value | LS 3 states | P-value | LS 2 states | P-value | |
| a | 0.2342 | 0.0026 (iv) | 0.3496 | 0.0003 (i) | 0.2314 | 0.0036 (vi) | 0.2842 | 0.0016 (iii) |
| b | 0.0233 | 0.0868 | 0.0482 | 0.1115 | ||||
| c | 0.0662 | 0.4067 (9) | 0.0863 | 0.2711 (7) | 0.0413 | 0.624 (15) | 0.0616 | 0.4455 (11) |
| d | 0.6763 | 0.4773 | 0.6791 | 0.5427 | ||||
Habitat is treated as the dependent variable and some parameters (selected by either a backward-elimination or a forward-selection procedure) and the three first principal coordinates as independent variables. Arabic numbers in parentheses next to the probabilities indicate the rank in the family of tests of phylogenetic signal; Roman numbers in parentheses indicate rank in the family of tests of ecological signal (see Table 5). a, Fraction related to phenotype (body size and bone microanatomy); b, fraction related to phenotype and phylogeny; c, fraction related to phylogeny; d, residual variation (unexplained fraction). Abbreviations as in Table 1.
Table 3.
Mean and standard variation of parameters of the femur in lissamphibians of each lifestyle
| Lifestyle | LN(PLg) | LN(10*MD) | S | P | Prad | Min | Max | Cc | Cp | Cg3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Aquatic | Mean | 2.3858 | 2.9835 | 0.0328 | 0.2995 | 0.3012 | 0.0270 | 0.9910 | 0.0353 | 0.9854 | 0.6689 |
| SD | 0.9191 | 0.8373 | 0.0465 | 0.1962 | 0.1957 | 0.1458 | 0.0563 | 0.1417 | 0.0473 | 0.2426 | |
| Amphibious | Mean | 1.4770 | 2.6390 | 0.0293 | 0.4605 | 0.4679 | −0.0096 | 0.9960 | −0.0085 | 0.9960 | 0.4817 |
| SD | 0.4907 | 0.6078 | 0.0112 | 0.1639 | 0.1586 | 0.0160 | 0.0184 | 0.0133 | 0.0185 | 0.2526 | |
| Terrestrial | Mean | 1.6629 | 2.6657 | 0.0257 | 0.4834 | 0.4851 | −0.0056 | 0.9960 | −0.0043 | 0.9960 | 0.4390 |
| SD | 0.5313 | 0.7877 | 0.0135 | 0.1623 | 0.1637 | 0.0116 | 0.0099 | 0.0078 | 0.0098 | 0.2310 |
Abbreviations as in Table 1.
The FIC analysis using the PDAP module of Mesquite (Midford et al. 2003; Maddison & Maddison, 2008) showed that presacral length is correlated with lifestyle (Table 4) and that aquatic lissamphibians are generally larger than amphibious and terrestrial taxa. Note that, even though the habitat has been coded as a discrete variable, it can be analysed by independent contrasts (at least when considered the independent variable) because it can be conceptualized as a continuous variable (proportion of time spent in water or on land) that has been coded as a binary or ternary variable for lack of more detailed information (Al-kahtani et al. 2004).
Table 4.
Relationship between lifestyle and phenotypic characters (body size and bone microanatomical characters) assessed through phylogenetically independent contrasts analysis [Felsenstein independent contrasts (FIC)] and T-tests
| Method | Lifestyle (binary coding) vs.: | LN(PLg) | LN(10*MD) | Cg3 | P | Prad | S0.1 |
|---|---|---|---|---|---|---|---|
| FIC | R2 | 0.2248 | 0.0542 | 0.0503 | 0.0653 | 0.0345 | 0.0025 |
| P-value | 0.0008 (ii) | 0.1193 (xi) | 0.1338 (xii) | 0.0866 (x) | 0.2164 (xiv) | 0.7435 | |
| T-test | P-value | 0.0120 (viii) | 0.1846 (xiii) | 0.0131 (ix) | 0.0092 (vii) | 0.0035 (v) | 0.3051 (xv) |
| Equality of variance (for T-test) | No | Yes | Yes | Yes | Yes | No |
For the FIC analyses, proportion of explained variance and associated probabilities are shown. Only characters whose contrasts were adequately standardized are included. T-tests were conducted in Microsoft Excel, using formulae for samples of equal or unequal variances, depending on the character. For each character, equality of variances was tested through an F-test. Abbreviations as in Table 1.
Non-phylogenetic T-tests showed that, in addition to presacral length, medullary size (P, Prad) and global compactness (Cg) vary depending on the lifesyle, when coded as a binary variable.
