Abstract
The role of the flexible loop 1 in protein conformational motion and the dissociation of enzymatic product from Ribonuclease A (RNase A) was investigated by creation of a chimeric enzyme in which a six residue loop 1 from the RNase A homolog, eosinophil cationic protein (ECP) replaced the twelve residue loop in RNase A. The chimera (RNase AECP) experiences only local perturbations in NMR backbone chemical shifts compared to WT RNase A. Many of the flexible residues that were previously identified in WT as involved in an important conformational change now experience no NMR-detected millisecond motions in the chimera. Likewise, binding of the product analog, 3′-CMP to RNase AECP results in only minor chemical shift changes in the enzyme similar to what is observed for the H48A mutant of RNase A and in contrast to WT enzyme. For both RNase AECP and H48A there is a 10-fold decrease in the product release rate constant, koff compared to WT and in agreement with previous studies indicating the importance of flexibility in RNase A in the overall rate-limiting product release step. Together these NMR and biochemical experiments provide additional insight into the mechanism of millisecond motions in the RNase A catalytic cycle.
Keywords: Ribonuclease, NMR relaxation dispersion, protein dynamics, flexibility, loops
Protein motions are an essential aspect of enzyme function. These motions can enable proper positioning of catalytic residues, sequester reactive intermediates, allow access of substrate to the enzyme’s active site, and facilitate the release of product(s) (1–10). Ribonuclease A (RNase A) is one example in which the backbone and side chain positions change conformation as the enzyme interacts with substrate, product and transition-state analogs (3, 11–28), indicating the close relationship between the chemical nature of the bound ligand and the structure of the protein. Solution NMR relaxation experiments have characterized these motions in RNase A, localizing them to a specific region of the enzyme including the active site and a distant loop (loop 1) as well as some surrounding residues (3, 13, 21, 22, 25, 27–30). There is increasing evidence for the importance of this motion in RNase A function including the similarity of the timescales for protein motion and the catalytic turnover rate (kcat) (13, 29), a similar pH dependence of RNA transphosphorylation and protein motion (13), nearly identical kinetic solvent isotope effects (KSIE) on kcat and on motion (27, 28), and the observation that site-directed mutations that decrease kcat also cause a reduction or abolition of protein motion (3, 28).
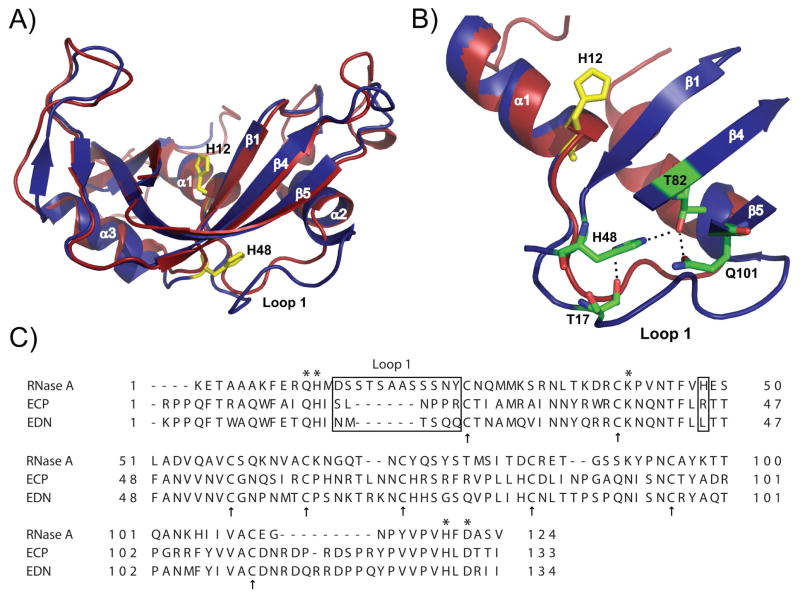
Many of these motions in RNase A are localized in a distinct area, approximately 20 Å from the active-site histidine residues (His12 and His119), that encompasses loop 1 (Figure 1) and several surrounding residues. In RNase A, this loop extends from residues 14 to 25 and contains the sequence, DSSTSAASSSNY. An important interaction identified in previous work involves His48, which makes a hydrogen bond via its Nε2 atom with the Oγ of T17 in loop 1. Mutation of His48 to alanine results in a loss of motion in and surrounding loop 1, a 10-fold decrease in catalytic activity, and a loss of the KSIE on the transphosphorylation of RNA (28). These experiments suggested that the motion of loop 1 was somehow coupled via His48 to the active site and that disruption of these interactions leads to less than optimal enzymatic function. Loop 1 in RNase A assumes different conformations in the various liganded states (29–32) and NMR data indicates that this loop is flexible and moves on the timescale of the rate-limiting product release step (13, 29). These experimental data implicate motion of this loop as an essential component of the product release step. Two other interesting observations suggest that the flexibility of this loop is essential for its function in this capacity. First, RNase A structural homologs, eosinophil cationic protein (ECP) and eosinophil derived neurotoxin (EDN) that have low ribonuclease activity have a much shorter loop 1 (Figure 1), which would restrict its motion and thus its ability to access conformations available to loop 1 in RNase A. Also, unlike RNase A, which shows distinct loop 1 conformations in the apo and bound states, there are insignificant differences in the ligated and unligated conformations of ECP and EDN (33, 34), suggesting that loop 1 in the eosinophil enzymes is immobile or unresponsive to different ligand structures. However, the details of the role of loop 1 motion in RNase A function remain unclear.
Figure 1.
Structural comparison of bovine ribonuclease A (RNase A) and human Eosinophil Cationic Protein (ECP). A) Superposition of RNase A (blue structure, PDB code 1FS3 (35)) and ECP (red structure, PDB code 1DYT (33)). H12 highlights the position of the active site relative to the position of His48 and loop 1 in RNase A. B) Zoomed view of a few atomic interactions between His48 and loop 1 in RNase A. Note the longer loop 1 in RNase A (D14SSTSAASSSNY25) relative to the shorter loop 1 of ECP (S17LNPPR22, ECP numbering). C) Primary sequence alignment of RNase A, ECP and EDN. Active site positions Gln11, His12, Lys41, His119 and Asp121 are marked with stars while cysteine residues forming the strictly conserved four disulfide bridges are represented by arrows. Loop 1 and position 48 are boxed. Alignment was performed with TCoffee Expresso (36) using PDB coordinates 7RSA (32), 1DYT (37) and 1GQV (38).
In this work we investigate the effects of swapping the short ECP loop 1 into the RNase A enzyme by creating a recombinant chimera (RNase AECP). NMR dynamics experiments indicate a similar abolition of motion in this region of the enzyme upon loop swapping. The product release step as measured by NMR line shape analysis indicates a significant decrease in the koff value in the chimeric enzyme compared to WT RNase A, implicating the motion of loop 1 in enabling efficient release of product.
