Abstract
The plant-like, bifunctional dihydrofolate reductase-thymidylate synthase (DHFR-TS) from malaria parasites has been a good target for drug development. Dihydrofolate reductase (DHFR) is inhibited by clinically established antimalarials, pyrimethamine and cycloguanil. Thymidylate synthase (TS) is the target of potent experimental antimalarials such as 5-fluoroorotate and 1843U89. Another enzyme in folate recycling, serine hydroxymethyltransferase (SHMT), produces 5,10-methylenetetrahydrofolate which, in many cells, is required for the de novo, biosynthesis of thymidine and methionine. Thus, the biochemical characterization of malarial SHMT was of interest. The principle, active P. falciparum SHMT (PfSHMT) was expressed in E. coli and purified using an N-terminal histidine tag. Unlike the plant enzyme, but like the host enzyme, PfSHMT requires the cofactor pyridoxal 5’-phosphate for enzymatic activity. The substrate specificities for serine, tetrahydrofolate, and pyridoxal 5’-phosphate were comparable to those for SHMT from other organisms. Antifolates developed for DHFR and TS inhibited SHMT in the mid-micromolar range, offering insights into the binding preferences of SHMT but clearly leaving room for improved new inhibitors. As previously seen with P. falciparum DHFR-TS, PfSHMT bound its cognate mRNA but not control RNA for actin. RNA-binding was not reversed with enzyme substrates. Unlike DHFR-TS, the SHMT RNA-protein interaction was not tight enough to inhibit translation. Another gene PF14_0534, previously proposed to code for an alternate mitochondrial SHMT, was also expressed in E. coli but found to be inactive. This protein, nor DHFR-TS, enhanced the catalytic activity of PfSHMT. The present results set the stage for developing specific, potent inhibitors of SHMT from P. falciparum.
Keywords: Malaria, inhibitor, enzyme, kinetics, RNA-binding
1. Introduction
Malaria infects approximately 300 to 600 million people [1]. Of these malaria cases, approximately 1.5 to 2.7 million result in death [2]. It is estimated that 70% of the malaria deaths occur in Sub-Saharan Africa and 25% of the cases worldwide occur in Southeast Asia [1]. In the absence of an effective vaccine, one of the few methods of combating malaria is chemotherapy. Unfortunately, resistance to most commonly used antimalarials is becoming widespread [3–7]. This highlights the pressing need for the development of new antimalarials.
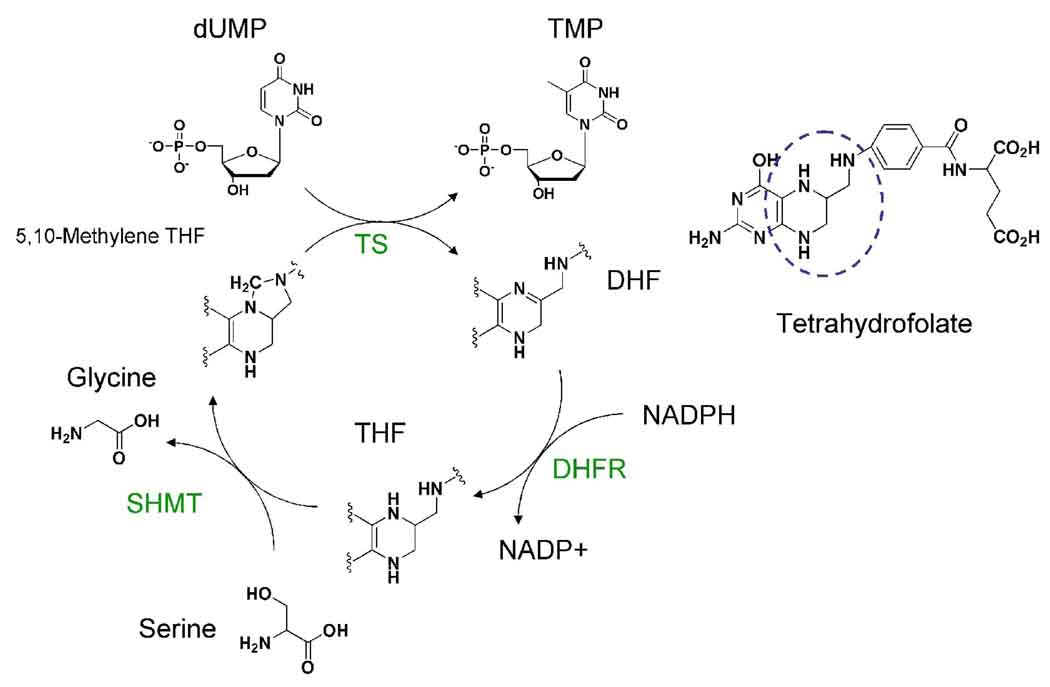
Enzymes of the folate cycle include dihydrofolate reductase (DHFR), thymidylate synthase (TS), and serine hydroxymethyltransferase (SHMT) (Fig 1). Two of these enzyme activities of the folate cycle are clearly “drugable” for malaria as well as other human diseases, including cancer: DHFR reduces dihydrofolate (DHF) to tetrahydrofolate (THF) using NADPH as a cofactor in the reduction. DHFR is the target of the antimalarials pyrimethamine and cycloguanil (the active metabolite of proguanil) [8–10], as well as the target of the anticancer agents methotrexate and pemetrexed [11, 12]. TS methylates the 5-carbon of 5’-deoxyuridine monophosphate (dUMP) to form thymidine monophosphate (TMP) using 5,10-methylene THF as a cofactor. TS is the target for many anticancer agents including pemetrexed, ZD1694, 1843U89, AG331, and AG337 [11, 13–18].
Fig 1.
The folate cycle. DHFR: dihydrofolate reductase, SHMT: serine hydroxymethyltransferase, TS: thymidylate synthase, dUMP: deoxyuridine monophosphate, TMP: deoxythymidine monophosphate, DHF: dihydrofolate, THF tetrahydrofolate.
Since SHMT also plays a key role in the folate cycle, it also has potential as an attractive drug target. SHMT transfers a methylene group from serine to THF to form 5,10-methylene THF and glycine, using pyridoxal 5’-phosphate (PLP) as a cofactor in the mammalian reaction [19–22]. SHMT protein has previously been purified from many non-malaria organisms, including human, rabbit, E. coli, L. donovani, and T. cruzi [24–35]. SHMT has also been partially purified from the avian malaria Plasmodium lophurae and the rodent malaria P. chabaudi [36, 37] but not from P. falciparum. Though PLP is required by most SHMTs for catalytic activity, mung bean SHMT is active in the absence of PLP [50]. Due to the plant-like origins of Plasmodium, it would be of interest to know if P. falciparum SHMT functions in the absence of PLP. Cloning of the P. falciparum SHMT gene [38, 39] indicates that the gene has three exons, the first comprised of only the methionine start codon. The identity of the gene was confirmed by expression of functionally active protein in E. coli [38]. In yeast and humans, in addition to the cytoplasmic SHMT, there is a mitochondrial isoform of SHMT that is involved in one-carbon metabolism in this subcellular organelle. A hypothetical protein (PF14_0534) has been annotated as a mitochondrial SHMT in P. falciparum due to its homology to a protein annotated as mitochondrial SHMT in P .yoelii. PF14_0534 contains a mitochondrial signal sequence, however, the gene sequence lacks a highly conserved active site motif found in most SHMTs [23].
The crystal structure of SHMTs from human, E. coli, and B. stearothermophilus have previously been solved [40–43]. In the absence of a P. falciparum SHMT crystal structure, a homology model has been constructed [44]. P. falciparum SHMT is highly conserved. A comparison of active site residues shows that there are three residues that differ at the active site of human and P. falciparum SHMT. Active site residues that have been shown to be critical for catalytic activity are conserved in P. falciparum. These residues include lysine 237 which forms a Schiff base with PLP [45, 46] and glutamic acid 56 which is involved in several proton transfers throughout the catalytic mechanism [47]. Structurally, most eukaryotic SHMTs are tetramers while most prokaryotic SHMTs are dimers. The tetramer formed by eukaryotic SHMTs can be better described as a dimer of dimers, with just a few contacts between the two dimers. Eukaryotic SHMTs have conserved residues that provide important contacts for tetramer formation. The conserved histidine and the insertions that provide contacts for tetramer formation are missing in the P. falciparum enzyme [44].
Human DHFR and TS have been shown to bind to their cognate mRNA and inhibit translation in vitro [48]. In the presence of substrates and inhibitors, RNA binding and translational inhibition is released. P. falciparum DHFR-TS was also shown to bind to its cognate mRNA and inhibit translation. However, unlike the human enzymes, RNA binding and translational inhibition was not released in the presence of substrates or inhibitors of DHFR or TS. The experimental DHFR inhibitor WR99210 gains selectivity through the differences in RNA-protein interactions of human and P. falciparum DHFR [49]. . Thus differential expression and regulation of human and malaria DHFR and TS provide a unique opportunity for selectivity [48, 49, 51]. It is not known if PfSHMT also binds its cognate RNA and creates opportunities for selective vulnerabilities through host-parasite differences in gene regulation.
The present study was undertaken to express PfSHMT in its functional form and to characterize its basal enzymatic properties. This lays the ground work for future drug development projects directed at malarial SHMT.