False discovery rate analysis did not modify our conclusions, in this case. By coincidence, all results deemed significant above (at a 0.05 type I error rate threshold) remained significant after the false discovery rate analysis (Table 5). Note that, for the phylogenetic signal, three tests yielded exactly the same probability (0.0004); we suggest using the mean ranked probability as a threshold, in this case (0.0059 + 0.088 + 0.0118)/3 = 0.0088. In our case, these three identical probabilities were unproblematic because all three were smaller than the threshold.
Table 5.
False discovery rate analysis (Benjamini & Hochberg, 1995; Curran-Everett, 2000) of the statistical results about the phylogenetic and ecological signals
| Phylogenetic signal detection tests |
Ecological signal detection test |
||||||
|---|---|---|---|---|---|---|---|
| Rank | Variable concerned | Probability | Threshold | Rank | Variable concerned | Probability | Threshold |
| 1 | Cg3 | 0.0001 | 0.0029 | i | Backward model, binary | 0.0003 | 0.0031 |
| 2 | LN(10*MD) | 0.0004 | 0.0059 | ii | LN(PLg), FIC | 0.0008 | 0.0063 |
| 3 | P | 0.0004 | 0.0088 | iii | Forward model, binary | 0.0016 | 0.0094 |
| 4 | Prad | 0.0004 | 0.0118 | iv | Backward model, ternary | 0.0026 | 0.0125 |
| 5 | S0.1 | 0.0555 | 0.0147 | v | Prad, T-test | 0.0035 | 0.0156 |
| 6 | Srad | 0.2110 | 0.0176 | vi | Forward model, ternary | 0.0036 | 0.0188 |
| 7 | Backward model, binary | 0.2711 | 0.0206 | vii | P, T-test | 0.0092 | 0.0219 |
| 8 | Maxrad | 0.3093 | 0.0235 | viii | LN(PLg), T-test | 0.0120 | 0.0250 |
| 9 | Backward model, ternary | 0.4067 | 0.0265 | ix | Cg3, T-test | 0.0131 | 0.0281 |
| 10 | Cp | 0.4099 | 0.0294 | x | P, FIC | 0.0866 | 0.0313 |
| 11 | Forward model, binary | 0.4455 | 0.0324 | xi | LN(18*MD), FIC | 0.1193 | 0.0344 |
| 12 | Cc | 0.4461 | 0.0353 | xii | Cg3, FIC | 0.1338 | 0.0375 |
| 13 | Min | 0.5395 | 0.0382 | xiii | LN(18*MD), T-test | 0.1846 | 0.0406 |
| 14 | Max | 0.5807 | 0.0412 | xiv | Prad, FIC | 0.2164 | 0.0438 |
| 15 | Forward model, ternary | 0.6240 | 0.0441 | xv | S0.1, T-test | 0.3051 | 0.0469 |
| 16 | Minrad | 0.6748 | 0.0471 | xvi | S0.1, FIC | 0.7435 | 0.0500 |
| 17 | LN(PLg) | 0.7550 | 0.0500 | ||||
| Critical probability (α) | 0.05 | 0.05 | |||||
| Number of tests (m) in each family | 17 | 16 | |||||
| α/m | 0.00294 | 0.00313 | |||||
This analysis was performed separately for tests of phylogenetic signal and for those of ecological signal, as the tested hypotheses differ. Probabilities are given in increasing order and are numbered in Arabic or Roman numerals for phylogenetic and ecological signal, respectively, as in Tables 1, 2, 4. Note that, in this case, every probability considered significant when taken in isolation remains significant after false discovery rate analysis. FIC, Felsenstein independent contrasts.
We performed two main discriminant analyses to obtain inference models of habitat. First, the parameters selected in the variance partitioning method by a backward elimination procedure, (LN[PLg]), Max and Cp, Table 2) were included as independent variables. When lifestyle is coded as a binary variable, the discriminant function correctly attributed the lifestyle of 38 species (82.6%, Table 6; Appendix 2). The habitat of only 50% of the aquatic animals was correctly inferred but the lifestyles of nearly 94% of amphibious and terrestrial taxa were properly attributed. Second, the parameters selected in the variance partitioning method by a forward-selection procedure (PLg, Min and Minrad; Table 2) were included as independent variables (Table 6; Appendix 2). In that case, we found the same proportion (82.6%) of correctly inferred habitats. Nevertheless, the errors were not identically distributed; this model does not reasonably discriminate aquatic taxa from others because only one-third of these animals were inferred aquatic, whereas all amphibious and terrestrial lissamphibians were accurately modeled.
Table 6.