MATERIALS AND METHODS
RNase AECP DNA Construct
Oligonucleotide synthesis was performed by the W. M. Keck Foundation Biotechnology Resource Laboratory (Yale University, New Haven, CT) and by Integrated DNA Technologies (Coralville, IA). The wild-type, 372-bp bovine RNase A gene was codon-optimized for better expression in E. coli by GenScript Co. (Piscataway, NJ), generating construct pUC57-RNaseA(CO). The chimeric RNase AECP gene was PCR-amplified from that construct using the two following terminal primers: NdeI-RNaseA-13ECP26-F (5′-CACACACATATGAAAGAAACCGCGGCGGCCAAATTTGAACGTCAGCACATGAGCCTGAAT CCACCGCGTTGCAATCAGATGATGAAAAGCCGTAATCTG-3′) and RACOHINDR (5′-CACACAAAGCTTTTATTACACGCTCGCATCAAAATGCAC-3′). The 99-bp forward primer was designed so as to replace the 12-residue loop 1 of RNase A (D14SSTSAASSSNY25) by the 6-residue loop 1 of ECP (S17LNPPR22 (ECP numbering)). An additional TAA stop codon was also engineered in the reverse primer. The recombinant RNase AECP gene was digested with NdeI and HindIII, subcloned into NdeI/HindIII-digested pET22b(+) and transformed into E. coli XL10-Gold cells (Stratagene, La Jolla, CA). Vector pET22b(+) was obtained from an NdeI/HindIII digestion of the pBXR construct (39), which was a gift from Professor Ronald T. Raines (University of Wisconsin, Madison, WI). Cloning of the codon-optimized RNase AECP gene was designed so as to remove the original pELB leader sequence present in the pBXR construct. Colonies were individually picked after selection on a Luria-Bertani (LB) medium containing 100 μg/mL ampicillin, and the sequence of the RNase AECP gene was confirmed by DNA sequencing. For protein expression purposes, the resulting pET22b(+)-RNase AECP construct was transformed into E. coli BL21 (DE3) cells (Lucigen Co., Middleton, WI).
Protein expression
Isotopically 15N- or 15N-13C-labeled RNase AECP was expressed and purified according to the following protocol based on previously published methods (39). All chemicals were obtained from Sigma (St. Louis, MO), except for isotopes, which were obtained from Cambridge Isotope Laboratories (Andover, MA). All cell growths were performed in M9 minimal medium supplemented with MEM vitamins (Invitrogen, Carlsbad, CA), trace metals, 2.5 g/L of 15NH4Cl, 4 g/L of 13C-glucose (for 15N-13C-labeled enzymes) or 12C-glucose (for 15N-labeled enzymes) and 100 μg/mL ampicillin. Overnight cultures of BL21(DE3)/pET22b(+)-RNase AECP grown in M9 media were used to inoculate a total of 3 liters of M9 media at 37°C. These were grown until the A600nm = 0.8. Protein expression was then induced by the addition of IPTG to a final concentration of 1 mM, the cultures were grown for an additional 3 hours and harvested by centrifugation (4000 rpm, 20 min, 4°C).
Cell lysis and RNase AECP refolding
The E. coli cell pellet was resuspended in osmotic shock lysis buffer (20 mM Tris, 1 mM EDTA, pH 8.0) and the cells were sonicated on ice for 5 × 40 s intervals using an S-450A ultrasonic cell disruptor from Branson Ultrasonics Co. (Danbury, CT). The sonicated cell pellets were centrifuged for 1 hour at 20,000 g. The pellet from this centrifugation step (which contains the insoluble RNase AECP chimera) was resolubilized in 400 mL of urea solubilization buffer (20 mM Tris, pH 8.0, 1 mM EDTA, 400 mM NaCl, 6 M Urea, 20 mM DTT) and briefly sonicated again to resuspend cell debris. This was allowed to stir overnight at room temperature. Insoluble debris was removed by an additional centrifugation step (4°C, 45 minutes, 20,000 g). The soluble proteins were refolded by performing three sequential 24-h dialysis steps at 4°C against a refolding buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM reduced glutathione, 0.2 mM oxidized glutathione, 0.5 mM PMSF).
Protein concentration and purification
The supernatant from dialysis was collected, centrifuged (4°C, 30 minutes, 20,000 g), filtered through a 0.22 μm membrane filter and concentrated down to ~30–40 mL using an Amicon 402 stirred-cell concentrator and a 3,000 NMWL regenerated cellulose ultrafiltration membrane from Millipore (Billerica, MA). The concentrated supernatant was dialyzed overnight against 20 mM Tris (pH 8.0) and loaded on a 10 mL prepacked anion-exchange HiTrap Q HP column (GE Healthcare, Piscataway, NJ) using an ÄKTA FPLC system pre-equilibrated with the same buffer. The flow-through was collected and further purified with a 1-mL Mono S 5/50 GL cation-exchange column (GE Healthcare) and RNase AECP was eluted by a salt gradient using a 20 mM Tris (pH 8.0) buffer containing 1 M NaCl. The elution peak of RNase AECP was typically observed at a conductivity of 12–14 mS/cm. Fractions containing the pure protein were combined and concentrated to ~350 μl using a 3,000 NMWL Centricon® filter device (Millipore). Finally, the buffer was changed to 5 mM MES-NaOH, 7 mM NaCl, 0.01 % NaN3, 5 % D2O, pH 6.4 by performing multiple dilution/concentration steps with the same Centricon filter device. Protein purity was estimated by SDS-PAGE to be higher than 98 % and protein concentration was calculated by UV absorbance using ε277.5 nm = 9800 M−1cm−1 (40).
Solution NMR Experiments
All NMR experiments were performed at 298 K as calibrated with a standard methanol sample (41) on Varian 11.7 T, 14.1 T and/or 18.8 T NMR instruments equipped with triple-resonance probes and pulsed-field gradients. A 160 μM 15N-labeled RNase AECP sample was used to perform all the relaxation experiments and the 3′-CMP titrations experiments. A 410 μM 15N-13C-labeled RNase AECP sample was used for all the assignment experiments. Backbone and side chain resonances were assigned by using standard multidimensional experiments (41). The kinetics of the 3′-CMP binding interaction with RNase AECP were measured by titration of 3′-CMP as described (3, 29), where 15N-HSQC spectra were recorded for titration points corresponding to 3′-CMP/RNase AECP mole ratios of 0, 0.174, 0.393, 0.691, 1.31, 2.71, 6, and 12. NMR line-shape analysis was performed using the software LineShapeKin (version 3.1) developed by Professor Evgenii L. Kovrigin (Medical College of Wisconsin, Milwaukee, WI, http://lineshapekin.net/) (7). Because it is difficult to obtain true estimates of fully formed protein-ligand complexes when binding constants are not very tight, titrations for RNase AECP and H48A were considered incomplete and titration end points were adjusted manually to optimally fit models to the experimental data. The 3′-CMP titrations were performed as 1H-15N sensitivity-enhanced HSQC experiments (42–45) at 11.7 T using spectral widths (points) of 1600 Hz (256) and 7000 Hz (8192) in the t1 and t2 dimensions respectively.
Relaxation compensated Carr-Purcell-Meiboom-Gill (rcCPMG) experiments (46) were performed at 18.8 T and 14.1 T on 15N-labeled wild-type and mutant RNases. The NMR relaxation data were typically acquired with spectral widths in the t1 and t2 dimensions of 2700 and 10000 Hz with a digital resolution of 0.1 and 0.2 points/Hz respectively. The proton carrier frequency was coincident with the H2O resonance and the 15N frequency was placed in the middle of the amide region at 120 ppm. Spin-relaxation rate constants at each CPMG pulse repetition time (τcp) were acquired in a constant time manner (47) using a total relaxation delay of 40 ms. Transverse Relaxation rates were determined for CPMG delays (τcp) = 0.625, 0.714 (× 2), 1.0, 1.25, 1.67, 2.0, 2.50 (× 2), 3.33, 5.0, and 10 ms. The individual spectra were acquired as an interleaved three-dimensional experiment in which the two-dimensional planes were extracted and peak intensities determined from a 3 × 3 grid using in-house written software. The CPMG dispersion data were fit with the fast-limit equation (48):
| (1) |
in which φex = pApBΔω2, pA/B are the equilibrium site populations for the two (A and B) magnetically distinct conformers, Δω is the chemical shift difference between conformations A and B, kex is the is the exchange rate constant and is the sum of the forward and reverse rate constants, is the‘exchange-free’ transverse relaxation rate and τcp is the time delay between 180° CMPG pulses. Amino acid residues were considered for further analysis if the difference in measured R2 (ΔR2(1/τcp)) values at fast (τcp = 0.625 ms) and slow (τcp = 10 ms) pulsing rates was greater than 1.5 s−1. The parameter α (49) that determines the chemical exchange time regime was estimated from
| (2) |
in which B01(2) refers to the static magnetic field strength, 14.1(18.8) T respectively and Rex1(2) is the exchange contribution due to conformational exchange and is determined from fits of equation (1) to the CPMG dispersion data.
