Abstract
Sirtuin activators, including small molecules such as polyphenols and resveratrol, are much desired due to their potential to ameliorate metabolic disorder and delay or prevent aging. In contrast, recent studies demonstrate that targeted silencing of sirtuin 1 (SIRT1) expression or activity by the deleted in breast cancer 1 (DBC1) may be beneficial by promoting p53-induced apoptosis in cancer cells, and by sensitizing cancerous cells to radiation therapy. Negative SIRT1 regulation also alleviates gene-repression associated with fragile X mental retardation syndrome. The targeted activation or inhibition of SIRT1 activity therefore emerges as a critical point of regulation in disease pathogenesis.
Sirtuin 1 (SIRT1), mammalian homologue of the silencing information regulator 2 (Sir2) yeast gene, is a class III nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylase that is conserved across species and plays an important role in extending lifespan [1]. Located within the nucleus, SIRT1 regulates cell survival by modulating cellular stress, metabolic pathways and apoptosis. Although the numerous targets and functions of SIRT1 are constantly being unraveled (as summarized in Figure 1 and reviewed in [2–6]), cell survival was initially demonstrated to involve the SIRT-associated deacetylation of tumor suppressor gene, p53 [7–9]. Intriguingly, overexpression of SIRT1 was observed in colon, prostate, breast [10] and lung [11] cancers, and was correlated with silenced tumor suppressor genes (TSGs) [12] as well as resistance to chemotherapy and ionizing radiation [13,14]. Knockdown of SIRT1 expression induced a p53-dependent apoptosis in response to DNA damage, whereas inhibition of SIRT1 deacetylating activity by small molecule inhibitors was insufficient to induce apoptosis, suggesting that catalytic activity alone may not be adequate to influence carcinogenesis [15]. It was therefore hypothesized that the down-regulation of SIRT1 would be beneficial for cancer treatment and prevention.
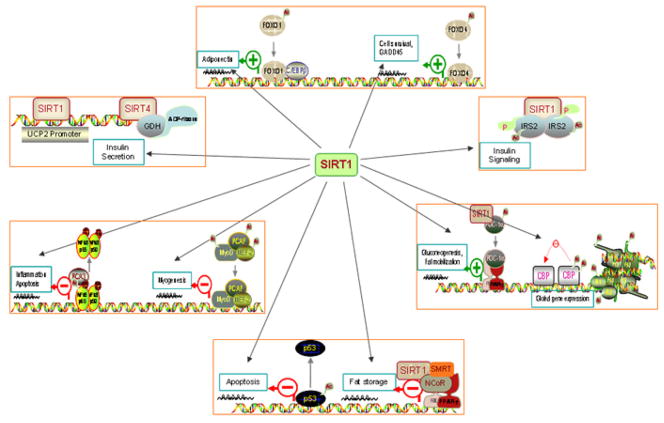
Figure 1. Targets and functions of SIRT1.
Numerous studies have now unraveled the role of SIRT1 not only in histone deacetylation, but also in the deacetylation and ADP-ribosylation of proteins involved in cell survival (p53, FOXO), adipogenesis (PPARγ), glucose and lipid metabolism (GDH, IRS2, PGC1-α, UCP2), peripheral insulin resistance (PTP-1B, adiponectin), inflammation and apoptosis (NF-κB) as well as myogenesis (MyoD).
Promoter hypermethylation, critical in gene silencing, is implicated in the regulation of genes which are involved in cellular pathways such as cell cycle regulation (CDKN2B, CDKN2A, TP73, PTEN, NES-1, LATS-1), apoptosis (p14, ASPP-1, APAF1) or cell adhesion (CDH13, CDH1; please see [16] and references within). The deleted in breast cancer 1 (DBC1) gene was identified as a potential tumor suppressor gene that is either found hypermethylated or deleted in cancer [16–18], and is associated with antiproliferative properties leading to cell cycle arrest in the G1 phase [19,20]. In the January 2008 issue of Nature Zhao et al. demonstrated an association between p53-mediated apoptosis and the inhibition of SIRT1 by DBC1 [21]. Using affinity chromatography purification and mass spectrometry, they identified SIRT1-containing protein complexes from native HeLa cells and demonstrated that DBC1 was not only strongly associated with SIRT1 but also co-immunoprecipitated with it [21]. The importance of DBC1-SIRT1 interactions was further emphasized by the fact that the p53 deacetylating capacity of Fag-tagged SIRT1 is strongly repressed by Flag-tagged DBC1 in a dose dependent manner, suggesting that DBC1 is a negative regulator of SIRT1 activity [21]. RNAi knockdown of SIRT1 abolished the protein-protein binding of DBC1 and increased both p53 acetylation and BAX expression, whereas RNAi knockdown of DBC1 expression did not affect p53 stability, but significantly reduced the expression of its downstream transcriptional targets, PUMA and BAX [21]. The inhibition of DBC1 expression by RNAi also rendered the DNA-damaged human osteosacrcoma cells (U2OS ) resistant to p53-mediated apoptosis, but otherwise had no effect upon the deacetylation of p53 by histone deacetylase complex 1 (HDAC1 [21]).