Values of the constants of the discriminant functions used to infer the lifestyle (0, aquatic; 1, amphibious or terrestrial) of lissamphibians
| Discriminant function obtained with LN(PLg), Max, Cp | Discriminant function obtained with PLg, Min, Minrad | ||||
|---|---|---|---|---|---|
| 0 | 1 | 0 | 1 | ||
| Y intersection | −790.40 | −817.06 | Y intersection | −2.87679 | −0.47783 |
| LN(PLg) | 2.67 | −0.15 | PLg | 0.17292 | 0.05540 |
| Max | −1969.48 | −2182.07 | Min | 6.11333 | −1.03903 |
| Cp | 3575.56 | 3822.33 | Minrad | 0.00077 | 0.01348 |
For each habitat, the discriminant function can be used to infer the habitat (by multiplying the constant of each parameter by the value of the parameter of a given species). The habitat that has the highest score is the inferred lifestyle (Appendix 2). Abbreviations as in Table 1.
As in Canoville & Laurin (2009), to make our inference models useful, we produced spreadsheets that allow anyone to infer a lissamphibian lifestyle solely from body size and femoral compactness characters (Appendix 2).
Discussion
Ecological and taxonomic differences in bone microanatomy
The histological descriptions above largely confirm earlier works in this field (e.g. de Ricqlès, 1995) but with a denser taxonomic sample of lissamphibians. Species of larger body size seem to show a greater variety of tissue types, with primary osteons and vascular spaces being present in some species of anurans. Urodeles may be less vascularized and no osteons were found in them, which may reflect their low metabolic rate (Pough et al. 2004, p. 13), which may be linked to their large genome (Sessions et al. 2008, p. 572). The absence of osteons may also be a primitive character or be linked to the low mechanical stress experienced by the skeleton; there is at least circumstantial evidence that Haversian remodeling is linked to bone repair (Castanet et al. 2001).
The ecological signal appears more clearly at the microanatomical level. The medullary cavity is smaller in aquatic taxa than in terrestrial and amphibious species. However, it is difficult to separate amphibious and terrestrial taxa. Aquatic anurans and urodeles have a thicker cortex, which may act as ballast, as in many aquatic amniotes (Germain & Laurin, 2005; Kriloff et al. 2008).
The medullary cavity of Bombina orientalis (an aquatic species) is not particularly small (P = 0.54) but it is smaller than that of its nearest studied relative, the amphibious Discoglossus sp. Otth, 1837 (P = 0.71). Optimization of the lifestyle does not resolve the habitat of the last common ancestor of these species. The divergence between these taxa may (at the latest) have taken place a little before 23 Ma ago, which is the age of the oldest known Discoglossus (Marjanović & Laurin, 2007: fig. 5). The lineage leading to B. orientalis may have become aquatic subsequent to this divergence.
Aquatic ambystomatids may also not show a marked increase in femoral compactness. Indeed, the aquatic Ambystoma andersoni has a larger medullary cavity (higher P value) than the terrestrial A. opacum,contrary to the general pattern observed in other lissamphibians. However, A. andersoni probably belongs to a group that became aquatic (and neotenic) fairly recently because its divergence from A. opacum may date from about 10 Ma (Estes, 1981), and the evolutionary radiation of the Mexican species of Ambystoma apparently began less than 5 Ma ago (Shaffer & McKnight, 1996, p. 429). Furthermore, most adults of ambystomatids are terrestrial (Goin et al. 1978), so this lifestyle is probably primitive for this clade and, even in the A. tigrinum Green, 1825 group that includes A. andersoni(Shaffer & McKnight, 1996), many adults, such as Ambystoma mexicanum Shaw & Nodder, 1798 populations that live in the eastern United States, are terrestrial (Goin et al. 1978). Even Ambystoma velasci Dugès, 1888, which is closely related to A. andersoniand probably diverged from it less than 5 Ma ago (Schaffer & McKnight, 1996), has terrestrial adults. Thus, A. andersoni probably returned to an aquatic lifestyle less than 5 Ma ago. Finally, comparisons of the compactness profiles within Ambystoma are hampered by a two-fold size difference in linear dimensions (eight-fold in body mass) between the species of this genus included in this study.