Differential multiple-quantum relaxation experiments were performed as described by Kloiber and Konrat (50) in an identical manner to earlier work on RNase A (28). Peak intensities for a 10 ms relaxation delay (Tc) were measured for two experiments, a reference (Iauto) and a cross-relaxation (Icross) experiment. The intensities of each experiment represent the decay of multiple quantum coherence (2IxSx) and the conversion of 2IxSx to 2IySy respectively. The conformational exchange modulated isotropic chemical shifts differentially affects these terms. The ratio of peak intensities for the relaxation of 2IxSx (Icross) and relaxation of 2IxSx to IySy (Iauto) were used to determine the differential multiple quantum relaxation rate ΔRMQ from:
| (3) |
RESULTS
Chemical shift assignments
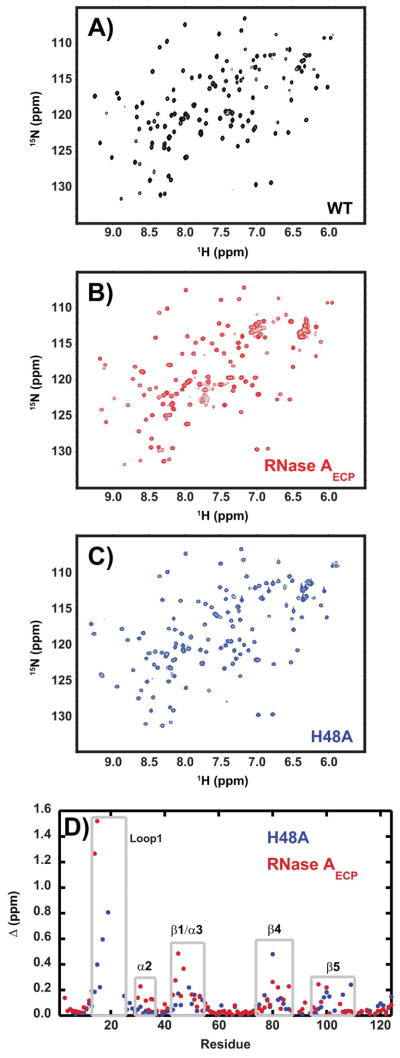
Both H48A and RNase AECP are well-folded enzymes as evidenced by the chemical shift dispersion in Figure 2A–C. Upon mutation to form the H48A enzyme and the RNase AECP chimera, 1H and 15N chemical shifts are perturbed for several similar regions in these proteins (Figure 2). For H48A, the largest changes in chemical shifts are located in loop 1 and in β-sheet 4, with more minor effects observed for the β1/α3 region and β5 residues (Figure 2). For the loop 1 chimera, the regions that are most significantly affected are β1/α3 and α2, with more minor perturbations occurring in β4 and β5 (Figure 1). Not surprisingly, the residues Met13, Asp14, and Ser15, which are just N-terminal to the site of loop swapping display the largest changes in chemical shifts in RNase AECP (Figure 2).
Figure 2.
Chemical shift changes caused by mutation. 1H-15N HSQC spectra for (A) wild-type, (B) RNase AECP, and (C) H48A enzymes. The spectra were acquired at 14.1 T and 298K, pH = 6.4. In (D) chemical shift differences for WT vs. H48A (blue) and WT vs. RNase AECP are shown as a function of amino acid sequence. Gray boxed areas depict protein regions with larger than average chemical shift disturbances. (Δ (ppm) = [(Δδ2HN + Δδ2 N/25)/2]1/2) in which δ is the difference in chemical shift between the two proteins for NH and HN nuclei (52).
3′-CMP Ligand Interaction
To further investigate the kinetics of the rate-limiting product release step of this reaction, we performed a steady-state NMR line shape analysis (3) by following the titration of the product analog 3′-CMP in WT RNase A, RNase AECP and in the previously-characterized point mutant H48A (28). In addition to following the various chemical shift effects observed upon ligand binding, this technique allows for the calculation of the dissociation constant (Kd) and the dissociation rate constant (koff) of 3′-CMP from RNase A.
A series of two-dimensional HSQC experiments were used to follow the 1H-15N chemical shifts observed for each enzyme upon progressive increases in [3′-CMP] (Figure 3 and Figure 4). As shown by the chemical shift variations observed in Figure 3, binding of 3′-CMP to WT alters the chemical environment of a number of residues throughout the enzyme, mainly in regions encompassing residues 10–15, 40–50, 80–90, 95–102, and 119–123. The protein-wide average chemical shift changes per amino acid residue upon binding of 3′-CMP are 0.071, 0.053, and 0.040 ppm for WT, RNase AECP, and H48A respectively. Many of the residues displaying the largest chemical shift changes (Δ > 0.15 ppm) between the apo (mole ratio, 3′-CMP:RNase A = 0) and 3′-CMP bound (mole ratio, 3′-CMP:RNase A = 12) forms for WT are either in direct contact or in close proximity to the ligand (Lys7, Gln11, Lys41, Val43, Asn44, His119, Asp121 and Ala122) (51). A number of residues located farther away from the CMP binding site also display chemical shift variations greater than 0.15 ppm in WT, including Phe46, Asp83, Arg85, Glu86, Thr100 and Gln101, all of which are located in the adjacent β1-sheet that separates the active site cavity from loop 1. In contrast to WT, the chemical shift differences observed between the unbound and the 3′-CMP-bound forms of both RNase AECP and mutant H48A are significantly smaller (Figure 3). For example, the only residues displaying a Δ > 0.15 ppm in these two mutant enzymes are Val43, Asp83, His119, Asp121 for RNase AECP and Val43, Asn44 and Glu86 for H48A, demonstrating that mutation of either loop 1 or His48 alters the mode of ligand interaction with RNase A.
Figure 3.
1H-15N chemical shift differences upon 3′-CMP binding to WT RNase A, RNase AECP, and H48A. (A) 1H-15N chemical shift differences Δ (ppm) mapped on the primary sequence of WT, RNase AECP, and H48A. The value of Δ was calculated between a ligand-free 1H-15N HSQC (mole ratio, 3′-CMP/Enzyme = 0) and a 3′-CMP-bound 1H-15N HSQC (mole ratio, 3′-CMP/Enzyme = 12) according to the following equation: (Δ (ppm) = [(Δδ2 HN + Δδ2 N/25)/2]1/2) (52). (B) Mapping of Δ (ppm) on the three-dimensional structure of RNase A (PDB code 1RPF (51)). The white to red gradient corresponds to Δ values ranging from 0 ppm (white) to > 0.15 ppm (dark red). (C) Same view as in (B) showing residues with Δ > 0.15 ppm as green spheres. 3′-CMP is shown as orange sticks representation.