2. Materials and Methods
2.1 Cloning of PfSHMT and PF14_0534
Genes for PfSHMT and PF14_0534 were cloned from total RNA from malaria parasites strain 3D7. First, reverse transcription was performed using the Retroscript kit (Ambion) using the random decamer as a primer, followed by 30 cycles of PCR. The forward primer used to amplify SHMT was 5’ ATCTAGCGATTCCATGGCTATGTTTAACAACGACCCTTTGC 3’, the forward primer used to amplify SHMT with an N-terminal histidine tag was 5’ ATCTAGCGATTCCATGGCTCACCACCACCACCACCACATGTTTAA CAACGACCCTTTGC 3’, and the reverse primer used to amplify SHMT was 5’ TTAGCCATTAAGGATCCGTTAGGCAAATGGTAAGTTTTTTGC 3’. The forward primer used to amplify PF14_0534 was 5’ ATCTAGCGATTCCATGGCTATGCTGAAGGAGTTTGTTAAAAATG 3’, the forward primer used to amplify PF14_0534 with an N-terminal histidine tag was 5’ ATCTAGCGATTCCATGGCTCACCACCACCACCACCACATGCTGAAG GAGT TTGTTAAAAATG 3’, and the forward primer used to amplify PF14_0534 without the predicted mitochondrial signal sequence was 5’ ATCTAGCGATTCCATGGCTCAGAGCCGTTTAAATGATATAG 3’. The reverse primer used for amplification of PF14_0534 was 5’ TTAGCCATTAAGGATCCGTTATTCATTTGTGTATGGAGATGG 3’. SHMT was cloned into the vector pET-9d+ (Novagen) between the Nco1 and BamH1 restriction sites. PF14_0534 was cloned into the vector pET-3d+ (Novagen) between the Nco1 and BamH1 restriction sites. The plasmids were transformed into the SHMT-negative, glycine auxotroph E. coli strain, GS245(DE3)pLysS (E. coli stock center, Yale University) [38] by electroporation.
2.2 Expression of PfSHMT
A single colony of GS245(DE3)pLysS pET-9d+NhisSHMT3D7 was picked and used to inoculate 3 ml of LB supplemented with 50 µg/ml phenylalanine, 10 µg/ml thiamine, 50 µg/ml glycine, 30 µg/ml of kanamycin and 35 µg/ml of chloramphenicol. The culture was incubated at 37 °C with shaking at 200 rpm for 16 hours. The 3 ml culture was used to inoculate 200 ml of LB supplemented as previously described, the 200 ml culture was incubated as previously described [38]. The 200 ml culture was centrifuged at 5000xg at 4 °C for 5 minutes. The supernatant was poured off, and the pellet was resuspended in 1 L of LB supplemented as previously described. The 1 L culture was incubated at 37 °C with shaking at 200 rpm, when the OD600nm reached 0.6, the culture was induced with 1 mM IPTG. The culture was incubated at 37 °C with shaking at 200 rpm for an additional 6 hours. The cells were centrifuged at 8000xg for 5 minutes at 4 °C and the pellet was stored at −80°C.
2.3 Purification of PfSHMT
The bacterial pellet was removed from the −80 °C freezer and allowed to reach room temperature. The pellet was resuspended in 40 ml of lysis buffer (50 mM Tris-Cl pH 8.0, 500 mM NaCl, 20 mM imidazole, 2 mM DTT and 100 µg/ml lysozyme) and incubated on ice for 30 minutes. The cells were then sonicated at 30 W for 6x15 seconds on ice. The lysate was clarified by centrifugation at 8000xg for 30 minutes at 4 °C. The clarified lysate was loaded onto a His GraviTrap column (GE Healthcare) that was preequilibrated with 5 ml of elution buffer (50 mM Tris-Cl pH 8.0, 500 mM NaCl, 500 mM imidazole, and 2 mM DTT) and 10 ml of wash buffer (50 mM Tris-Cl pH 8.0, 500 mM NaCl, 20 mM imidazole, and 2 mM DTT). Once the lysate had been loaded onto the column, the column was washed with 10 ml of wash buffer and eluted with 6 ml of elution buffer.
The eluted protein was dialyzed 2 times against 1 L of SHMT buffer A (50 mM Tris-Cl pH 8.0, 2 mM EDTA, 1 M ammonium sulfate, and 2 mM DTT) for at least 5 hours each time. The protein was loaded onto a 10 ml Phenyl Sepharose column connected to an AKTA FPLC (GE Healthcare) at 4 ml/minute. Once the OD280nm returned to zero, the protein was eluted by running a gradient from 0 to 100% SHMT buffer B (50 mM Tris-Cl pH 8.0, 2 mM EDTA, and 2 mM DTT) over 45 minutes at 4 ml/minute. The gradient was held constant once the protein began to elute from the column. The protein eluted between 25 and 35% SHMT buffer B. The fractions containing the highest OD280nm were resolved by SDS-PAGE. Pure fractions were pooled and concentrated to a final volume of 1 ml using a Vivaspin 20 concentrator (Vivascience).
The protein was further purified by size exclusion chromatography. The protein was passed over a 60 ml Superdex 200 size exclusion column (GE Healthcare) at a flow rate of 1 ml/minute in SHMT gel filtration buffer (50 mM Tris-Cl pH 8.0, 150 mM NaCl, 2 mM EDTA, and 2mM DTT). The fractions containing the highest OD280nm were resolved by SDS-PAGE. The purified proteins were applied to a Vivaspin 20 concentrator and centrifuged at 3500xg at 4°C until about 1 ml of liquid remained. The protein was desalted using 15 ml of no salt buffer (50 mM Tris-Cl pH 8.0, 2 mM EDTA, 20% glycerol, and 2 mM DTT) in a diafiltration cup (Vivascience Inc.). The protein was concentrated to a final volume of 1 ml and stored at −20 °C. The concentration of protein was determined using the Bio-Rad protein microassay.
2.4 Native Molecular Weight of PfSHMT
The molecular weight of SHMT was determined by passing the protein over a 100 ml Superdex 200 size exclusion column in SHMT gel filtration buffer at a flow rate of 0.7 ml/min. The elution volume of SHMT was compared to the elution volume of standard proteins to determine the molecular weight. The standard proteins were purchased from Sigma-Aldrich, and were: cytochrome c (12.4 KDa), carbonic anhydrase (29 KDa), alcohol dehydrogenase (150 KDa), β-amylase (200 KDa), apoferritin (443 KDa), and thyroglobulin (670 KDa). Blue dextran (2000 KDa) was used to determine the void volume of the column. 5 ml fractions were collected. Each fraction was assayed for SHMT activity to determine the elution profile of active enzyme.
2.5 Preparation of Apo and Holo PfSHMT
The apo enzyme was prepared following the method of Brahatheeswaran et al. [19] with minor modifications. PLP was removed from SHMT by treatment with 100 mM L-cysteine. SHMT was dialyzed 2 times against 1 L of SHMT buffer A containing 100 mM L-cysteine for at least 5 hours each time. The dialyzed protein was passed over a 10 ml Phenyl Sepharose column that had been equilibrated with SHMT buffer A containing 100 mM L-cysteine. Once the protein was loaded onto the column, and the OD280nm returned to zero, the protein was eluted by running a gradient of 0 to 100% SHMT buffer B over 45 minutes at 4 ml/minute. The fractions containing the highest OD280nm were resolved by SDS-PAGE. Pure fractions were pooled and concentrated to a final volume of 1 ml using a Vivaspin 20 concentrator. L-Cysteine was removed by passage of the protein over a Superdex 200 size exclusion column as described in the protein purification section.
The holo enzyme was prepared by incubating the apo enzyme with 250 µM PLP at 4 °C for 16 hours. SHMT was separated from unbound PLP by passage over a 60 ml Superdex 200 size exclusion column in SHMT gel filtration buffer. The fractions containing the highest OD280nm were resolved by SDS-PAGE. Fractions containing protein were pooled and concentrated using a Vivaspin 20 concentrator.
The apo and holo enzymes were concentrated to a final concentration of 50 µM on a microcon YM-10 concentrator (Millipore). The concentrators were centrifuged at 14,000xg for 30 minutes at 4 °C. The final concentration of the proteins was measured using the Bio-Rad protein microassay. The UV-vis spectra of each protein was measured on a ND-1000 spectrophotometer (NanoDrop Technologies Inc.).
2.6 PfSHMT Activity and its inhibition
SHMT activity was measured by following the conversion of radioactive serine to 5,10-methylene THF [38, 52]. 1 µg (200nM final concentration) of SHMT was added to each 50 µl reaction containing 50 mM Tris-Cl pH 8.0, 2.5 mM EDTA, 10 mM 2-mercaptoethanol, 125 µM PLP, 1 mM THF, and 0.2 mM serine (0.05 Ci/mmol, GE Healthcare). The reaction was allowed to proceed for 20 minutes at 37 °C. The reaction was stopped by spotting 12.5 µl onto a 2.3 cm Whatman DE-81 paper. Unreacted serine was removed by washing the paper for 30 minutes in 10 mM Tris-Cl pH 8.0. The amount of radioactive product remaining on the paper was determined by liquid scintillation counting. Counts per minute were converted to pmol of product, based on the specific activity and counting efficiency of our system.
IC50 values of antifolates were determined by performing enzyme activity assays in the presence of varying concentrations of antifolates. Michaelis-Menten constants for each substrate were determined by performing enzyme activity assays using saturating concentrations of the substrates that were not being tested and varying the concentration of the substrate that was being tested. Serine was varied between 0.1 and 5 mM, THF was varied between 0.1 and 10 mM, and PLP was varied between 0.5 and 125 µM. The data were fitted to the equation v=Vmax[S]/(Km+[S]) using the program Sigmaplot.
2.7 RNA-Protein Interactions
Labeled SHMT RNA, DHFR-TS RNA, or actin RNA (0.5 nM, 100,000 cpm) was incubated with 500 nM SHMT protein in 10 mM HEPES pH 7.5, 3 mM MgCl2, 40mM KCl, 5% glycerol, 200mM 2-mercaptoethanol, and 1 unit of RNasin (Ambion) at 37 °C for 15 minutes. 1 unit of RnaseT1 (Ambion) was added for 10 minutes at room temperature. Heparin (5 mg/ml) was then added for an additional 10 minutes at room temperature. Following incubation, 1 µl of gel loading buffer (30% glycerol, 0.3% bromophenol blue) was added to the solution. The RNA was loaded onto a 5% TBE polyacrylamide gel (Bio-rad) and resolved at 120 V in 1X TBE (89 mM Tris, 89 mM boric acid, and 2 mM EDTA, pH 8.3). Electrophoresis was stopped when the dye reached the bottom of the gel. The gel was dried on a Bio-rad gel dryer and visualized by autoradiography. A storage phosphor screen (GE Healthcare) was exposed to the gel for 3 hours at room temperature. The image was visualized on a Storm 860 phosphorimager (GE Healthcare). Competition assays were performed by preincubating the protein with unlabeled RNA or substrates for 15 minutes at room temperature prior to addition of labeled RNA [49].