A few terrestrial Caudata have a small medullary cavity, like their aquatic relatives. However, these taxa, such as Salamandrella keyserlingii and some species of Triturus, such as Tristurus marmoratusLatreille, 1800 and T. montandoni Boulenger, 1880, have a small body size (presacral length 3.3–6.2 cm; maximal femoral section diameter ranging from 0.6 to 1.3 mm). At these size ranges, there seems to be little difference in compactness (and parameter P) between aquatic and terrestrial caudates. However, nearly all caudates in these size ranges are amphibious or terrestrial. The smallest aquatic caudate in our sample is Onychodactylus fischeri Boulenger, 1886 and it is much larger (with a presacral length about 50% greater) than its closest terrestrial relative S. keyserlingii. Thus, among the smallest caudates, changes of lifestyle may have more repercussions on body size than on femoral compactness (and the parameter P). Among anurans, size may not discriminate aquatic from amphibious and terrestrial taxa, although the two smallest studied anurans are terrestrial (Pelobates fuscus Laurenti, 1768) and amphibious (Ascaphus truei Stejneger, 1899).
Statistical analyses
The tree used for phylogenetically independent contrast analyses and variance partition using PVR (Fig. 1B) had branch lengths fairly different from the initial tree (Fig. 1A) but it remains biologically plausible to the extent that a literal interpretation implies an origin of Lissamphibia in the Early Carboniferous (near the Viséan/Serpukhovian boundary, about 325 Ma ago). However, our data do not imply that lissamphibians originated that early because FIC analyses are not meant to date trees and because many characters were adequately standardized using shorter branches; the tree that we used was only the shortest of the tested trees that could standardize the greatest set of characters. Nevertheless, many recent molecular dating studies (San Mauro et al. 2005; Zhang et al. 2005; Roelants et al. 2007) suggest an even earlier origin of this taxon, in the Late Devonian to Early Carboniferous (369–337 Ma ago). The date of lissamphibian origin in our tree (Fig. 1B) is slightly older than the age (292–323 Ma) recently inferred on the basis of the nuclear gene RAG-1 (Hugall et al. 2007, p. 554). However, the paleontological record suggests a much later, Permian origin (Marjanović & Laurin, 2008), which is congruent with our original time-calibrated tree (Fig. 1A). Other clades in the tree used for our analyses (Fig. 1B) are older than recently proposed. For instance, the divergence between Triturus cristatus and T. marmoratus must date from more than 23 Ma, if the affinities of the fossil Triturus were correctly established (Estes, 1981, p. 87). Our starting tree (Fig. 1A) implies a slightly older age (about 28 Ma) but our transformed tree, because of the large minimal values, implies a much older age of more than 70 Ma for this clade, which is much older than recently suggested by Zhang et al. (2008). Clearly, such dates must be used with caution.
Phylogenetically independent contrasts using the PDAP module of Mesquite (Midford et al. 2003; Maddison & Maddison, 2008) demonstrate a significant correlation between body size and habitat, contrary to previous analyses (Laurin et al. 2004, p. 597) of a very similar dataset using CAIC (‘comparative analysis by independent contrasts’; Purvis & Rambaut, 1995). A few reasons can explain this difference: first, the topology and initial branch lengths were modified to incorporate recent phylogenetic and paleontological work, which probably resulted in a more reliable phylogeny; second, the branch lengths were manipulated using the Stratigraphic Tools (Josse et al. 2006) to obtain adequate standardization of contrasts; and third, the algorithm used in this analysis, which consists of considering that the discrete variable (habitat) is an approximation of an intrinsically continuous character (proportion of time of activity spent on land), is probably more powerful than the algorithm used by CAIC to deal with discrete characters (in that case, very few contrasts are used, which should reduce power). It is likely that all of these factors have combined to yield the more significant results reported above.
Body size and bone microanatomical data appear to contain a significant ecological signal, as shown by the variance partition analyses with PVR (Table 2) and T-tests. The fact that independent contrasts could not corroborate this (except for body size) is probably linked to several factors. First, the characters most correlated to the habitat and selected by the models (Table 2) could not be subjected to independent-contrast analysis (because statistical artifacts remained, despite branch length and data transformations). Second, the relatively small number of transitions between habitats (six independent appearances of an aquatic habitat from the probably primitive amphibious habitat are shown in Fig. 1B) significantly reduces the power of methods that take the phylogeny into consideration through the use of contrasts. As terrestrial and amphibious lissamphibians do not differ significantly from each other in most bone microanatomical features (Laurin et al. 2004), only transitions to a fully aquatic lifestyle are expected to leave a signature on bones, and very recent transitions, which may represent half of sampled events (Fig. 1B), may leave little or no trace. The lack of power of FIC in this context can be illustrated by comparisons with Student's T-tests (for samples of equal or unequal variances, depending on the data), which show a significant effect of four out of the six microanatomical and size variables tested, when lifestyle is coded as a binary variable. FIC analysis of medullary size (P) vs. binary habitat is not significant (Table 4), although a simple (non-phylogenetic) T-test gives significant results (P = 0.0092). The variance partition with PVR showed an ecological signal in microanatomy but it was performed as a multiple regression, whereas contrasts have been performed (as usually done) as simple linear regressions, and it incorporates the phylogeny in a different way than contrasts. The fact that medullary cavity size (P) was not retained by the forward- or backward-selection procedures is surprising because this character seems to show the greatest habitat-related difference, after presacral length (Table 4). This probably results from the correlation between this character and presacral length (log-transformed); an FIC analysis shows that presacral length explains 12.5% of the variance in medullary cavity size, and that this result is significant (P = 0.016). Simple linear regressions show that the other characters selected by the models (Table 2) are not correlated with presacral length. We realize that comparative data should preferably be analysed with methods that use phylogenetic data but, in some cases, other methods may be useful (at least when used together with more sophisticated methods) because the true evolutionary model of our data is unknown and appears in many cases to depart quite strongly from pure Brownian motion and, in such cases, simple linear regressions may perform as well as, or even better than, methods using phylogenetic data (Martins et al. 2002).