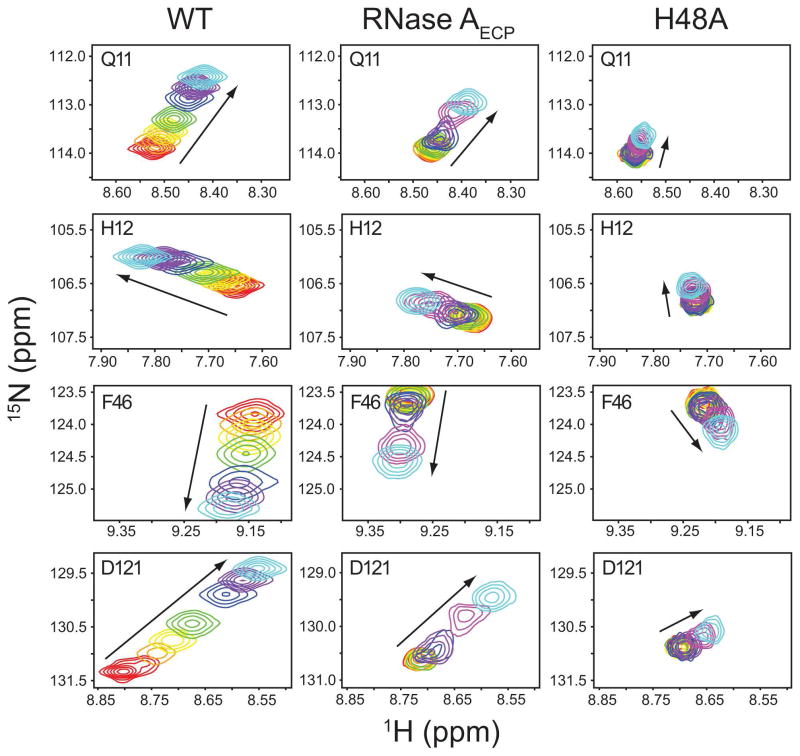
Figure 4.
Kinetics of 3′-CMP binding to WT RNase A, RNase AECP and mutant H48A. Titration of 3′-CMP is followed by monitoring the changes in backbone chemical shifts observed in 1H-15N-HSQC spectra for WT RNase A, RNase AECP and mutant H48A. Titration points are shown for residues Gln11, His12, Phe46 and Asp121 for [3′-CMP]:[RNase A] mole ratios of 0 (red) 0.174 (orange), 0.393 (yellow), 0.691 (green), 1.31 (blue), 2.71 (purple), 6 (magenta), and 12 (cyan). Arrows indicate the direction of resonance shift with increasing [3′-CMP]. The magnitude of 3′-CMP binding-induced chemical shifts is smaller in mutant H48A than in RNase AECP.
The extent of the attenuation of the chemical shift changes upon binding of 3′-CMP is illustrated in Figure 4. The magnitude of the chemical shift changes is greatly diminished in both RNase AECP and H48A, yet the direction of the changes in chemical shift for both is similar to WT RNase A.
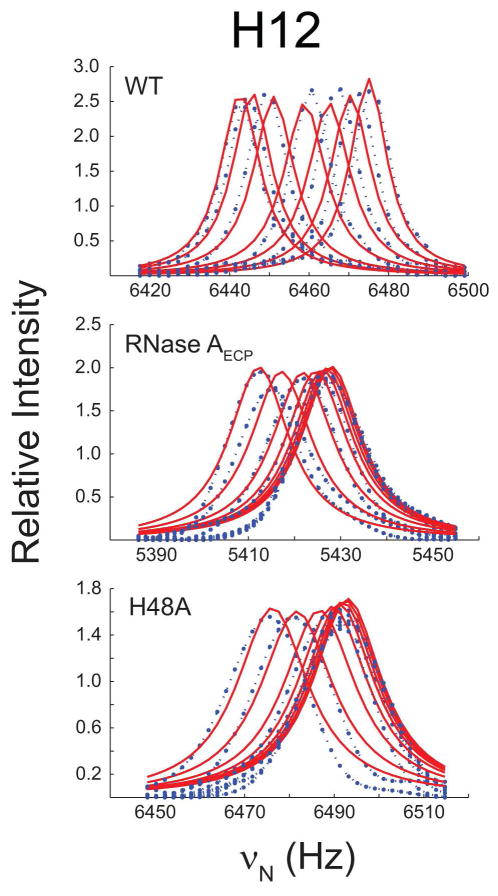
Among all the residues with chemical shifts in the 3′-CMP titration experiment, only a restricted set (Gln11, His12, Arg39, Lys41, Phe46, Ser75, Tyr97 and Ser123) showed significant shifts and were also sufficiently resolved for line shape quantitation in all three enzymes (Figure 5). Whether NMR line shape analysis was independently performed on individual residues or simultaneously fitted using a global, two-state model, there were few variations in the calculated binding constants, indicating that all residues likely report on the same binding event. 3′-CMP binding values are therefore reported for a global fit of residues and are presented in Table 1.
Figure 5.
3′-CMP titration of RNase A. The titration curves of residue His12 are shown for WT, RNase AECP and H48A. Fitted line shapes (red) are superimposed on the experimental NMR data (blue).
Table 1.
3′-CMP binding constants for WT RNase A, RNase AECP and mutant H48A.
| Enzyme | Kd (μM) | koff (s−1) | kon (M−1s−1)a |
|---|---|---|---|
| WT Rnase A | 74 ± 2 | 1155 ± 120 | 1.56 × 107 ± 1.67 × 106 |
| RNase AECP | 300 ± 16 | 96 ± 9 | 3.20 × 105 ± 3.45 × 104 |
| RNase A (H48A) | 895 ± 50 | 142 ± 19 | 1.59 × 105 ± 2.30 × 104 |
kon determined from koff/Kd.
The 3′-CMP dissociation constant (Kd) for WT RNase A determined from these NMR titrations is 74 ± 2 μM. This value is indicative of tight binding between 3′-CMP and WT RNase A and is in good agreement with dissociation constants previously determined for this ligand (11, 18, 53). In contrast, the resulting 3′-CMP dissociation constants (Kd) for RNase AECP and mutant H48A are respectively 300 ± 16μM and 895 ± 50 μM. Replacement of the distal 12-residue loop 1 of RNase A by the shorter 6-residue loop 1 of ECP in the RNase AECP chimera reduces binding affinity for 3′-CMP ~4-fold relative to WT. Interestingly, despite the fact that it is more than 18 Å away from the catalytic center and that it is in direct contact with loop 1 (Figure 1), mutation at position His48 reduces the binding affinity to a greater extent (~12-fold relative to WT) than does swapping the entire loop 1 sequence.
The product release (koff) is the rate-limiting step of the reaction in WT RNase A (29, 54–56). In previous work it was demonstrated that koff and the conformational exchange rate constant kex were equivalent (28) and in combination with other experimental evidence suggested that motion in RNase A, particularly loop 1, was part of the rate-limiting step (3, 13, 27–29). As shown in Table 1, the NMR line shape analysis of the 3′-CMP titration series yields a WT koff value of ~1200 s−1, a result in good agreement with previous calculations (3). Replacement of loop 1 or mutation of H48 in RNase A has a significant effect on the rate of product release, as koff values for 3′-CMP are decreased ~10-fold in both RNase AECP (koff = 96 ± 9 s−1) and mutant H48A (koff = 142 ± 19 s−1) relative to WT (Table 1).