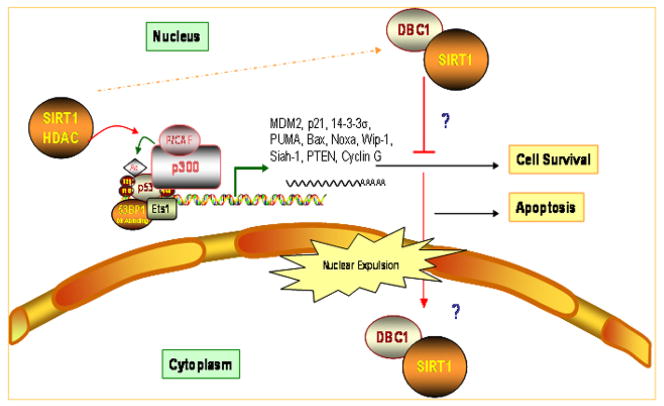
Within the same issue of Nature, Kim et al. concurrently demonstrated a co-immunoprecipitation of DBC1 with SIRT1 in 293T cells and an associated inhibition of SIRT activity [22]. Both Zhao et al. [21] and Kim et al. [22] demonstrate that DBC1 specifically binds to SIRT1, but not SIRT6 or SIRT7, which co-localize in the nucleus with SIRT1. Regardless of its nuclear localization, SIRT1 is subject to nucleocytoplasmic shuttling, and the nuclear localization of SIRT1 was associated with the inhibition of oxidative stress-induced cell death in C2C12 mouse muscle cells [23]. In contrast, the nuclear expulsion and cytoplasmic localization of SIRT1 appears critical in muscle development through an amelioration of MyoD repression [24]. Studies by Zhao et al. were conducted using whole cell extracts and therefore the precise location of DBC1-SIRT1 interactions is unclear [21]. Similar to SIRT1, DBC1 is also localized to the nucleus, but upon cleavage by caspases becomes localized within the cytoplasm and functions to promote apoptosis [25]. Is it then possible that DBC1-SIRT interactions are cytoplasmic, as SIRT1 is highly expressed within the cytoplasm of cancerous cells [15]? If so, future studies are warranted to identify the triggers that co-ordinately sequester DBC1 and SIRT1 within the cytoplasm (Figure 2). The binding of DBC1 to SIRT1 was abolished by deleting the leucine zipper motif of DBC1 and the catalytic domain of SIRT1, implicating the importance of these two regions in the regulation of SIRT1 by DBC1 [22]. Moreover, co-expression of DBC1 significantly reduced the binding of SIRT1 to its substrates, p53 and forkhead box transcription factor (FOXO) by blocking substrate access [22]. Functional studies reveal that genotoxic stress stimulates DBC1 binding to SIRT1 and increased expression of FOXO target genes, manganese superoxide dismutase (MnSOD) and Gadd45α, with a concomitant reduction in Bim expression [22]. It is also predicted that the Nudix hydroxylase domain of DBC1 may bind NAD metabolites such as ADP-ribose [25]. Overall, although inhibition of apoptosis in response to cellular oxidative stress is critical to retard/prevent ageing, the promotion of apoptosis is also desirable to prevent carcinogenesis.
Figure 2. Depicts the inhibitory effects of protein-protein interaction between DBC1 and SIRT1.
SIRT1 is highly expressed in the cytoplasm of cancer cells, while nuclear expulsion of DBC1 induces apoptosis. Binding of DBC1 to SIRT catalytic domain was demonstrated to induce apoptosis in cancer cells and sensitize them to ionizing radiation. It is however unknown whether binding of DBC1 triggers cytoplasmic translocation of SIRT or whether the nuclear export of the DBC1/SIRT1 complex is triggered by factors regulating DBC1.