2.8 In vitro Translation
Before in vitro translation, a Poly A tail was added to the 3’ end of RNA by treatment with poly A polymerase (Ambion). Each 100 µl reaction contained 1x poly A polymerase buffer, 2.5 mM MnCl2, 1 mM ATP, 40 µg RNA, and 4 units of poly A polymerase. The reaction was incubated for 1 hour at 37 °C. Following incubation, 35 µl of DEPC treated water and 15 µl of 5 M NH4OAc was added to the reaction. The RNA was purified by extraction with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1). The reactions were mixed and centrifuged at 14000xg for 4 minutes. The aqueous layer was added to an equal volume of chloroform:isoamyl alcohol (25:1) for further purification. The reaction was mixed and centrifuged at 14000xg for 4 minutes. The aqueous layer was added to 2.5 volumes of 100% ethanol and the RNA was precipitated by incubation at −20 °C for 30 minutes. The RNA was centrifuged at 14000xg for 15 minutes and the supernatant was removed. Salt was removed from the RNA by washing with 100 µl of 70% ethanol. The RNA was centrifuged at 14000xg for 5 minutes and the supernatant was removed. The purified RNA was allowed to dry for 15 minutes and was resuspended in 20 µl of DEPC treated water. The concentration of RNA was determined by measuring the OD260nm on a ND-1000 spectrophotometer. The size and integrity of the RNA was determined by formaldehyde agarose gel electrophoresis.
In vitro translation experiments were performed using the nuclease-treated rabbit reticulocyte lysate system (Promega) following the manufacturer’s protocol. First, 5.7 nM poly A tailed SHMT, DHFR-TS or luciferase mRNA was added to 50 µl of 45% rabbit reticulocyte lysate containing 20 µM amino acid mixture (minus methionine), 20 units of Superasein (Ambion), and 10 µCi of 35S-methionine (1000 Ci/mmol, American Radiolabeled Chemicals). Reactions were incubated at 30 °C for 90 minutes. Following incubation, 3 µl of the reaction was added to 27 µl of SDS-PAGE sample buffer (62.5 mM Tris-Cl pH 6.8, 2% SDS, 25% glycerol, .01% bromophenol blue, and 5% 2-mercaptoethanol) and heated at 95 °C for 5 minutes. Next, 20 µl of the SDS-PAGE sample buffer/reaction mixture was resolved on a 10% Tris-HCl gel (Biorad). The gel was fixed for 30 minutes in fixing solution A (50% methanol, 10% acetic acid). To prevent the gel from cracking, the gel was soaked in fixing solution B (7% acetic acid, 7% methanol, and 1% glycerol) for an additional 30 minutes. The gel was then dried on a gel dryer for 2 hours. A storage phosphor screen (GE Healthcare) was exposed to the gel for 16 hours at room temperature. The image was visualized on a Storm 860 phosphorimager (GE Healthcare). Translational inhibition experiments were performed by adding an appropriate amount of SHMT or DHFR-TS protein to the reaction.
3. Results and Discussion
3.1 Expression of SHMT and PF14_0534
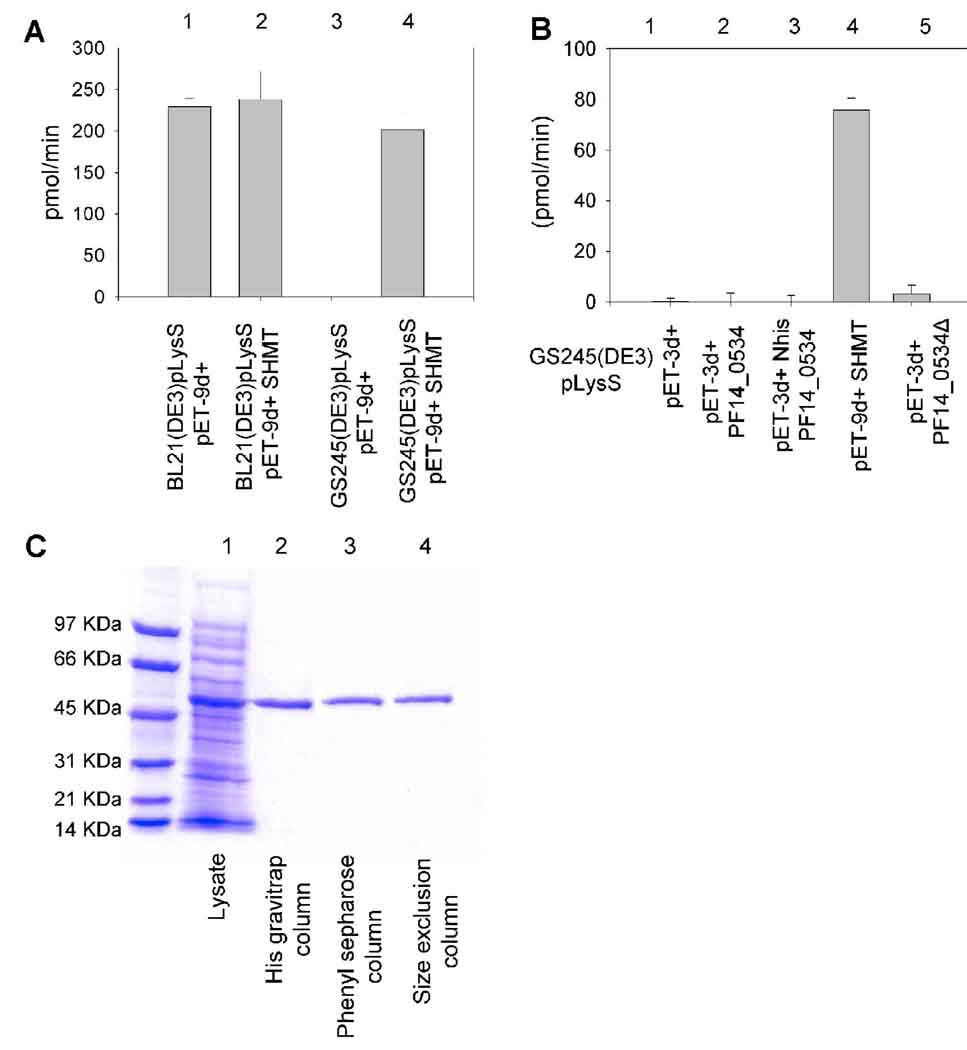
P. falciparum SHMT had previously been cloned and expressed in our laboratory [38], however, the protein lysate did not show an increase in activity over the host BL21(DE3)pLysS (Fig 2A reactions 1 and 2). The plasmid DNA was transformed into GS245(DE3)pLysS E. coli [38], which lacks endogenous SHMT. The GS245(DE3)pLysS E. coli lysate expressing P. falciparum SHMT displayed significant SHMT activity while the lysate expressing the empty expression vector had no enzyme activity at all (Fig 2A reactions 3 and 4). Although more protein was expressed by the BL21(DE3)pLysS E. coli than the GS245(DE3)pLysS E. coli, the absence of bacterial enzyme activity in the GS245(DE3)pLysS E. coli strain makes this a superior strain for studying functional PfSHMT.
Fig 2.
PfSHMT protein expression and purification. A) Enzyme activity of bacterial cell lysates expressing SHMT (lanes 2 and 4) or the empty expression vector (lanes 1 and 3). Lanes 1 and 2 were expressed in BL21(DE3)pLysS, lanes 3 and 4 were expressed in GS245(DE3)pLysS E. coli. B) Enzyme activity of bacterial cell lysates expressing the empty expression vector (lane 1), PF14_0534 (lane 2), PF14_0534 with a N-terminal histidine-tag (lane 3), P. falciparum SHMT (lane 4), and PF14_0534 without the predicted mitochondrial signal sequence. All lysates were from GS245(DE3)pLysS E. coli. C) SDS-PAGE analysis of aliquots from each step of the purification. Lysate (lane 1), nickel column (lane 2), phenyl sepharose column (lane 3), size exclusion column (lane 4).
In many cell types there are cytoplasmic and mitochondrial isoforms of SHMT. It has been suggested that the hypothetical protein PF14_0534 could be the mitochondrial isoform of SHMT [23]. The gene PF14_0534 was cloned in its full native state, with an N-terminal histidine-tag, and in the absence of the predicted mitochondrial signal sequence. Plasmid DNA was transformed into GS245(DE3)pLysS E. coli to test the expression and activity of the different versions of the gene product of PF14_0534. SHMT enzyme assays showed that lysates of bacteria expressing the gene PF14_0534 and the histidine-tagged PF14_0534 had no SHMT activity, even in the presence of high concentrations of serine, THF, and PLP (Fig 2B reactions 2 and 3). Bacteria cell lysates expressing PF14_0534 without the predicted mitochondrial signal sequence showed some SHMT activity, however this activity was barely over background and thus considered insignificant (Fig 2B reaction 5). Bacterial cell lysate of GS245(DE3)pLysS E. coli expressing P. falciparum SHMT was used as a positive control and showed significant amounts of SHMT enzyme activity (Fig 2B reaction 4). This data suggests that the hypothetical protein, PF14_0534, that is annotated as a mitochondrial SHMT does not contain SHMT catalytic activity. In reterospect, the sequence of this gene had little homology to SHMTs from other organisms and did not contain the highly conserved regions deemed critical for SHMT function [23]. Its mitochondrial SHMT designation came from the fact that it had high homology to other genes annotated as SHMT in Plasmodium species (Plasmodb) and PF14_0534 contained a predicted mitochondrial signal sequence at the N-terminus of the gene [23].