15N Backbone Dynamics
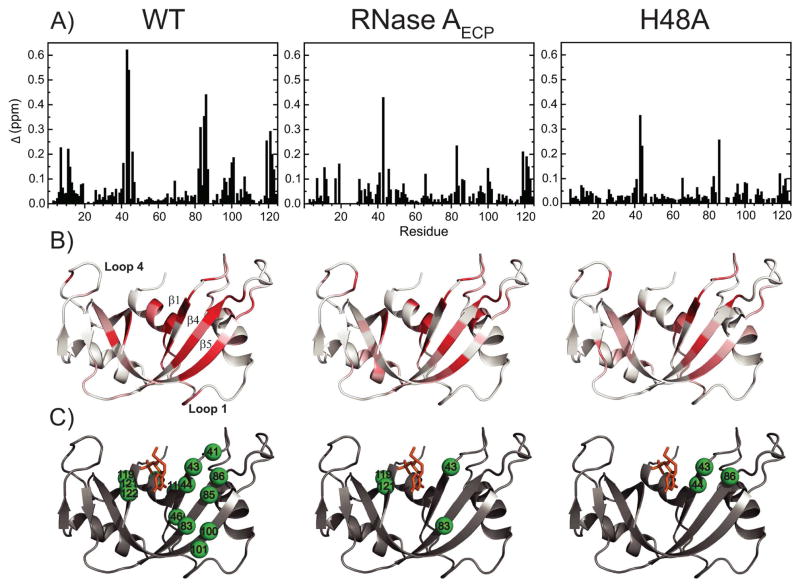
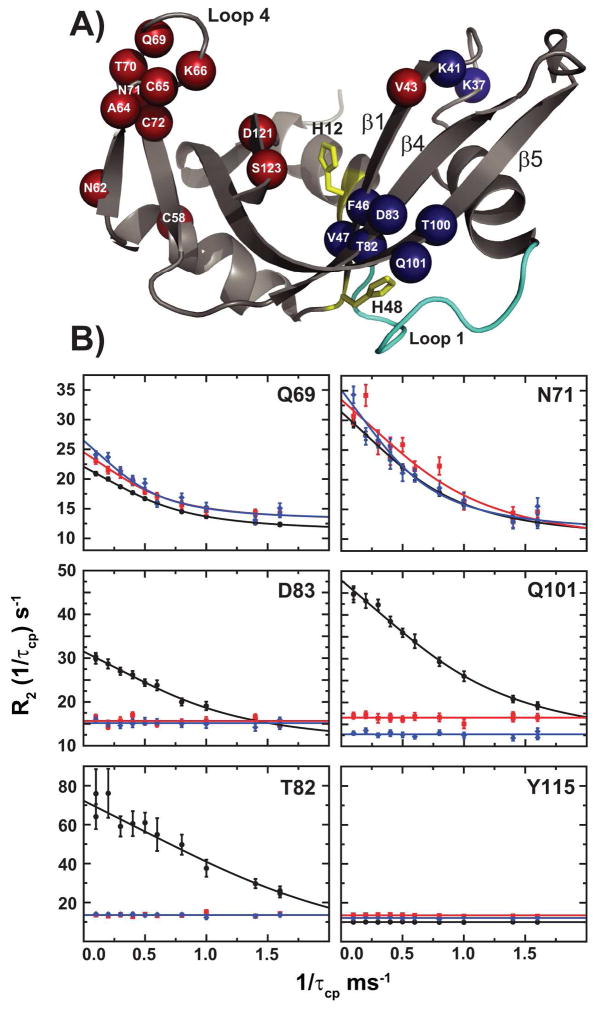
A relaxation-compensated CPMG dispersion experiment was performed on apo 15N-labeled RNase AECP for comparison with WT and H48A to investigate the effects of the loop 1 swap on the important millisecond motions of the backbone amide groups. As evidenced by the characteristic difference in R2 values measured at increasing values of the CPMG 180° pulse delays (τcp), a number of residues display conformational exchange throughout the structure of RNase AECP (Figure 6). Among the well-resolved 1H-15N resonances that show good signal-to-noise values, twelve residues experience a conformational exchange process with a ΔR2 (1/τcp) value > 1.5 s−1 (Val43, Cys58, Asn62, Ala64, Cys65, Lys66, Gln69, Thr70, Asn71, Cys72, Asp121 and Ser123). A few of those residues are amenable to quantitative analysis (Ala64, Cys65, Gln69 and Asn71) and are shown in Figure 6B. These dynamic positions are mainly clustered in a surface loop adjacent to the active site of RNase A (loop 4), which contains the highly conserved P0 and B2 subsite residues K66 and N71, respectively (57). Interestingly, those residues also display similar dispersion profiles both in WT RNase A and in the previously characterized mutant H48A (Figure 6B) (28).
Figure 6.
Conformational dynamics in RNase A. (A) Residues in RNase AECP undergoing conformational exchange with a ΔR2 (1/τcp) greater than 1.5 s−1 are plotted on the structure of RNase A (PDB code 7RSA (32)). Red spheres = similar rate constants (kex) in WT and RNase AECP; Blue spheres = Decreased kex values in RNase AECP relative to WT. H12 and H48 are shown as yellow sticks and loop 1 is shown in cyan. B) CPMG dispersion curves for WT RNase A (black circles), RNase AECP (red squares) and mutant H48A (blue diamonds). 15N relaxation dispersion curves are shown for two residues displaying similar conformational exchange in the three enzymes (Gln69 and Asn71), and three residues displaying significantly affected kex in both RNase AECP and mutant H48A (Thr82, Asp83 and Gln101). Y115 is represented as a motionless control. Fitted lines to data points are from single-field fits at 14.1 T using equation (1).
Comparison with WT dynamics also shows a number of residues displaying loss of CPMG relaxation dispersion, indicative of a loss of μs-ms protein motion for RNase AECP. Among the many residues experiencing chemical exchange in WT, several (Lys41, Phe46, Val47, Ser80, Thr82, Asp83, Thr100 and Gln101) show no NMR evidence of any conformational exchange motion in RNase AECP (Figure 6B). Surprisingly, every residue that displays a flat dispersion profile in RNase AECP also exhibits the same flat profile in the H48A mutant (Figure 6B), indicating that both alterations to RNase A result in the loss of a similar conformational exchange process. The residues common to both RNase AECP and WT that experience measurable CPMG dispersion curves (Cys65, Gln69, Asn71, and Asp121; Figure 6B) also exist in similar exchange time regimes as estimated from the α parameter (49), with the exception of Asn71 (Supporting Information). Residues 65, 69, and 121 all have α values greater than 1.2 indicating conformational exchange that is fast on the chemical shift time scale. For Asn71 in RNase AECP α = 0.7 ± 0.1 and is 1.6 ± 0.1 WT RNase A. The lower value of α for Asn71 is most likely due to underestimation of Rex at 18.8 T (Supporting Information).
The flat 15N dispersion curves for both H48A and RNase AECP indicate that in these two proteins the amide nitrogen atoms are not moving, do not experience significant Δω values, and/or exists in a highly skewed equilibrium (> 0.995:0.005). To address this further, differential multiple quantum relaxation rates (ΔRMQ) (50) were measured (Supporting Information). The ΔRMQ values depend on chemical shift differences experienced by both 15N and 1H and can therefore provide an additional assessment of the presence of millisecond motions. For all assignable residues, the ΔRMQ rates mirror those of the 15N single quantum relaxation dispersion measurents. Residues that have measurable 15N-single quantum dispersion curves also have non-zero differential multiple quantum relaxation rates. Therefore from the perspectives of the 1H and the 15N nuclei Lys37, Lys41, Phe46 Val47, Thr82, Asp83, Thr100 and Gln101 are no longer flexible on the μs-ms timescale in H48A and RNase AECP (Supporting Information).