To recapitulate, recent studies by Zhao et al. [21] and Kim et al. [22] appear to support the old adage that ‘one man’s food is another man’s poison’. The very unique feature and ability of SIRT1 to promote cell survival has proven detrimental to cancerous cells, as SIRT1 overexpression is often associated with deacetylation of TSG, p53, leading to survival of tumor cells and possible resistance to cancer therapy [15,16,26]. New studies reveal an additional function of p53 in regulating not only mitochondrial respiration, but also secondary changes in glycolysis and glucose metabolism [27,28]. Since SIRT1 also regulates glucose metabolism and p53 functions [29–31], it would be of further interest to identify the exact role of SIRT1/p53 in the regulation of glucose metabolism and apoptosis in tumor cells. Calorie restriction (CR) is a robust phenomenon and is the only known intervention which increases lifespan across species via an augmentation of SIRT1 activity [32–35]. Long-term CR is an extremely challenging task, and therefore the quest for CR mimetic and SIRT activators has captured undivided attention from both the scientific and pharmaceutical communities. SIRT seems to stand at the nexus of two cross roads: cellular senescence, a potent mechanism of tumor suppression; and cell survival or retardation of apoptosis involved in aging. It is therefore of concern that ubiquitous up-regulation may not be always desirable, as exemplified further by the fact that inhibition of SIRT1 alleviates gene silencing detrimental in diseases such as fragile X mental retardation syndrome [36], and various models of neurodegeneration including Parkinson’s disease [37]. In summary, it seems that it is not only SIRT1 activators, but also inhibitors, which offer therapeutic potential in the treatment of human health and disease.
Acknowledgments
This work was supported partially by U.S. Public Health Service grants from the National Institutes of Health (NIH), National Center for Complementary and Alternative Medicine (NCCAM, R21AT003719), Research Centers in Minority Institutions (RCMI, G12 RR003061), National Center for Research Resources (NCRR, P20 RR011091), National Center on Minority Health and Health Disparities (NCMHD, P20 MD 000173) and the Hawaii Community Foundation (20041652).
Abbreviations
- CR
calorie restriction
- DBC1
deleted in breast cancer 1
- FOXO
Forkhead box O
- GDH
glutamate dehydrogenase
- IRS2
insulin receptor substrate-2
- NAD
nicotinamide adenine dinucleotide
- PPARγ
peroxisome proliferator-activated receptor gamma
- PGC1-α
PPAR coordinator 1α
- TSG
tumor suppressor gene
- UCP2
uncoupling protein 2
References
- 1.Porcu M, Chiarugi A. The emerging therapeutic potential of sirtuin-interacting drugs: from cell death to lifespan extension. Trends Pharmacol Sci. 2005;26:94–103. doi: 10.1016/j.tips.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol Endocrinol. 2007;21:1745–55. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- 3.Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann Med. 2007;39:335–45. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- 4.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–21. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 5.Anastasiou D, Krek W. SIRT1: linking adaptive cellular responses to aging-associated changes in organismal physiology. Physiology (Bethesda) 2006;21:404–10. doi: 10.1152/physiol.00031.2006. [DOI] [PubMed] [Google Scholar]
- 6.Nerurkar PV, Nerurkar VR. Respected Sir(2): Magic target for diabetes. CellScience. 2008:4. [PMC free article] [PubMed] [Google Scholar]
- 7.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–48. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 8.Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. Embo J. 2002;21:2383–96. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith J. Human Sir2 and the ‘silencing’ of p53 activity. Trends Cell Biol. 2002;12:404–6. doi: 10.1016/s0962-8924(02)02342-5. [DOI] [PubMed] [Google Scholar]
- 10.Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859–64. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu F, Chou PM, Zheng X, Mirkin BL, Rebbaa A. Control of multidrug resistance gene mdr1 and cancer resistance to chemotherapy by the longevity gene sirt1. Cancer Res. 2005;65:10183–7. doi: 10.1158/0008-5472.CAN-05-2002. [DOI] [PubMed] [Google Scholar]