Analysing comparative data remains a challenging task but we hope that the method presented above will facilitate the task somewhat by making it easier to compile time-calibrated supertrees using a combination of paleontological and molecular dates, and by providing new ways to perform branch length transformation. Our data provide moderate additional support for the long-established view that bone microanatomical data reflect the habitat in lissamphibians (Leclair et al. 1993; Castanet & Caetano, 1995; Laurin et al. 2004) and in tetrapods in general (de Buffrénil & Buffetaut, 1981; Wall, 1983; Fish & Stein, 1991; Germain & Laurin, 2005; Scheyer & Sander, 2007; Kriloff et al. 2008).
Acknowledgments
We thank all individuals who contributed material for this study. This includes H. Francillon-Vieillot (formerly at Université Paris 7), J. Castanet (Université Paris 6), C. Miaud (Université de Savoie), F. Renoult and D. Robineau (both in the Laboratoire d’Anatomie Comparée, Muséum National d’Histoire Naturelle, Paris), V. H. Reynoso-Rosales (Universidad Nacional Autónoma de México) and A. S. Severtsov (Moscow State University). D. Marjanovic and V. de Buffrénil made numerous suggestions that significantly improved the draft.
Supporting Information
Additional Supporting Information may be found in the online version of this article
Appendix 1 Cross-sections of femora of lissamphibians studied for this analysis (46 species). The lifestyle had either a ternary (0, aquatic; 1, amphibious; 2, terrestrial) or a binary (0, aquatic; 1, amphibious to terrestrial) coding. These states are defined by the relative amount of time spent in water: > 90% for aquatic taxa, between 20% and 90% for amphibious taxa and < 20% for terrestrial taxa. LS, lifestyle; PLg, presacral length (in cm); LN(PLg), natural logarithm of presacral length; MD, maximal diameter of the cross-section (in mm); LN(10*MD), natural logarithm of 10*maximal diameter; S, reciprocal of the slope at the inflection point that generally reflects the width of the transition zone between the cortical compacta and medulla; Srad, radial values of parameter S; P is proportional to the size of the medullary cavity; Prad, radial value of P; Min, compactness in the center of the medullary region; Minrad, radial values of parameter Minrad; Max, compactness in the outermost cortex; Maxrad, radial values of parameter Maxrad; Cc, compactness in the center of the cross-section; Cp, compactness in the periphery of the cross-section; Cg, global compactness.
Appendix 2 (a) Discriminant function 1; (b) detailed formula 1; (c) discriminant function 2; (d) detailed formula 2.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Al-kahtani MA, Zuleta C, Caviedes V, Garland TJ. Kidney mass and relative medullary thickness of rodents in relation to habitat, body size, and phylogeny. Physiol Biochem Zool. 2004;77:346–365. doi: 10.1086/420941. [DOI] [PubMed] [Google Scholar]
- Arntzen JW, Themudo GE, Wielstra B. The phylogeny of crested newts (Triturus cristatussuperspecies): nuclear and mitochondrial genetic characters suggest a hard polytomy, in line with the paleogeography of the centre of origin. Contrib Zool. 2007;76:261–278. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Bininda-Emonds ORP, Sanderson MJ. Assessment of the accuracy of matrix representation with parsimony analysis. Syst Biol. 2001;50:565–579. [PubMed] [Google Scholar]
- Blomberg SP, Garland TJ, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- Britton T. Estimating divergence times in phylogenetic trees without a molecular clock. Syst Biol. 2005;54:500–507. doi: 10.1080/10635150590947311. [DOI] [PubMed] [Google Scholar]
- Brochu CA. Calibration age and quartet divergence date estimation. Evolution. 2004;58:1375–1382. doi: 10.1111/j.0014-3820.2004.tb01715.x. [DOI] [PubMed] [Google Scholar]
- Cannatella DC, Trueb L. Evolution of pipoid frogs: intergeneric relationships of the aquatic frog family Pipidae (Anura) Zool J Linn Soc. 1988;94:1–38. [Google Scholar]
- Canoville A, Laurin M. Microanatomical diversity of the humerus and lifestyle in lissamphibians. Acta Zool. 2009;90:110–122. [Google Scholar]
- Casgrain P. Permute! Montreal. http://www.bio.umontreal.ca/Casgrain/prog/labo/permute/index.html.