DISCUSSION
Loops are typically tethered to the main body of the protein via rigid secondary structure elements and therefore can be envisioned as structural units moving on protein hinges. This simplified view of loop function often explains a great deal regarding their biochemical role (8, 58, 59). This inherent flexibility of loop regions has been harnessed to provide proteins and enzymes with a mechanism to alter ligand-binding specificity (60–67). In an extreme example, a loop distant from the protease active site modulates the differential specificity between trypsin and chymotrypsin (63). Loop regions in RNase A have also been the subject of loop-swapping experiments. Benner and coworkers exchanged three loops from the RNase homolog Angiogenin into bovine pancreatic RNase A (60). Loop residues 63–74 contact the purine portion of the substrate. When these residues from Angiogenin were placed into RNase A, the resulting enzyme had purine substrate specificity resembling that of Angiogenin, not that of RNase A. Unlike loop 1 in ECP, which is six residues long, loop 1 in Angiogenin is nine residues long. Exchange of the longer Angiogenin loop 1 into RNase A had a modest 2-fold effect on catalytic activity (60). Here we show that the flexibility offered by the longer loop 1 in RNase A is closely associated with the product release step despite not directly interacting with substrate or product.
We observe an important similarity between the millisecond dynamical behavior of RNase AECP and the previsouly characterized mutant H48A (28). Both mutants display flexible residues involved in conformational exchange at similar atomic positions throughout the enzyme (Figure 6 and Figure 2 of ref. (68)), and both enzymes display two independent clusters of flexible residues that are affected in a similar manner when His48 or loop 1 are mutated. The first cluster is close to the active site of RNase A and is centered around the flexible loop 4 (Figure 6, red spheres). This cluster is insensitive to solvent isotope effects in H48A and displays WT-like motional behavior in both mutants. The second cluster of residues, centered about the adjacent β1-sheet (formed by β1, β4 and β5 strands) (Figure 6A, blue spheres), experiences an important kinetic solvent isotope effect in WT RNase A, which is absent in H48A. In addition, this second group of residues display significantly reduced dynamics in both RNase AECP and in H48A noted by the lack of CPMG dispersion. The fact that His48 is hydrogen bonded to loop 1 through T17 (Figure 1B) and that both mutants show a similar motional behavior upon replacement of either one of these structural elements suggests that both loop 1 and His48 are dynamically linked by a network of coupled motions on the catalytically relevant millisecond time scale. It is reasonable to assume that these concerted motions are at least partly propagated through Thr17 (Figure 1B), which was shown to display conformational exchange in WT but not in H48A (68).
The fact that mutation of His48 or swapping of loop 1 affects the dynamics of many residues of the β1-sheet in a similar fashion relative to WT (e.g. Thr82, Asp83, Thr100, Gln101) (Figure 6) also demonstrates that the β1-sheet, His48 and loop 1 are all part of a global concerted dynamical event that propagates between the β1, β4 and β5 strands through His48 to loop 1 on the time scale of catalytic turnover. Mutating either His48 or loop 1 results in a similar loss of motion in the β1-sheet, further demonstrating the importance of His48 in acting as a relay of the dynamical changes from loop 1 to the the β1-sheet, and/or vice versa. Threonine 17 and Thr82 form hydrogen bonds to His48 and may be an important mechanism of connectivity of loop 1 dynamics to the active site of RNase A. Interestingly, we also observe a significant correlation between residues of the second cluster (Figure 6) and many residues showing altered dynamics upon 3′-CMP binding.
Whether the millisecond dynamics of RNase AECP and H48A are retained with respect to WT (as with residues Gln69 and Asn71) or significantly reduced (as with residues Thr82, Asp83 and Gln101), these two mutant enzymes are affected in the same way at the same amino acid positions. This result further supports the existence of a network of coupled motions in RNase A that would propagate dynamics from loop 1 to His48 and other residues of the adjacent β1-sheet, extending all the way to the ligand in the active site of the enzyme. Additional experiments are currently underway to further address this hypothesis.
Supplementary Material
Acknowledgments
ND acknowledges a postdoctoral fellowship from the Fonds Québécois de la Recherche sur la Nature et les Technologies (FQRNT) from the Government of Québec. EDW acknowledges NIH training grant 5T32GM0082383. JPL acknowledges NSF awards MCB-0236966 and MCB-0744161.
The authors thank Professor Evgenii L. Kovrigin (Medical College of Wisconsin, Milwaukee, WI) for helpful discussions. This manuscript is dedicated to Professor Fred Richards.
ABBREVIATIONS
- RNase A (or WT)
wild-type ribonuclease A
- ECP
eosinophil cationic protein
- EDN
eosinophil-derived neurotoxin
- RNase AECP (or chimera)
RNase A with loop 1 from ECP
- H48A
RNase A (H48A)
- 3′-CMP
cytidine-3′-monophosphate
- rcCPMG
relaxation compensated Carr-Purcell-Meiboom-Gill
Footnotes
SUPPORTING INFORMATION AVAILABLE
Experimental results for the differential multiple-quantum relaxation experiments (Figure S1, Table S1) and field dependent Rex values for WT and RNase AECP (Table S2). This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Daniel RM, Dunn RV, Finney JL, Smith JC. The role of dynamics in enzyme activity. Annu Rev Biophys Biomol Struct. 2003;32:69–92. doi: 10.1146/annurev.biophys.32.110601.142445. [DOI] [PubMed] [Google Scholar]
- 2.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 3.Kovrigin EL, Loria JP. Enzyme dynamics along the reaction coordinate: critical role of a conserved residue. Biochemistry. 2006;45:2636–2647. doi: 10.1021/bi0525066. [DOI] [PubMed] [Google Scholar]
- 4.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 5.Blacklow SC, Raines RT, Lim WA, Zamore PD, Knowles JR. Triosephosphate isomerase catalysis is diffusion controlled. Biochemistry. 1988;27:1158–1167. doi: 10.1021/bi00404a013. [DOI] [PubMed] [Google Scholar]
- 6.Guallar V, Jacobson M, McDermott A, Friesner RA. Computational modeling of the catalytic reaction in triosephosphate isomerase. J Mol Biol. 2004;337:227–239. doi: 10.1016/j.jmb.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 7.Jogl G, Rozovsky S, McDermott AE, Tong L. Optimal alignment for enzymatic proton transfer: structure of the Michaelis complex of triosephosphate isomerase at 1.2-Å resolution. Proc Natl Acad Sci USA. 2003;100:50–55. doi: 10.1073/pnas.0233793100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joseph D, Petsko GA, Karplus M. Anatomy of a conformational change: hinged “lid” motion of the triosephosphate isomerase loop. Science. 1990;249:1425–1428. doi: 10.1126/science.2402636. [DOI] [PubMed] [Google Scholar]