- 14.Matsushita N, Takami Y, Kimura M, Tachiiri S, Ishiai M, Nakayama T, Takata M. Role of NAD-dependent deacetylases SIRT1 and SIRT2 in radiation and cisplatin-induced cell death in vertebrate cells. Genes Cells. 2005;10:321–32. doi: 10.1111/j.1365-2443.2005.00836.x. [DOI] [PubMed] [Google Scholar]
- 15.Stunkel W, Peh BK, Tan YC, Nayagam VM, Wang X, Salto-Tellez M, Ni B, Entzeroth M, Wood J. Function of the SIRT1 protein deacetylase in cancer. Biotechnol J. 2007;2:1360–8. doi: 10.1002/biot.200700087. [DOI] [PubMed] [Google Scholar]
- 16.San Jose-Eneriz E, Agirre X, Roman-Gomez J, Cordeu L, Garate L, Jimenez-Velasco A, Vazquez I, Calasanz MJ, Heiniger A, Torres A, Prosper F. Downregulation of DBC1 expression in acute lymphoblastic leukaemia is mediated by aberrant methylation of its promoter. Br J Haematol. 2006;134:137–44. doi: 10.1111/j.1365-2141.2006.06131.x. [DOI] [PubMed] [Google Scholar]
- 17.Nishiyama H, Hornigold N, Davies AM, Knowles MA. A sequence-ready 840-kb PAC contig spanning the candidate tumor suppressor locus DBC1 on human chromosome 9q32-q33. Genomics. 1999;59:335–8. doi: 10.1006/geno.1999.5891. [DOI] [PubMed] [Google Scholar]
- 18.Habuchi T, Luscombe M, Elder PA, Knowles MA. Structure and methylation-based silencing of a gene (DBCCR1) within a candidate bladder cancer tumor suppressor region at 9q32-q33. Genomics. 1998;48:277–88. doi: 10.1006/geno.1997.5165. [DOI] [PubMed] [Google Scholar]
- 19.Izumi H, Inoue J, Yokoi S, Hosoda H, Shibata T, Sunamori M, Hirohashi S, Inazawa J, Imoto I. Frequent silencing of DBC1 is by genetic or epigenetic mechanisms in non-small cell lung cancers. Hum Mol Genet. 2005;14:997–1007. doi: 10.1093/hmg/ddi092. [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama H, Gill JH, Pitt E, Kennedy W, Knowles MA. Negative regulation of G(1)/S transition by the candidate bladder tumour suppressor gene DBCCR1. Oncogene. 2001;20:2956–64. doi: 10.1038/sj.onc.1204432. [DOI] [PubMed] [Google Scholar]
- 21.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–90. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 23.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–32. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 24.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 25.Anantharaman V, Aravind L. Analysis of DBC1 and its homologs suggests a potential mechanism for regulation of Sirtuin domain deacetylases by NAD metabolites. Cell Cycle. 2008:7. doi: 10.4161/cc.7.10.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim CS. SIRT1: tumor promoter or tumor suppressor? Med Hypotheses. 2006;67:341–4. doi: 10.1016/j.mehy.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 27.Ma W, Sung HJ, Park JY, Matoba S, Hwang PM. A pivotal role for p53: balancing aerobic respiration and glycolysis. J Bioenerg Biomembr. 2007;39:243–246. doi: 10.1007/s10863-007-9083-0. [DOI] [PubMed] [Google Scholar]
- 28.Bensaad K, Vousden KH. p53: new roles in metabolism. Trends in cell biology. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Tikoo K, Tripathi DN, Kabra DG, Sharma V, Gaikwad AB. Intermittent fasting prevents the progression of type I diabetic nephropathy in rats and changes the expression of Sir2 and p53. FEBS Lett. 2007;581:1071–8. doi: 10.1016/j.febslet.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Yang T, Fu M, Pestell R, Sauve AA. SIRT1 and endocrine signaling. Trends Endocrinol Metab. 2006;17:186–91. doi: 10.1016/j.tem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–8. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 32.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–2. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 33.Couzin J. Research on aging. Gene links calorie deprivation and long life in rodents. Science. 2004;304:1731. doi: 10.1126/science.304.5678.1731a. [DOI] [PubMed] [Google Scholar]
- 34.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–6. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guarente L, Picard F. Calorie restriction--the SIR2 connection. Cell. 2005;120:473–82. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 36.Biacsi R, Kumari D, Usdin K. SIRT1 Inhibition Alleviates Gene Silencing in Fragile X Mental Retardation Syndrome. PLoS Genet. 2008;4:e1000017. doi: 10.1371/journal.pgen.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, Young AB, Abagyan R, Feany MB, Hyman BT, Kazantsev AG. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317:516–9. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]