- Casgrain P, Legendre P, Vaudor A. The R Package for multidimensional and spatial analysis (version 4.0d10 for the Macintosh). Montreal. http://www.bio.umontreal.ca/casgrain/en/telecharger/
- Castanet J, Caetano MH. Influence du mode de vie sur les caractéristiques pondérales et structurales du squelette chez les amphibiens anoures. Can J Zool. 1995;73:234–242. [Google Scholar]
- Castanet J, Cubo J, de Margerie E. Signification de l’histodiversité osseuse: le message de l’os. Biosystema. 2001;19:133–147. [Google Scholar]
- Che J, Pang J, Zhao H, Wu G-F, Zhao E-M, Zhang Y-P. Phylogeny of Raninae (Anura: Ranidae) inferred from mitochondrial and nuclear sequences. Mol Phyl Evol. 2007;43:1–13. doi: 10.1016/j.ympev.2006.11.032. [DOI] [PubMed] [Google Scholar]
- Cubo J, Ponton F, Laurin M, de Margerie E, Castanet J. Phylogenetic signal in bone microstructure of sauropsids. Syst Biol. 2005;54:562–574. doi: 10.1080/10635150591003461. [DOI] [PubMed] [Google Scholar]
- Curran-Everett D. Multiple comparisons: philosophies and illustrations. Am J Physiol Regul Integrat Comp Physiol. 2000;279:1–8. doi: 10.1152/ajpregu.2000.279.1.R1. [DOI] [PubMed] [Google Scholar]
- de Buffrénil V, Buffetaut E. Skeletal growth lines in an Eocene crocodilian skull from Wyoming as an indicator of ontogenic age and paleoclimatic conditions. J Vertebr Paleontol. 1981;1:57–66. [Google Scholar]
- de Buffrénil V, Rage J-C. La ‘pachyostose’ vertébrale de Simoliophis(Reptilia, Squamata): données comparatives et considérations fonctionnelles. Ann Paléont. 1993;79:315–335. [Google Scholar]
- de Ricqlès A. Recherches paléohistologiques sur les os longs des tétrapodes VII. Sur la classification, la signification fonctionnelle et l’histoire des tissus osseux des tétrapodes. Deuxième partie, suite. Ann Paléont. 1977;63:33–56. [Google Scholar]
- de Ricqlès A. Les tissus squelettiques. In: Delsol M, editor. Amphibiens. Paris: Masson; 1995. pp. 49–96. [Google Scholar]
- de Ricqlès A, de Buffrénil V. Bone histology, heterochronies and the return of tetrapods to life in water: w[h]ere are we? In: Mazin JM, de Buffrénil V, editors. Secondary Adaptation of Tetrapods to Life in Water. München: Dr Friedrich Pfeil; 2001. pp. 289–310. [Google Scholar]
- de Ricqlès A, Castanet J, Francillon-Vieillot H. The ‘message’ of bone tissue in paleoherpetology. Ital J Zool. 2004;71:3–12. [Google Scholar]
- Desdevises Y, Legendre P, Azouzi L, Morand S. Quantifying phylogenetically structured environmental variation. Evolution. 2003;57:2467–2652. doi: 10.1111/j.0014-3820.2003.tb01508.x. [DOI] [PubMed] [Google Scholar]
- Díaz-Uriarte R, Garland TJr. Effects of branch length errors on the performance of phylogenetically independent contrasts? Syst Biol. 1998;47:654–672. doi: 10.1080/106351598260653. [DOI] [PubMed] [Google Scholar]
- Diniz-Filho JAF, de Sant’Ana CER, Bini LM. An eigenvector method for estimating phylogenetic inertia. Evolution. 1998;52:1247–1262. doi: 10.1111/j.1558-5646.1998.tb02006.x. [DOI] [PubMed] [Google Scholar]
- Duellman WE, Trueb L. Biology of Amphibians. New York: McGraw-Hill; 1986. [Google Scholar]
- Estes R. Gymnophiona, Caudata. In: Wellnhofer P, editor. Encyclopedia of Paleoherpetology. Munich: Dr Friedrich Pfeil; 1981. pp. 1–115. [Google Scholar]
- Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. A mitochondrial DNA phylogeny of African clawed frogs: phylogeography and implications for polyploid evolution. Mol Phyl Evol. 2003;33:197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Faure E, Lony E, Lovigny R, Menegoz A, Ting Y, Laurin M. StratAdds module for Mesquite. Paris. http://mesquiteproject.org/packages/stratigraphicTools/