- 9.Rozovsky S, Jogl G, Tong L, McDermott AE. Solution-state NMR investigations of triosephosphate isomerase active site loop motion: ligand release in relation to active site loop dynamics. J Mol Biol. 2001;310:271–280. doi: 10.1006/jmbi.2001.4673. [DOI] [PubMed] [Google Scholar]
- 10.Sampson NS, Knowles JR. Segmental movement: definition of the structural requirements for loop closure in catalysis by triosephosphate isomerase. Biochemistry. 1992;21:8482–8487. doi: 10.1021/bi00151a014. [DOI] [PubMed] [Google Scholar]
- 11.Cathou RE, Hammes GG. Relaxation spectra of ribonuclease. I The interaction of ribonuclease with cytidine 3′-phosphate. J Am Chem Soc. 1965;86:3240–3245. doi: 10.1021/ja00949a003. [DOI] [PubMed] [Google Scholar]
- 12.Cathou RE, Hammes GG. Relaxation spectra of ribonuclease. III Further investigation of the interaction of ribonuclease and cytidine 3′-phosphate. J Am Chem Soc. 1965;87:4674–4680. doi: 10.1021/ja00949a003. [DOI] [PubMed] [Google Scholar]
- 13.Cole R, Loria JP. Evidence for flexibility in the function of ribonuclease A. Biochemistry. 2002;41:6072–6081. doi: 10.1021/bi025655m. [DOI] [PubMed] [Google Scholar]
- 14.de Mel VS, Doscher MS, Martin PD, Edwards BF. The occupancy of two distinct conformations by active-site histidine-119 in crystals of ribonuclease is modulated by pH. FEBS Lett. 1994;349:155–160. doi: 10.1016/0014-5793(94)00664-4. [DOI] [PubMed] [Google Scholar]
- 15.Deng H, Brugner JW, Callender R. Structure of the ribonuclease-uridine-vanadate transition state analogue complex by raman difference spectroscopy: mechanistic implications. J Am Chem Soc. 1998;120:4717–4722. [Google Scholar]
- 16.Erman JE, Hammes GG. Relaxation spectra of ribonuclease. IV The interaction of ribonuclease with cytidine 2′:3′-cyclic phosphate. J Am Chem Soc. 1966;88:5607–5614. doi: 10.1021/ja00975a046. [DOI] [PubMed] [Google Scholar]
- 17.Erman JE, Hammes GG. Relaxation spectra of ribonuclease. V The interaction of ribonuclease with cytidylyl-3′:5′-cytidine. J Am Chem Soc. 1966;88:5614–5617. doi: 10.1021/ja00975a047. [DOI] [PubMed] [Google Scholar]
- 18.French TC, Hammes GG. Relaxation spectra of ribonuclease. II Isomerization of ribonuclease at neutral pH values. J Am Chem Soc. 1965;87:4669–4673. doi: 10.1021/ja00949a002. [DOI] [PubMed] [Google Scholar]
- 19.Glushko V, Lawson PJ, Gurd FRN. Conformational states of bovine pancreatic ribonuclease A observed by normal and partially relaxed carbon 13 nuclear magnetic resonance. J Biol Chem. 1972;247:3176–3185. [PubMed] [Google Scholar]
- 20.Hammes GG, Walz FG. Relaxation spectra of ribonuclease. VI Interaction of ribonuclease with uridine 3′-monophosphate. J Am Chem Soc. 1969;91:7179–7186. [Google Scholar]
- 21.Howarth OW, Lian LY. Ribonuclease A: carbon-13 nuclear magnetic resonance assignments, binding sites, and conformational flexibility. Biochemistry. 1984;23:3515–3521. doi: 10.1021/bi00310a020. [DOI] [PubMed] [Google Scholar]
- 22.Hughes LT, Cohen JS, Szabo A, Niu C, Matsuura S. 13C NMR studies of the molecular dynamics of selectively 13C-enriched ribonuclease complexes. Biochemistry. 1984;23:4390–4394. doi: 10.1021/bi00314a023. [DOI] [PubMed] [Google Scholar]
- 23.Leonidas DD, Chavali GB, Oikonomakos NG, Chrysina ED, Kosmopoulou MN, Vlassi M, Frankling C, Acharya KR. High-resolution crystal structures of ribonuclease A complexed with adenylic and uridylic nucleotide inhibitors. Implications for structure-based design of ribonucleolytic inhibitors. Protein Sci. 2003;12:2559–2574. doi: 10.1110/ps.03196603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonidas DD, Shapiro R, Irons LI, Russo N, Acharya KR. Crystal structures of ribonuclease A complexes with 5′-diphosphoadenosine 3′-phosphate and 5′-diphosphoadenosine 2′-phosphate at 1.7 Å resolution. Biochemistry. 1997;36:5578–5588. doi: 10.1021/bi9700330. [DOI] [PubMed] [Google Scholar]
- 25.Markley JL. Correlation proton magnetic resonance studies at 250 MHz of bovine pancreatic ribonuclease. II pH and inhibitor-induced conformational transitions affecting histidine-48 and one tyrosine residue of ribonuclease A. Biochemistry. 1975;14:3554–3561. [PubMed] [Google Scholar]
- 26.Tilton RF, Dewan JC, Petsko GA. Effects of temperature on protein structure and dynamics: x-ray crystallographic studies of the protein ribonuclease-A at nine different temperatures from 98 to 320 K. Biochemistry. 1992;31:2469–2481. doi: 10.1021/bi00124a006. [DOI] [PubMed] [Google Scholar]
- 27.Kovrigin EL, Loria JP. Characterization of the transition state of functional enzyme dynamics. J Am Chem Soc. 2006;128:7724–7725. doi: 10.1021/ja061435a. [DOI] [PubMed] [Google Scholar]
- 28.Watt ED, Shimada H, Kovrigin EL, Loria JP. The mechanism of rate-limiting motions in enzyme function. Proc Natl Acad Sci USA. 2007;104:11981–11986. doi: 10.1073/pnas.0702551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beach H, Cole R, Gill M, Loria JP. Conservation of mus-ms enzyme motions in the apo- and substrate-mimicked state. J Am Chem Soc. 2005;127:9167–9176. doi: 10.1021/ja0514949. [DOI] [PubMed] [Google Scholar]
- 30.Kovrigin EL, Cole R, Loria JP. Temperature dependence of the backbone dynamics of Ribonuclease A in the ground state and bound to the inhibitor 5′-phosphothymidine (3′–5′) pyrophosphate adenosine 3′-phosphate. Biochemistry. 2003;42:5279–5291. doi: 10.1021/bi034027h. [DOI] [PubMed] [Google Scholar]
- 31.Wladkowski BD, Svensson LA, Sjolin L, Ladner JE, Gilliland GL. Structure (1.3 Å) and charge states of ribonuclease A-Uridine vanadate complex: implications for the phosphate ester hydrolysis mechanism. J Am Chem Soc. 1998;120:5488–5498. [Google Scholar]
- 32.Wlodawer A, Svensson LA, Sjolin L, Gilliland GL. Structure of phosphate-free ribonuclease A refined at 1.26 Å. Biochemistry. 1988;27:2705–2717. doi: 10.1021/bi00408a010. [DOI] [PubMed] [Google Scholar]
- 33.Mallorqui-Fernandez G, Pous J, Peracaula R, Aymami J, Maeda T, Tada H, Yamada H, Seno M, de Llorens R, Gomis-Ruth FX, Coll M. Three-dimensional crystal structure of human eosinophil cationic protein (RNase 3) at 1.75 angstrom resolution. J Mol Biol. 2000;300:1297–1307. doi: 10.1006/jmbi.2000.3939. [DOI] [PubMed] [Google Scholar]
- 34.Mohan CG, Boix E, Evans HR, Nikolovski Z, Nogues MV, Cuchillo CM, Acharya KR. The crystal structure of eosinophil cationic protein in complex with 2′,5′-ADP at 2.0 Å resolution reveals the details of the ribonucleolytic active site. Biochemistry. 2002;41:12100–12106. doi: 10.1021/bi0264521. [DOI] [PubMed] [Google Scholar]