3.2 Purification of SHMT
PfSHMT was purified to homogeneity using three columns, a nickel column, a phenyl sepharose column, and a size exclusion column (Fig 2C). This is the first time that P. falciparum SHMT has been purified to homogeneity. The purification procedure resulted in 1.9 mg of protein from 1 L of bacterial culture with a specific activity of 210 nmol/min/mg. SHMT was purified 21.5 fold over lysate and 58.5% of the enzymatic activity was recovered after purification.
3.3 Dimer form of PfSHMT
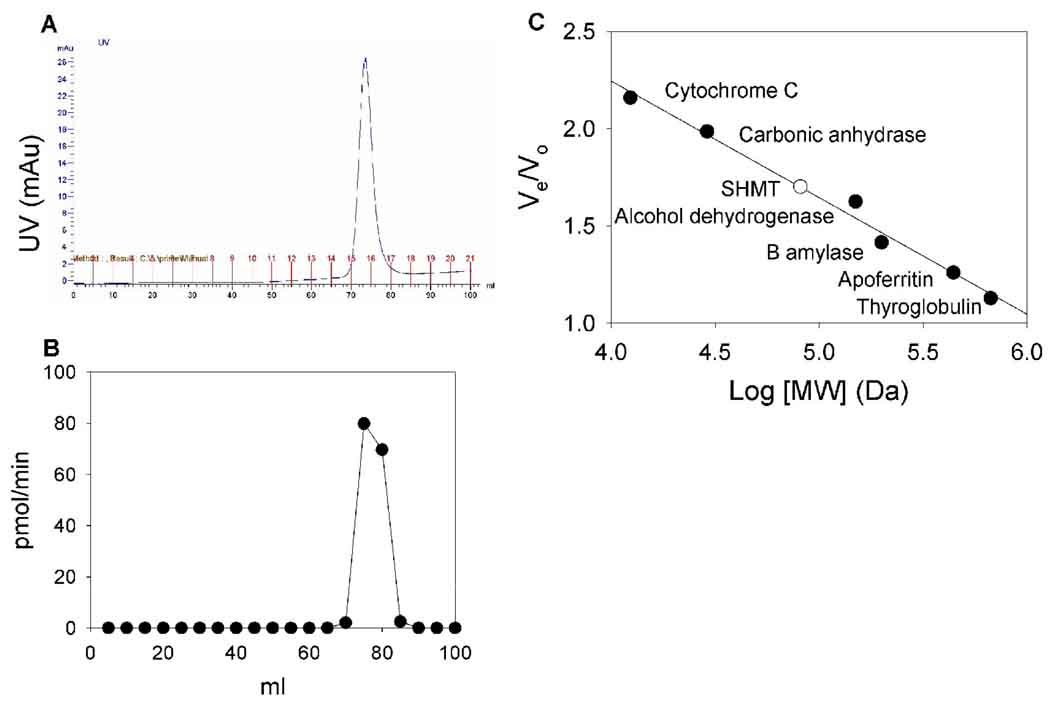
In order to determine the oligomeric status of P. falciparum SHMT, the protein was passed over a Superdex 200 size exclusion column and compared to the elution volume of standard proteins. Purified SHMT eluted from the size exclusion column as a single peak (Fig 3A). 76% of the protein was recovered from the column as determined by enzyme activity assays. The enzyme activity that eluted from the column corresponded with the elution of SHMT as determined by OD280nm (Fig 3B). A standard curve was constructed resulting in a straight line with a slope of −.597 and an r2 value of .985 (Fig 3C). Comparison of the Ve/Vo of P. falciparum SHMT to the standard curve gave a calculated molecular weight of 81.5 KDa. This value is lower than the expected molecular weight of 100 KDa for a dimer. Since most SHMTs found in nature are either a dimer or a tetramer, this data can confidently rule out that P. falciparum SHMT is a tetramer. In a homology model of P. falciparum SHMT and structures of other SHMTs, the active sites of each dimer are formed by residues of both monomers [41–44]. Because the enzyme activity elutes from the column in the same fractions as the UV absorbance, one can be confident that the protein is not a monomer. Thus P. falciparum SHMT appears to be a dimer in solution.
Fig 3.
Native molecular weight of SHMT. A) Size exclusion column trace of purified SHMT. B) Enzyme activity of fractions collected from the size exclusion column. C) Standard curve of elution volumes of molecular weight standards (●) and SHMT (○).
In tetrameric human SHMT [41] there are just a few contacts that hold two dimmers together. They include a histidine residue (his135 human SHMT) [41] that is conserved in all tetramers. This histidine residue forms hydrogen bond interactions with the same histidine of the opposing dimer. P. falciparum SHMT lacks these conserved histidine residue. E. coli SHMT, which is known to be a dimer, also lacks the conserved histidine residue. Residues 157–165 and 270–286 of human SHMT form additional supporting contacts for tetramer formation. Residues in P. falciparum SHMT corresponding to human SHMT residues 160–165 are not conserved. P. falciparum SHMT residues corresponding to human SHMT residues 270–286 are mostly missing, while P. falciparum SHMT residues corresponding to 274–278 of human SHMT are present in the malaria enzyme, they are not conserved. Thus P. falciparum SHMT was predicted to be a dimer [44].
3.4 Pyridoxal 5’-phosphate requirement
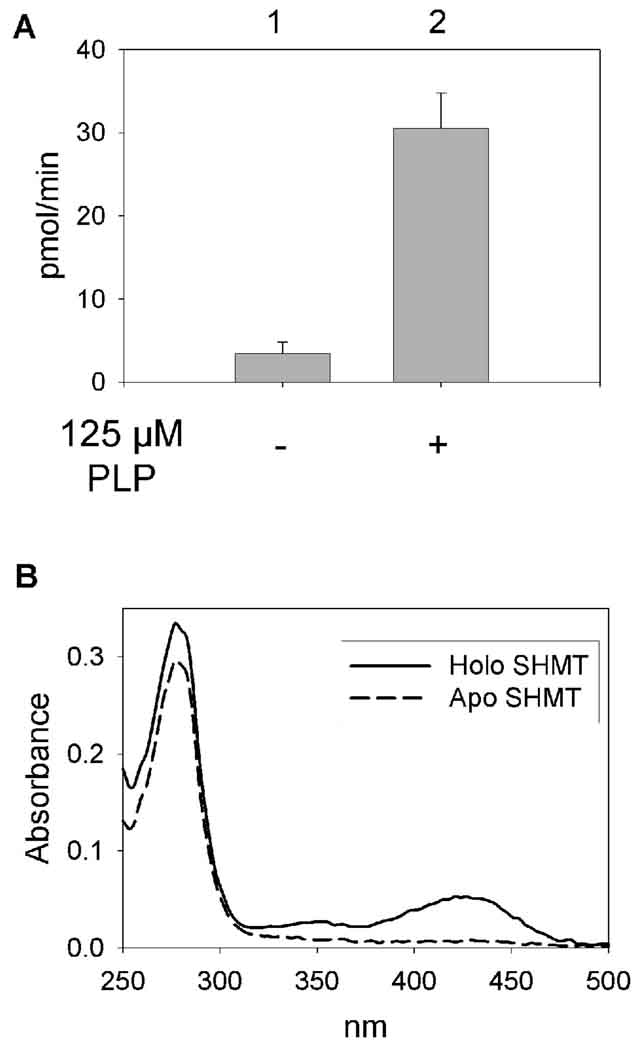
Almost all SHMTs found in nature require the cofactor PLP for enzymatic activity [19–22]. However, SHMT from mung bean is enzymatically active in the absence of PLP [50]. Therefore, it was of interest to determine if P. falciparum SHMT required PLP for catalysis. The apo enzyme was formed by passage of the protein over a phenyl sepharose column in the presence of L-cysteine. L-cysteine forms a thiazolidine complex with PLP, reducing its affinity to the protein active site. The protein sticks to the column while the L-cysteine in the buffer removes PLP from the protein. L-Cysteine remaining in the protein solution was removed by size exclusion chromatography. The SHMT activity of the apo enzyme was drastically reduced. The apo enzyme has some enzyme activity in the absence of PLP but this activity is only slightly above background and could be due to incomplete removal of PLP.
The holo enzyme was reconstituted by incubating the apo enzyme with PLP and separating the unbound PLP from the enzyme by size exclusion chromatography. With PLP added to the reaction, the enzyme activity increased by more than ten fold (Fig 4A). The UV-visible spectrum of each enzyme was measured (Fig 4B) to confirm that the holo enzyme did indeed contain PLP and the apo enzyme did not. The holo enzyme had a 278 nm/428 nm ratio of 6.3. This ratio has been observed in a range of 4.5–9.3 in other SHMTs [25, 26, 29,31, 32, 34]. The spectrum of the apo enzyme has a single peak at 280 nm corresponding to the protein. The spectrum of the holo enzyme has the same peak at 280 nm and a broad peak at 423 nm corresponding to PLP forming an internal aldimine with a conserved lysine (K237) at the SHMT active site.
Fig 4.
Pyridoxal 5’-phosphate is required for SHMT activity. A) Enzyme activity of the apo enzyme with (lane 2) and without (lanes 1) PLP. B) UV-vis spectra of apo (dashed line) and holo SHMT (solid line).
Taken together, these results demonstrate that P. falciparum SHMT binds PLP and that the cofactor is a definite requirement for enzymatic activity. While it should be possible to design inhibitors of SHMT targeted to the PLP binding site, more than 140 different enzymatic activities also utilize PLP [53], and thus extraordinary care will be required to avoid off target effects during design of SHMT inhibitors.