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- Fish FE, Stein BR. Functional correlates of differences in bone density among terrestrial and aquatic genera in the family Mustelidae (Mammalia) Zoomorphology. 1991;110:339–345. [Google Scholar]
- Foote JS. A contribution to the comparative histology of the femur. Smithsonian Contr Knowl. 1916;35:1–242. [Google Scholar]
- Frost DR, Grant T, Faivovich J, et al. The amphibian tree of life. Bull Am Mus Nat Hist. 2006;297:1–370. [Google Scholar]
- Garland TJr, Harvey PH, Ives AR. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst Biol. 1992;41:18–32. [Google Scholar]
- Germain D, Laurin M. Microanatomy of the radius and lifestyle in amniotes (Vertebrata, Tetrapoda) Zool Scr. 2005;34:335–350. [Google Scholar]
- Girondot M, Laurin M. Bone Profiler: a tool to quantify, model and statistically compare bone section compactness profiles. J Vert Paleontol. 2003;23:458–461. [Google Scholar]
- Goin CJ, Goin OB, Zug GR. Introduction to Herpetology. New York: W. H. Freeman; 1978. [Google Scholar]
- Grafen A. The phylogenetic regression. Phil Trans R Soc B. 1989;326:119–157. doi: 10.1098/rstb.1989.0106. [DOI] [PubMed] [Google Scholar]
- Graur D, Martin W. Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends Genet. 2004;20:80–86. doi: 10.1016/j.tig.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Hugall AF, Foster R, Lee MSY. Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Syst Biol. 2007;56:543–563. doi: 10.1080/10635150701477825. [DOI] [PubMed] [Google Scholar]
- Josse S, Moreau T, Laurin M. Stratigraphic tools for Mesquite. http://mesquiteproject.org/packages/stratigraphicTools/
- Kriloff A, Germain D, Canoville A, Vincent P, Sache M, Laurin M. Evolution of bone microanatomy of the tetrapod tibia and its use in palaeobiological inference. J Evol Biol. 2008;21:807–826. doi: 10.1111/j.1420-9101.2008.01512.x. [DOI] [PubMed] [Google Scholar]
- Laurin M. The evolution of body size, Cope's rule, and the origin of amniotes. Syst Biol. 2004;53:594–622. doi: 10.1080/10635150490445706. [DOI] [PubMed] [Google Scholar]
- Laurin M. The splendid isolation of biological nomenclature. Zool Scr. 2008;37:223–233. [Google Scholar]
- Laurin M, Girondot M, Loth M-M. The evolution of long bone microanatomy and lifestyle in lissamphibians. Paleobiology. 2004;30:589–613. [Google Scholar]
- Leclair RJr, Lamontagne C, Aubin A. Allométrie de la masse du squelette chez des amphibiens anoures. Can J Zool. 1993;71:352–357. [Google Scholar]
- Maddison WP. Squared-change parsimony reconstructions of ancestral states for continuous-valued characters on a phylogenetic tree. Syst Zool. 1991;40:304–314. [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: A Modular System for Evolutionary Analysis. Version 2.5. http://mesquiteproject.org.
- Marjanović D, Laurin M. Fossils, molecules, divergence times, and the origin of lissamphibians. Syst Biol. 2007;56:369–388. doi: 10.1080/10635150701397635. [DOI] [PubMed] [Google Scholar]
- Marjanović D, Laurin M. Assessing confidence intervals for stratigraphic ranges of higher taxa: the case of Lissamphibia. Acta Palaeont Pol. 2008;53:413–432. [Google Scholar]
- Martins EP, Diniz-Filho JAF, Housworth EA. Adaptative constraints and the phylogenetic comparative method: a computer simulation test. Evolution. 2002;56:1–13. [PubMed] [Google Scholar]
- Midford P, Garland TJ, Maddison WP. PDAP Package for Mesquite. http://mesquiteproject.org/pdap_mesquite/index.html.