- 35.Chatani E, Hayashi R, Moriyama H, Ueki T. Conformational strictness required for maximum activity and stability of bovine pancreatic ribonuclease A as revealed by crystallographic study of three Phe120 mutants at 1.4 Å resolution. Protein Sci. 2002;11:72–81. doi: 10.1110/ps.31102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallorqui-Fernandez G, Pous J, Peracaula R, Aymami J, Maeda T, Tada H, Yamada H, Seno M, de Llorens R, Gomis-Ruth FX, Coll M. Three-dimensional crystal structure of human eosinophil cationic protein (RNase 3) at 1.75 A resolution. J Mol Biol. 2000;300:1297–1307. doi: 10.1006/jmbi.2000.3939. [DOI] [PubMed] [Google Scholar]
- 38.Swaminathan GJ, Holloway DE, Veluraja K, Acharya KR. Atomic resolution (0.98 Å) structure of eosinophil-derived neurotoxin. Biochemistry. 2002;41:3341–3352. doi: 10.1021/bi015911f. [DOI] [PubMed] [Google Scholar]
- 39.delCardayre SB, Ribo M, Yokel EM, Quirk DJ, Rutter WJ, Raines RT. Engineering ribonuclease A: production, purification and characterization of wild-type enzyme and mutants at Gln11. Protein Eng. 1995;8:261–273. doi: 10.1093/protein/8.3.261. [DOI] [PubMed] [Google Scholar]
- 40.Sela M, Anfinsen CB. Some spectrophotometric and polarimetric experiments with ribonuclease. Biochim Biophys Acta. 1957;24:229–235. doi: 10.1016/0006-3002(57)90186-5. [DOI] [PubMed] [Google Scholar]
- 41.Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. Elsevier Academic; San Diego: 2007. [Google Scholar]
- 42.Cavanagh J, Palmer AG, Wright PE, Rance M. Sensitivity improvement in proton-detected two-dimensional heteronuclear relay spectroscopy. J Magn Reson. 1991;91:429–436. [Google Scholar]
- 43.Cavanagh J, Rance M. Sensitivity improvement in isotropic mixing (TOCSY) experiments. J Magn Res. 1990;88:72–85. [Google Scholar]
- 44.Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–10665. [Google Scholar]
- 45.Palmer AG, Cavanagh J, Wright PE, Rance M. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J Magn Reson. 1991;93:151–170. [Google Scholar]
- 46.Loria JP, Rance M, Palmer AG. A Relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc. 1999;121:2331–2332. [Google Scholar]
- 47.Mulder FA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE. Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15)N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- 48.Luz Z, Meiboom S. Nuclear magnetic resonance study of the protolysis of trimethylammonium ion in aqueous solution—order of the reaction with respect to solvent. J Chem Phys. 1963;39:366–370. [Google Scholar]
- 49.Millet OM, Loria JP, Kroenke CD, Pons M, Palmer AG. The static magnetic field dependence of chemical exchange linebroadening defines the NMR chemical shift time scale. J Am Chem Soc. 2000;122:2867–2877. [Google Scholar]
- 50.Kloiber K, Konrat R. Differential multiple-quantum relaxation arising from cross-correlated time-modulation of isotropic chemical shifts. J Biomol NMR. 2000;18:33–42. doi: 10.1023/a:1008317212558. [DOI] [PubMed] [Google Scholar]
- 51.Zegers I, Maes D, Dao-Thi MH, Poortmans F, Palmer R, Wyns L. The structures of RNase A complexed with 3′-CMP and d(CpA): active site conformation and conserved water molecules. Protein Sci. 1994;3:2322–2339. doi: 10.1002/pro.5560031217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 53.Hammes GG, Schimmel PR. Equilibrium measurements of the binding of cytidine 3′-phosphate to ribonuclease. J Am Chem Soc. 1965;87:4665–4669. doi: 10.1021/ja00949a001. [DOI] [PubMed] [Google Scholar]
- 54.Park C, Raines RT. Catalysis by ribonuclease A is limited by the rate of substrate association. Biochemistry. 2003;42:3509–3518. doi: 10.1021/bi026076k. [DOI] [PubMed] [Google Scholar]
- 55.Thompson JE, Kutateladze MC, Venegas FD, Messmore JM, Raines RT. Limits to catalysis by ribonuclease A. Bioorg Chem. 1995;23:471–481. doi: 10.1006/bioo.1995.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee GC, Chan SI. A 31P NMR study of the association of uridine-3′-monophosphate to ribonuclease A. Biochem Biophys Res Commun. 1971;43:142–148. doi: 10.1016/s0006-291x(71)80098-0. [DOI] [PubMed] [Google Scholar]
- 57.Raines R. Ribonuclease A. Chem Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 58.Gerstein M, Chothia C. Analysis of protein loop closure: two types of hinges produce one motion in lactate dehydrogenase. J Mol Biol. 1991;220:133–149. doi: 10.1016/0022-2836(91)90387-l. [DOI] [PubMed] [Google Scholar]
- 59.White JL, Hackert ML, Buehner M, Adams MJ, Ford GC, Lentz PJ, Jr, Smiley IE, Steindel SJ, Rossmann MG. A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes. J Mol Biol. 1976;102:759–779. doi: 10.1016/0022-2836(76)90290-4. [DOI] [PubMed] [Google Scholar]
- 60.Allemann RK, Presnell SR, Benner SA. A hybrid of bovine pancreatic ribonuclease and human angiogenin: an external loop as a module controlling substrate specificity? Prot Eng. 1991;4:831–835. doi: 10.1093/protein/4.7.831. [DOI] [PubMed] [Google Scholar]
- 61.Bentley ML, Gaweska H, Kielec JM, McCafferty DG. Engineering the substrate specificity of Staphylococcurs aureus sortase A - The beta 6/beta 7 loop from SrtB confers NPQTN recognition to SrtA. J Biol Chem. 2007;282:6571–6581. doi: 10.1074/jbc.M610519200. [DOI] [PubMed] [Google Scholar]
- 62.Han KH, Syi JL, Brooks BR, Ferretti JA. Solution conformations of the B-loop fragments of human transforming growth factor α and epidermal growth factor by 1H nuclear magnetic resonance and restrained molecular dynamics. Proc Natl Acad Sci USA. 1990;87:2818–2822. doi: 10.1073/pnas.87.7.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hedstrom L, Szilagyi L, Rutter WJ. Converting trypsin to chymotrypsin: the role of surface loops. Science. 1992;255:1249–1253. doi: 10.1126/science.1546324. [DOI] [PubMed] [Google Scholar]
- 64.Ma W, Tang C, Lai L. Specificity of trypsin and chymotrypsin: loop-motion-controlled dynamic correlation as a determinant. Biophys J. 2005;89:1183–1193. doi: 10.1529/biophysj.104.057158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siggers K, Soto C, Palmer AG., 3rd Conformational dynamics in loop swap mutants of homologous fibronectin type III domains. Biophys J. 2007;93:2447–2456. doi: 10.1529/biophysj.106.100578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peng T, Zintsmaster JS, Namanja AT, Peng JW. Sequence-specific dynamics modulate recognition specificity in WW domains. Nat Struct Mol Biol. 2007;14:325–331. doi: 10.1038/nsmb1207. [DOI] [PubMed] [Google Scholar]
- 67.Akke M, Liu J, Cavanagh J, Erickson HP, Palmer AG. Pervasive conformational fluctuations on microsecond time scales in a fibronectin type III domain. Nat Struct Biol. 1998;5:55–59. doi: 10.1038/nsb0198-55. [DOI] [PubMed] [Google Scholar]
- 68.Watt ED, Shimada H, Kovrigin EL, Loria JP. The mechanism of rate-limiting motions in enzyme function. Proc Natl Acad Sci. 2007;104:11981–11986. doi: 10.1073/pnas.0702551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.