3.5 Kinetic Constants of SHMT
Michaelis-Menten constants for each substrate were determined. Michaelis-Menten constants are defined as the substrate concentration which produces a reaction velocity that is half the maximum reaction velocity [54–56]. To simplify the assay, the enzyme reaction was turned into a pseudo first-order reaction by using saturating concentrations of the substrates that were not being tested and varying the concentration of the substrate that was being tested. When fitted to the equation for a single substrate reaction, the Km of serine was calculated to be 0.7 mM with a Vmax of 139 pmol/min and an r2 of .99. The Km of THF was calculated to be 3.4 mM with a Vmax of 188.5 pmol/min and an r2 of .95. The plot of velocity vs. THF concentration does not completely reach saturation. This probably occurs because THF is easily oxidized in solution and clean higher THF concentrations in the reaction were not feasible. The data fits well to the appropriate equation giving an r2 of .95, thus the Km calculated provides a reliable estimate. The Km of PLP was calculated to be 13 μM with a Vmax of 113 pmol/min and an r2 of .94.
The substrate Km values calculated were compared to literature Km values of SHMT from human [34], the bacteria E. coli [31], the protozoan parasite Leishmania donovani [33], and the rodent malaria Plasmodium chabaudi [37]. The human and L. donovani SHMTs form tetramers while the E. coli SHMT is a dimer. The Km value of serine was in the range of what was observed for the other enzymes. The Km value of THF was significantly higher than the human and E. coli SHMT, but in the same range as L. donovani and P. chabaudi SHMT. The Km value of PLP was not calculated for the other organisms and thus could not be compared. The kcat value provides information on an enzyme’s efficiency and is defined as the maximum velocity divided by the number of active sites. The maximum velocity was taken from the velocity vs. substrate plots and the number of active sites was estimated by the number of moles of protein that were added to each reaction. The kcat value of 9.4 min−1 that was calculated is significantly lower (60 to 70 fold) than what was reported for human and E. coli SHMT. The kcat value for L. donovani and P. chabaudi was not reported and thus could not be compared (Table 1).
Table 1.
Comparison of Michaelis-Menten constants and estimated kcat values.
| Humana | E. colib | L. donovanic | P. chabaudid | P falciparum | |
|---|---|---|---|---|---|
| L-Serine Km (µM) |
100 | 400 | 1600 | 1100 | 700 |
| THF Km (µM) |
20 | 20 | 2400 | 2900 | 3400 |
| PLP Km (µM) |
NA | NA | NA | NA | 13 |
| kcat (min−1) | 575 | 640 | NA | NA | 9.4 |
Kruschwitz et al., Protein Expression and Purification, 6, 411–416, (1995)
Schrich et al., Journal of Bacteriology, 163, 1–7, (1985)
Vatsyayan et al., Protein Expression and Purification, 52, 433–440 (2007)
Ruenwongsa et al., Molecular and Biochemical Parasitology, 33, 265–272 (1989)
There are several possible explanations for the large variations in Km and kcat values. First, it is possible that some of the purified SHMT is misfolded or somehow inactive causing the kcat to appear artificially lower than what it should be. Second, since the Km values of L. donovani, P. chabaudi and P. falciparum are similar and generally higher than those seen for mammalian or bacterial SHMTs, it is possible that the parasite enzymes have a lower affinity for the substrates and convert substrate to product at a much slower rate. Third, since the P. falciparum SHMT was purified from a bacterial source, it is possible that it is missing a factor from the parasite that is required for complete enzyme activity. The kcat of purified Trypanosoma brucei s-adenosylmethionine decarboxylase increases 1000 fold in the presence of its prozyme (second inactive copy of the enzyme)[57].
Potential protein-protein interactions between SHMT and PF14_0534, or SHMT and DHFR-TS, may occur in the parasite but not in bacteria expressing individual proteins. To test such a possibility, lysates from bacteria expressing a His-tagged SHMT, PF14_0534, and DHFR-TS were mixed and purified on a nickel column. In purified enzymes, there was no increase in SHMT activity upon mixing of the lysates (supplemental fig 1A). TS assays demonstrated that DHFR-TS was not copurified on a nickel column upon mixing with SHMT (supplemental fig 1B). PF14_0534 and DHFR-TS do not seem to form protein-protein interactions with SHMT that are strong enough to survive purification on a nickel column. These proteins were mixed following lysis of the bacteria, it is still possible that the protein-protein interactions form during protein expression and folding.
3.6 Inhibition of PfSHMT by Antifolates
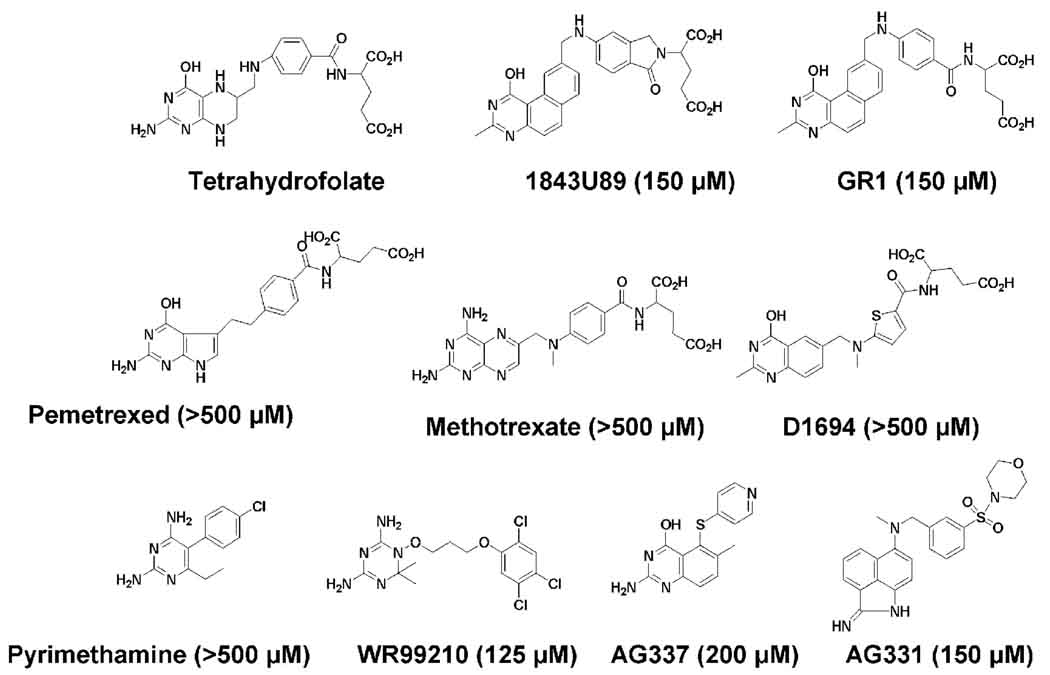
The substrate for SHMT, THF, is similar in structure to the substrate of DHFR, DHF, and the substrate of TS, 5,10-methyleneTHF. Given these structural similarities, it was possible that folate analogs that are designed to inhibit DHFR or TS could also inhibit SHMT. Inhibitors specific to P. falciparum DHFR, WR99210 and pyrimethamine, inhibited SHMT with an IC50 of 125 μM and >500 μM respectively. The DHFR inhibitor methotrexate had an IC50 of >500 μM. The dual DHFR and TS inhibitor pemetrexed had an IC50 of >500 μM. TS inhibitors 1843U89, GR1, and AG331 all had an IC50 of 150 μM. The TS inhibitor AG337 had an IC50 of 200 μM. The TS inhibitor D1694 had an IC50 of >500 μM (Fig 5). These results are not unexpected. Although the structures of the substrates are similar, each of the three enzymes catalyzes different reactions and bind unique forms of the folate substrates. Some of the better inhibitors in this study (WR99210, 1843U89, GR1, AG331, or AG337) could serve as a starting point for synthesizing new leads against PfSHMT. While an IC50 of 150 μM does not describe a potent inhibitor, a few critical modifications through chemical synthesis could turn these compounds into mid nM inhibitors of SHMT.
Fig 5.
Structures of DHFR or TS antifolates tested against PfSHMT. Values in brackets represent IC50 values for each compound.