- Pough FH, Andrews RM, Cadle JE, Crump ML, Savitzky AH, Wells K. Herpetology. Upper Saddle River, NJ: Prentice Hall; 2004. [Google Scholar]
- Pouydebat E, Laurin M, Gorce P, Bels V. Evolution of grasping among anthropoids. J Evol Biol. 2008;21:1732–1743. doi: 10.1111/j.1420-9101.2008.01582.x. [DOI] [PubMed] [Google Scholar]
- Purvis A, Rambaut A. Comparative analysis by independent contrasts (CAIC): an Apple Macintosh application for analysing comparative data. Comput Appl Biosci. 1995;11:247–251. doi: 10.1093/bioinformatics/11.3.247. [DOI] [PubMed] [Google Scholar]
- Rissler LJ, Taylor DR. The phylogenetics of desmognathine salamander populations across the southern Appalachians. Mol Phyl Evol. 2003;27:197–211. doi: 10.1016/s1055-7903(02)00405-0. [DOI] [PubMed] [Google Scholar]
- Roelants K, Gower DJ, Wilkinson M, et al. Global patterns of diversification in the history of modern amphibians. Proc Natl Acad Sci USA. 2007;104:887–892. doi: 10.1073/pnas.0608378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. r8 s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- San Mauro D, Vences M, Alcobendas M, Zardoya R, Meyer A. Initial diversification of living amphibians predated the breakup of Pangaea. Am Nat. 2005;165:590–599. doi: 10.1086/429523. [DOI] [PubMed] [Google Scholar]
- Scheyer TM, Sander PM. Shell bone histology indicates terrestrial palaeoecology of basal turtles. Proc R Soc Lond B. 2007;274:1885–1893. doi: 10.1098/rspb.2007.0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions SK, Stöck M, Vieites DR, Quarles R, Min M-S, Wake DB. Cytogenetic analysis of the Asian plethodontid salamander, Karsenia koreana: evidence for karyotypic conservation, chromosome repatterning, and genome size evolution. Chromosome Res. 2008;16:563–574. doi: 10.1007/s10577-008-1197-7. [DOI] [PubMed] [Google Scholar]
- Shaffer HB, McKnight ML. The polytypic species revisited: genetic differentiation and molecular phylogenetics of the tiger salamander Ambystoma tigrinum(Amphibia: Caudata) complex. Evolution. 1996;50:417–433. doi: 10.1111/j.1558-5646.1996.tb04503.x. [DOI] [PubMed] [Google Scholar]
- StatSoft France. STATISTICA (logiciel d’analyse de données) version 6 http://www.statsoft.com.
- Steinfartz S, Veith M, Tautz D. Mitochondrial sequence analysis of Salamandrataxa suggests old splits of major lineages and postglacial recolonizations of Central Europe from distinct source populations of Salamandra salamandra. Mol Ecol. 2000;9:397–410. doi: 10.1046/j.1365-294x.2000.00870.x. [DOI] [PubMed] [Google Scholar]
- Steinfartz S, Vicario S, Arntzen JW, Caccone A. A Bayesian approach on molecules and behavior: reconsidering phylogenetic and evolutionary patterns of the Salamandridae with emphasis on Triturusnewts. J Exp Zool B (Mol Dev Evol) 2007;308:139–162. doi: 10.1002/jez.b.21119. [DOI] [PubMed] [Google Scholar]
- Thorn R, Raffaëlli J. Les Salamandres de l’ancien monde. Paris: Société Nouvelles des Editions Boubée; 2001. [Google Scholar]
- Wall WP. The correlation between high limb-bone density and aquatic habits in Recent mammals. J Paleont. 1983;57:197–207. [Google Scholar]
- Weisrock DW, Papenfuss TJ, Macey JR, et al. A molecular assessment of phylogenetic relationships and lineage accumulation rates within the family Salamandridae (Amphibia, Caudata) Mol Phyl Evol. 2006;41:368–383. doi: 10.1016/j.ympev.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Bonett RM, Chippindale PT. Ontogeny discombobulates phylogeny: paedomorphosis and higher-level salamander relationships. Syst Biol. 2005;54:91–110. doi: 10.1080/10635150590906037. [DOI] [PubMed] [Google Scholar]
- Zhang P, Zhou H, Chen Y-Q, Liu Y-F, Qu L-H. Mitogenomic perspectives on the origin and phylogeny of living amphibians. Syst Biol. 2005;54:391–400. doi: 10.1080/10635150590945278. [DOI] [PubMed] [Google Scholar]
- Zhang P, Papenfuss TJ, Wake MH, Qu L, Wake DB. Phylogeny and biogeography of the family Salamandridae (Amphibia: Caudata) inferred from complete mitochondrial genomes. Mol Phyl Evol. 2008;49:586–597. doi: 10.1016/j.ympev.2008.08.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.