3.7 SHMT RNA-Protein Interactions
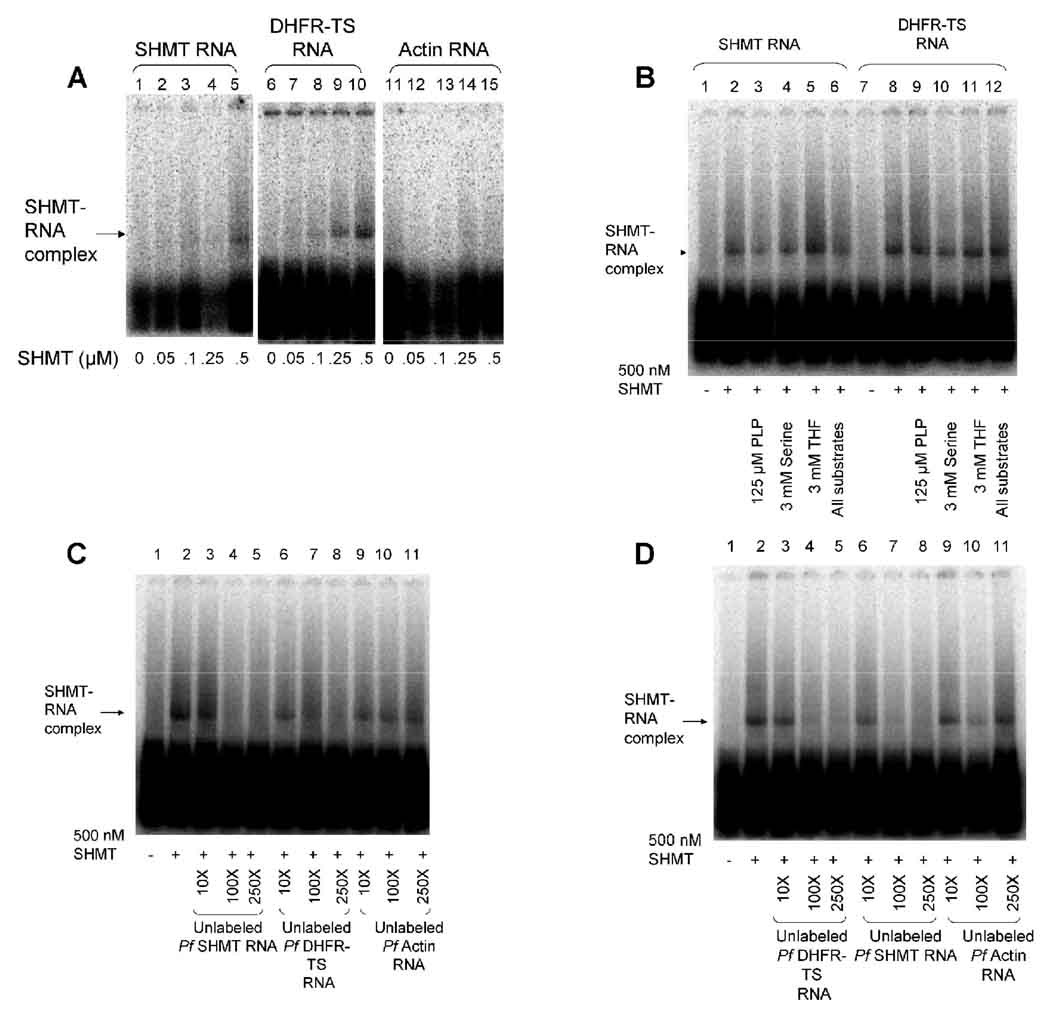
Given the differences in the RNA-protein interactions of human and malaria DHFR and TS [48, 49, 51], and the RNA-protein interaction observed in human SHMT [58], it was of interest to investigate if P. falciparum SHMT also bound to its cognate mRNA and if this RNA binding has an effect on translation. P. falciparum SHMT was found to bind to its cognate mRNA as well as DHFR-TS mRNA at a concentration of 500 nM. P. falciparum SHMT did not bind to the non-specific control actin mRNA (Fig 6A). P. falciparum SHMT binding to its cognate mRNA was abrogated by increasing amounts of unlabeled P. falciparum SHMT mRNA (Fig 6C lanes 3–5) and DHFR-TS mRNA (Fig 6C lanes 6–8). P. falciparum SHMT binding to its cognate mRNA was not abrogated by unlabeled actin mRNA (Fig 6C lanes 9–11). P. falciparum SHMT binding to DHFR-TS mRNA was abrogated by increasing amounts of unlabeled DHFR-TS mRNA (Fig 6D lanes 3–5) and P. falciparum SHMT mRNA (Fig 6D lanes 6–8). P. falciparum SHMT binding to DHFR-TS mRNA was not abrogated by unlabeled actin mRNA (Fig 6D lanes 9–12). These data confirms that the binding of P. falciparum SHMT protein to mRNA for SHMT and DHFR-TS is specific. The PfSHMT protein interaction with SHMT or DHFR-TS mRNA was not abrogated in the presence of SHMT substrates serine, THF, or PLP (fig 6B). This is similar to what has been observed for the P. falciparum DHFR-TS RNA-protein interaction [49].
Fig 6.
SHMT binds to its cognate RNA and DHFR-TS RNA. A) Labeled SHMT RNA (lanes 1–5), DHFR-TS RNA (lanes 6–10), and actin RNA (lanes 11–15), 0.5 nM, were incubated in the absence (lanes 1, 6, and 11) or presence of 50 nM (lanes 2, 7, and 12), 100 nM (lanes 3, 8, and 13), 250 nM (lanes 4, 9, and 14), and 500 nM SHMT protein (lanes 5, 10, and 15). B) SHMT RNA-protein interaction is not abrogated by substrates. Labeled SHMT RNA (lanes 1–6) or labeled DHFR-TS RNA (lanes 7–12), 0.5 nM, was incubated in the absence (lanes 1 and 7) or presence (lanes 2–6 and 8–12) of 500 nM SHMT protein. Reactions also included substrates for SHMT 125 μM PLP (lanes 3 and 9), 3 mM serine (lanes 4 and 10), 3 mM THF (lanes 5 and 11), or all three substrates (lanes 6 and 12). C) SHMT binds to its cognate RNA with specificity. Labeled SHMT RNA, 0.5 nM, was incubated in the absence (lane 1) or presence of 500 nM SHMT protein (lanes 2–11). Binding of labeled RNA to the protein was abrogated by competition with unlabeled SHMT RNA (lanes 3–5), unlabeled DHFR-TS RNA (lanes 6–8) but not unlabeled actin RNA (lanes 9–11). Reactions contained 10 fold (lanes 3, 6, and 9), 100 fold (lanes (4, 7, and 10), and 250 fold (lanes 5, 8, and 11) more unlabeled RNA than labeled RNA. D) SHMT binds to DHFR-TS RNA with specificity. Labeled DHFR-TS RNA, 0.5 nM, was incubated in the absence (lane 1) or presence of 500 nM SHMT protein (lanes 2–11). Binding of labeled RNA to the protein was abrogated by competition with unlabeled DHFR-TS RNA (lanes 3–5), unlabeled SHMT RNA (lanes 6–8) but not unlabeled actin RNA (lanes 9–11). Reactions contained 10 fold (lanes 3, 6, and 9), 100 fold (lanes (4, 7, and 10), and 250 fold (lanes 5, 8, and 11) more unlabeled RNA than labeled RNA.
The SHMT RNA-protein interactions offer some new potential possibilities for metabolic coordination. The fact that SHMT binding to DHFR-TS mRNA shows that although DHFR-TS protein can regulates expression of itself, there are possibilities for more complicated networks within the cell. SHMT binding to its cognate mRNA suggests that translational autoregulation could occur in many more metabolic genes in Plasmodium than just DHFR-TS. This idea is especially interesting given that only a few transcription factors and other regulatory elements have been identified in Plasmodium thus far.
3.8 Translational Inhibition by SHMT
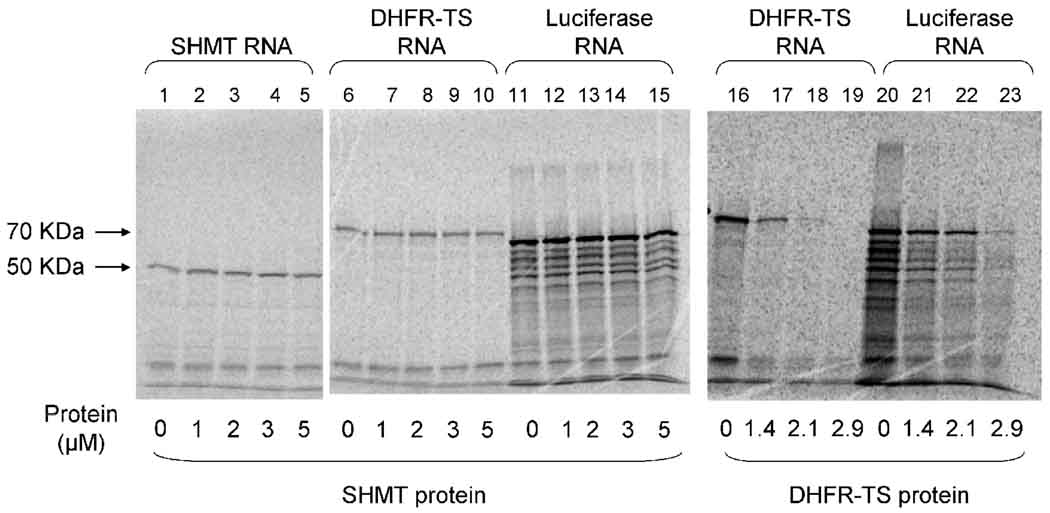
Given that SHMT protein binds to its cognate RNA and DHFR-TS RNA with specificity, it was of interest to determine if this RNA binding is able to inhibit translation. The SHMT RNA-protein interaction was found not to inhibit translation (figure 7). Incubation of SHMT or DHFR-TS RNA with increasing amounts of protein showed no decrease in the amount of protein produced in the in vitro translation assay. The DHFR-TS RNA-protein interaction was used as a positive control. Incubation of DHFR-TS RNA with DHFR-TS protein under the same conditions as with the SHMT protein showed a drastic decrease in the amount of protein translated in the reactions. The DHFR-TS protein also had a slight effect on the translation of the control luciferase RNA, however this effect was much smaller compared to the inhibition of DHFR-TS translation. This was expected as there are some non-specific interactions between DHFR-TS protein and RNA.
Fig 7.
SHMT RNA-protein interaction does not inhibit translation in vitro. About 5.7 nM poly A tailed SHMT RNA (lanes 1–5), DHFR-TS RNA (lanes 6–10), and luciferase RNA (lanes 11–15) were incubated with 45% rabbit reticulocyte lysate in the absence (lanes 1, 6, and 11) or presence (lanes 2–5, 7–10, and 12–15) of SHMT protein. Translational inhibition by DHFR-TS as a positive control. About 5.7 nM poly A tailed DHFR-TS RNA (lanes 1–4) or luciferase RNA (lanes 5–8) were incubated with 45% rabbit reticulocyte lysate in the absence (lanes 16 and 20) or presence (lanes 17–19 and 21–23) of DHFR-TS protein.
The DHFR-TS RNA-protein interaction is considerably stronger (requiring 100 fold more protein than RNA) than the SHMT RNA-protein interaction (requiring 1000 fold more protein than RNA). Nirmalan et al. [59] have shown that there is significant increase in SHMT mRNA as the erythrocytic cycle progresses, and that this increase is greater than the increase in RNA levels of other enzymes in the folate biosynthesis pathway, including DHFR-TS. It is possible that as the cell cycle progresses, the cell requires much more SHMT than other enzymes in the pathway and manages to do this by expressing higher levels of SHMT RNA and weaker feedback loops for translation of this message. The iron response protein, upon binding to the iron response element in the 3’ UTR of the transferrin mRNA stabilizes the RNA from degradation allowing increased expression of the RNA [60, 61]. It is also possible that because increased amounts of SHMT are required, the role of the SHMT RNA-protein interaction could be stabilization of mRNA from degradation.
Conclusion
In this report we demonstrate the biochemical characterization of Plasmodium falciparum SHMT, a pyridoxal 5’-phosphate-utilizing enzyme. Inhibitors of DHFR and TS were found to be modest inhibitors of SHMT. SHMT was found to bind to both its cognate mRNA and DHFR-TS mRNA although the detailed biological function of such RNA-protein interaction remains to be determined. The results presented here provide valuable reagents and chemical insights for future development of potent and selective inhibitors of SHMT.
Supplementary Material
Supplemental fig 1. PfSHMT is not potentiated by protein-protein interactions with DHFR-TS or a putative alternate SHMT. A) SHMT activity of nickel column purified protein. Lysates of histidine tagged SHMT (GS245(DE3)pLysSpET-9d+NhisSHMT) was mixed with lysates of GS245(DE3)pLysSpET-3d+PF14_0534 (lane 2) and lysates of Rue10(DE3)pET-23d+ DHFR-TS (lane 3) prior to purification on the nickel column. GS245(DE3)pLysSpET-9d+NhisSHMT (Lane 1), Rue10(DE3)pET-23d+ DHFR-TS (lane 4), and GS245(DE3)pLysSpET-3d+PF14_0534 (lane 5) were not mixed prior to purification on the nickel column. B) TS activity of nickel column purified protein. Lysates of histidine tagged SHMT (GS245(DE3)pLysSpET-9d+NhisSHMT) was mixed with lysates of Rue10(DE3)pET-23d+ DHFR-TS (lane 2) prior to purification on the nickel column. Rue10(DE3)pET-23d+ DHFR-TS lysates were tested for TS activity before (lane 4) and after (lane 1) purification on the nickel column. GS245(DE3)pLysSpET-9d+NhisSHMT lysates were tested for TS activity before (lane 5) and after (lane 3) purification on the nickel column.
Acknowledgment
This work was supported by the US National Institutes of Health (NIH grants AI26912 and AI60360). PKR is a recipient of a Senior Scholar Award in Global Infectious Diseases from the Ellison Medical Foundation.
Abbreviations
- DHFR
dihydrofolate reductase
- TS
thymidylate synthase
- mTHF
5,10-methylenetetrahydrofolate
- THF
tetrahydrofolate
- dUMP
2’-deoxyuridine monophosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Snow RW, Guerra CA, Noor AM, et al. The global distribution of clinical episodes Plasmodium falciparum malaria. Nature. 2005;434:214. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breman JG. The ears of the hippopotamus: manifestations, determinants, and estimates of the malaria burden. Am J Trop Med Hyg. 2001;64:1–11. doi: 10.4269/ajtmh.2001.64.1. [DOI] [PubMed] [Google Scholar]
- 3.Hyde JE. Mechanisms of resistance of Plasmodium falciparum to antimalarial drugs. Microbes Infect. 2002;4:165–174. doi: 10.1016/s1286-4579(01)01524-6. [DOI] [PubMed] [Google Scholar]
- 4.Le Bras J, Durand R. The mechanisms of resistance to antimalarial drugs in Plasmodium falciparum. Fundam Clin Pharmacol. 2003;17:147–153. doi: 10.1046/j.1472-8206.2003.00164.x. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez CP, McLean JE, Stein W, et al. Evidence for a substrate specific and inhibitable drug efflux system in chloroquine resistant Plasmodium falciparum strains. Biochemistry. 2004;43:16365–16373. doi: 10.1021/bi048241x. [DOI] [PubMed] [Google Scholar]
- 6.Su X, Kirkman LA, Fujioka H, et al. Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell. 1997;91:593–603. doi: 10.1016/s0092-8674(00)80447-x. [DOI] [PubMed] [Google Scholar]
- 7.Talisuna AO, Bloland P, D'Alessandro U. History, dynamics, and public health importance of malaria parasite resistance. Clin Microbiol Rev. 2004;17:235–254. doi: 10.1128/CMR.17.1.235-254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hitchings GH. The Use of an Antimetabolite in the Chemotherapy of Malaria and Other Infections. Clin Pharmacol Ther. 1960;1:570–589. [Google Scholar]
- 9.Hitchings GH. Folate antagonists as antibacterial and antiprotozoal agents. Ann N Y Acad Sci. 1971;186:444–451. doi: 10.1111/j.1749-6632.1971.tb31171.x. [DOI] [PubMed] [Google Scholar]
- 10.Russell PB, Hitchings GH. 2,4-Diaminopyrimidines as Antimalarials. III. 5-Aryl Derivatives. J Am Chem Soc. 1951;73:3763–3770. [Google Scholar]
- 11.Shih C, Chen VJ, Gossett LS, et al. LY231514, a pyrrolo[2,3-d]pyrimidine-based antifolate that inhibits multiple folate-requiring enzymes. Cancer Res. 1997;57:1116–1123. [PubMed] [Google Scholar]
- 12.Bertino JR. The Mechanism of Action of the Folate Antagonists in Man. Cancer Res. 1963;23:1286–1306. [PubMed] [Google Scholar]
- 13.Pendergast W, Dickerson SH, Dev IK, et al. Benzo[f]quinazoline inhibitors of thymidylate synthase: methyleneamino-linked aroylglutamate derivatives. J Med Chem. 1994;37:838–844. doi: 10.1021/jm00032a019. [DOI] [PubMed] [Google Scholar]
- 14.Varney MD, Marzoni GP, Palmer CL, et al. Crystal-structure-based design and synthesis of benz[cd]indole-containing inhibitors of thymidylate synthase. J Med Chem. 1992;35:663–676. doi: 10.1021/jm00082a006. [DOI] [PubMed] [Google Scholar]
- 15.Webber SE, Bleckman TM, Attard J, et al. Design of thymidylate synthase inhibitors using protein crystal structures: the synthesis and biological evaluation of a novel class of 5-substituted quinazolinones. J Med Chem. 1993;36:733–746. doi: 10.1021/jm00058a010. [DOI] [PubMed] [Google Scholar]
- 16.Ward WH, Kimbell R, Jackman AL. Kinetic characteristics of ICI D1694: a quinazoline antifolate which inhibits thymidylate synthase. Biochem Pharmacol. 1992;43:2029–2031. doi: 10.1016/0006-2952(92)90646-z. [DOI] [PubMed] [Google Scholar]
- 17.Jackman AL, Calvert AH. Folate-based thymidylate synthase inhibitors as anticancer drugs. Ann Oncol. 1995;6:871–881. doi: 10.1093/oxfordjournals.annonc.a059353. [DOI] [PubMed] [Google Scholar]
- 18.Taylor EC, Kuhnt D, Shih C, et al. A dideazatetrahydrofolate analogue lacking a chiral center at C-6, N-[4-[2-(2-amino-3,4-dihydro-4-oxo-7H-pyrrolo[2,3-d]pyrimidin-5- yl)ethyl]benzoyl]-L-glutamic acid, is an inhibitor of thymidylate synthase. J Med Chem. 1992;35:4450–4454. doi: 10.1021/jm00101a023. [DOI] [PubMed] [Google Scholar]
- 19.Brahatheeswaran B, Prakash V, Savithri HS, et al. Interaction of sheep liver aposerine hydroxymethyltransferase with pyridoxal-5'-phosphate: a physicochemical, kinetic, and thermodynamic study. Arch Biochem Biophys. 1996;330:363–372. doi: 10.1006/abbi.1996.0263. [DOI] [PubMed] [Google Scholar]
- 20.Cai K, Schirch D, Schirch V. The affinity of pyridoxal 5'-phosphate for folding intermediates of Escherichia coli serine hydroxymethyltransferase. J Biol Chem. 1995;270:19294–19299. doi: 10.1074/jbc.270.33.19294. [DOI] [PubMed] [Google Scholar]
- 21.Malerba F, Bellelli A, Giorgi A, et al. The mechanism of addition of pyridoxal 5'-phosphate to Escherichia coli apo-serine hydroxymethyltransferase. Biochem J. 2007;404:477–485. doi: 10.1042/BJ20061681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu TF, Boja ES, Safo MK, et al. Role of proline residues in the folding of serine hydroxymethyltransferase. J Biol Chem. 2003;278:31088–31094. doi: 10.1074/jbc.M303779200. [DOI] [PubMed] [Google Scholar]
- 23.Salcedo E, Sims PF, Hyde JE. A glycine-cleavage complex as part of the folate one-carbon metabolism of Plasmodium falciparum. Trends Parasitol. 2005;21:406–411. doi: 10.1016/j.pt.2005.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capelluto DG, Hellman U, Cazzulo JJ, et al. Purification and some properties of serine hydroxymethyltransferase from Trypanosoma cruzi. Eur J Biochem. 2000;267:712–719. doi: 10.1046/j.1432-1327.2000.01047.x. [DOI] [PubMed] [Google Scholar]
- 25.Sharma S, Bhakuni V. Cloning and structural analysis of Mycobacterium leprae serine hydroxymethyltransferase. Protein Expr Purif. 2007;55:189–197. doi: 10.1016/j.pep.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 26.Ulevitch RJ, Kallen RG. Purification and characterization of pyridoxal 5'-phosphate dependent serine hydroxymethylase from lamb liver and its action upon beta-phenylserines. Biochemistry. 1977;16:5342–5350. doi: 10.1021/bi00643a027. [DOI] [PubMed] [Google Scholar]
- 27.Chaturvedi S, Bhakuni V. Unusual structural, functional, and stability properties of serine hydroxymethyltransferase from Mycobacterium tuberculosis. J Biol Chem. 2003;278:40793–40805. doi: 10.1074/jbc.M306192200. [DOI] [PubMed] [Google Scholar]
- 28.Mukherjee M, Sievers SA, Brown MT, et al. Identification and biochemical characterization of serine hydroxymethyl transferase in the hydrogenosome of Trichomonas vaginalis. Eukaryot Cell. 2006;5:2072–2078. doi: 10.1128/EC.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schirch V. Purification of folate-dependent enzymes from rabbit liver. Methods Enzymol. 1997;281:146–161. doi: 10.1016/s0076-6879(97)81021-x. [DOI] [PubMed] [Google Scholar]
- 30.Schirch L, Peterson D. Purification and properties of mitochondrial serine hydroxymethyltransferase. J Biol Chem. 1980;255:7801–7806. [PubMed] [Google Scholar]
- 31.Schirch V, Hopkins S, Villar E, et al. Serine hydroxymethyltransferase from Escherichia coli: purification and properties. J Bacteriol. 1985;163:1–7. doi: 10.1128/jb.163.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang WN, Tsai JN, Chen BH, et al. Cloning, expression, purification, and characterization of zebrafish cytosolic serine hydroxymethyltransferase. Protein Expr Purif. 2006;46:212–220. doi: 10.1016/j.pep.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 33.Vatsyayan R, Roy U. Molecular cloning and biochemical characterization of Leishmania donovani serine hydroxymethyltransferase. Protein Expr Purif. 2007;52:433–440. doi: 10.1016/j.pep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 34.Kruschwitz H, Ren S, Di Salvo M, et al. Expression, purification, and characterization of human cytosolic serine hydroxymethyltransferase. Protein Expr Purif. 1995;6:411–416. doi: 10.1006/prep.1995.1055. [DOI] [PubMed] [Google Scholar]
- 35.di Salvo ML, Delle Fratte S, De Biase D, et al. Purification and characterization of recombinant rabbit cytosolic serine hydroxymethyltransferase. Protein Expr Purif. 1998;13:177–183. doi: 10.1006/prep.1998.0890. [DOI] [PubMed] [Google Scholar]
- 36.Platzer EG, Campuzano HC. The serine hydroxymethyltransferase of Plasmodium lophurae. J Protozool. 1976;23:282–286. doi: 10.1111/j.1550-7408.1976.tb03770.x. [DOI] [PubMed] [Google Scholar]
- 37.Ruenwongsa P, Luanvararat M, O'Sullivan WJ. Serine hydroxymethyltransferase from pyrimethamine-sensitive and -resistant strains of Plasmodium chabaudi. Mol Biochem Parasitol. 1989;33:265–271. doi: 10.1016/0166-6851(89)90088-1. [DOI] [PubMed] [Google Scholar]
- 38.Alfadhli S, Rathod PK. Gene organization of a Plasmodium falciparum serine hydroxymethyltransferase and its functional expression in Escherichia coli. Mol Biochem Parasitol. 2000;110:283–291. doi: 10.1016/s0166-6851(00)00282-6. [DOI] [PubMed] [Google Scholar]
- 39.Lee CS, Salcedo E, Wang Q, et al. Characterization of three genes encoding enzymes of the folate biosynthetic pathway in Plasmodium falciparum. Parasitology. 2001;122(Pt 1):1–13. doi: 10.1017/s0031182000006946. [DOI] [PubMed] [Google Scholar]
- 40.Renwick SB, Skelly JV, Chave KJ, et al. Purification, crystallization and preliminary X-ray analysis of human recombinant cytosolic serine hydroxymethyltransferase. Acta Crystallogr D Biol Crystallogr. 1998;54:1030–1031. doi: 10.1107/s0907444998004272. [DOI] [PubMed] [Google Scholar]
- 41.Renwick SB, Snell K, Baumann U. The crystal structure of human cytosolic serine hydroxymethyltransferase: a target for cancer chemotherapy. Structure. 1998;6:1105. doi: 10.1016/s0969-2126(98)00112-9. [DOI] [PubMed] [Google Scholar]
- 42.Scarsdale JN, Radaev S, Kazanina G, et al. Crystal structure at 2.4 A resolution of E. coli serine hydroxymethyltransferase in complex with glycine substrate and 5-formyl tetrahydrofolate. Journal of Molecular Biology. 2000;296:155–168. doi: 10.1006/jmbi.1999.3453. [DOI] [PubMed] [Google Scholar]
- 43.Trivedi V, Gupta A, Jala VR, et al. Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism. J Biol Chem. 2002;277:17161–17169. doi: 10.1074/jbc.M111976200. [DOI] [PubMed] [Google Scholar]
- 44.Franca TC, Pascutti PG, Ramalho TC, et al. A three-dimensional structure of Plasmodium falciparum serine hydroxymethyltransferase in complex with glycine and 5-formyl-tetrahydrofolate. Homology modeling and molecular dynamics. Biophys Chem. 2005;115:1–10. doi: 10.1016/j.bpc.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Bhavani S, Trivedi V, Jala VR, et al. Role of Lys-226 in the catalytic mechanism of Bacillus stearothermophilus serine hydroxymethyltransferase--crystal structure and kinetic studies. Biochemistry. 2005;44:6929–6937. doi: 10.1021/bi047800x. [DOI] [PubMed] [Google Scholar]
- 46.Schirch D, Delle Fratte S, Iurescia S, et al. Function of the active-site lysine in Escherichia coli serine hydroxymethyltransferase. J Biol Chem. 1993;268:23132–23138. [PubMed] [Google Scholar]
- 47.Szebenyi DM, Musayev FN, di Salvo ML, et al. Serine hydroxymethyltransferase: role of glu75 and evidence that serine is cleaved by a retroaldol mechanism. Biochemistry. 2004;43:6865–6876. doi: 10.1021/bi049791y. [DOI] [PubMed] [Google Scholar]
- 48.Chu E, Takimoto CH, Voeller D, et al. Specific binding of human dihydrofolate reductase protein to dihydrofolate reductase messenger RNA in vitro. Biochemistry. 1993;32:4756–4760. doi: 10.1021/bi00069a009. [DOI] [PubMed] [Google Scholar]
- 49.Zhang K, Rathod PK. Divergent Regulation of Dihydrofolate Reductase Between Malaria Parasite and Human Host. Science. 2002;296:545–547. doi: 10.1126/science.1068274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sukanya N, Vijaya M, Savithri HS, et al. Serine Hydroxymethyltransferase from Mung Bean (Vigna radiata) Is Not a Pyridoxal-5'-Phosphate-Dependent Enzyme. Plant Physiol. 1991;95:351–357. doi: 10.1104/pp.95.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu E, Koeller DM, Casey JL, et al. Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc Natl Acad Sci U S A. 1991;88:8977–8981. doi: 10.1073/pnas.88.20.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geller AM, Kotb MY. A binding assay for serine hydroxymethyltransferase. Anal Biochem. 1989;180:120–125. doi: 10.1016/0003-2697(89)90098-5. [DOI] [PubMed] [Google Scholar]
- 53.Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003;4:850–854. doi: 10.1038/sj.embor.embor914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piszkiewicz D. Kinetics of chemical and enzyme-catalyzed reactions. New York: Oxford university press; 1977. [Google Scholar]
- 55.Webb JL. Enzyme and Metabolic Inhibitors. New York and London: Academic Press; 1963. [Google Scholar]
- 56.Cleland WW. Steady State Kinetics. In: Boyer PD, editor. The Enzymes. New York and London: Academic Press; 1970. pp. 1–65. [Google Scholar]
- 57.Willert EK, Fitzpatrick R, Phillips MA. Allosteric regulation of an essential trypanosome polyamine biosynthetic enzyme by a catalytically dead homolog. Proc Natl Acad Sci U S A. 2007;104:8275–8280. doi: 10.1073/pnas.0701111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X, Reig B, Nasrallah IM, et al. Human Cytoplasmic Serine Hydroxymethyltransferase Is an mRNA Binding Protein. Biochemistry. 2000;39:11523–11531. doi: 10.1021/bi000665d. [DOI] [PubMed] [Google Scholar]
- 59.Nirmalan N, Wang P, Sims PF, et al. Transcriptional analysis of genes encoding enzymes of the folate pathway in the human malaria parasite Plasmodium falciparum. Mol Microbiol. 2002;46:179–190. doi: 10.1046/j.1365-2958.2002.03148.x. [DOI] [PubMed] [Google Scholar]
- 60.Theil EC. Regulation of ferritin and transferrin receptor mRNAs. J Biol Chem. 1990;265:4771–4774. [PubMed] [Google Scholar]
- 61.Theil EC, Eisenstein RS. Combinatorial mRNA regulation: iron regulatory proteins and iso-iron-responsive elements (Iso-IREs) J Biol Chem. 2000;275:40659–40662. doi: 10.1074/jbc.R000019200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental fig 1. PfSHMT is not potentiated by protein-protein interactions with DHFR-TS or a putative alternate SHMT. A) SHMT activity of nickel column purified protein. Lysates of histidine tagged SHMT (GS245(DE3)pLysSpET-9d+NhisSHMT) was mixed with lysates of GS245(DE3)pLysSpET-3d+PF14_0534 (lane 2) and lysates of Rue10(DE3)pET-23d+ DHFR-TS (lane 3) prior to purification on the nickel column. GS245(DE3)pLysSpET-9d+NhisSHMT (Lane 1), Rue10(DE3)pET-23d+ DHFR-TS (lane 4), and GS245(DE3)pLysSpET-3d+PF14_0534 (lane 5) were not mixed prior to purification on the nickel column. B) TS activity of nickel column purified protein. Lysates of histidine tagged SHMT (GS245(DE3)pLysSpET-9d+NhisSHMT) was mixed with lysates of Rue10(DE3)pET-23d+ DHFR-TS (lane 2) prior to purification on the nickel column. Rue10(DE3)pET-23d+ DHFR-TS lysates were tested for TS activity before (lane 4) and after (lane 1) purification on the nickel column. GS245(DE3)pLysSpET-9d+NhisSHMT lysates were tested for TS activity before (lane 5) and after (lane 3) purification on the nickel column.