Abstract
Aberrant signaling through G-protein coupled receptors promotes metastasis, the major cause of breast cancer death. We identified regulator of G-protein signaling 4 (RGS4) as a novel suppressor of breast cancer migration and invasion, important steps of metastatic cascades. By blocking signals initiated through Gi-coupled receptors, such as protease-activated receptor 1 and CXC chemokine receptor 4, RGS4 disrupted Rac1-dependent lamellipodia formation, a key step involved in cancer migration and invasion. RGS4 has GTPase-activating protein (GAP) activity, which inhibits G-protein coupled receptor signaling by deactivating G-proteins. An RGS4 GAP-deficient mutant failed to inhibit migration and invasion of breast cancer cells in both in vitro assays and a mouse xenograft model. Interestingly, both established breast cancer cell lines and human breast cancer specimens showed that the highest levels of RGS4 protein were expressed in normal breast epithelia and that RGS4 down-regulation by proteasome degradation is an index of breast cancer invasiveness. Proteasome blockade increased endogenous RGS4 protein to levels that markedly inhibit breast cancer cell migration and invasion, which was reversed by an RGS4-targeted short hairpin RNA. Our findings point to the existence of a mechanism for posttranslational regulation of RGS4 function, which may have important implications for the acquisition of a metastatic phenotype by breast cancer cells. Preventing degradation of RGS4 protein should attenuate aberrant signal inputs from multiple Gi-coupled receptors, thereby retarding the spread of breast cancer cells and making them targets for surgery, radiation, and immune treatment.
Introduction
Breast cancer remains the second largest killer of women by cancer (1). Patients who cannot be cured are those in whom breast cancer has metastasized, which refers to the migration and invasion of breast cancer cells to other organs such as lung and bone. Evidence indicates that chemoattractants such as chemokines, growth factors, and matrix metalloproteases are important in breast cancer migration and invasion during metastasis (2).
These ligands bind to membrane receptors on cancer cells, including G-protein coupled receptors (GPCR; ref. 3) and receptor tyrosine kinases (RTK; refs. 4, 5), thereby activating signaling pathways that regulate actin cytoskeleton dynamics, critical to breast cancer metastasis (6, 7). Knowledge of activation mechanisms and signaling pathways controlled by RTKs has allowed the development of target-specific drugs and new therapies for breast cancer (8). In contrast, knowledge of the GPCR-dependent signaling pathways involved in acquisition of migratory and invasive abilities by breast cancer cells lags far behind, which presents a continuing therapeutic challenge.
Evidence from preclinical and clinical studies shows excessive activation of GPCRs in breast cancer due to overexpression of receptors (3) and/or abnormally elevated ligands for GPCRs (9–11). Signals initiated by various activated GPCRs, including protease-activated receptor 1 (PAR1) and CXC chemokine receptor 4 (CXCR4), drive breast cancer cells to migrate and invade through surrounding tissues and spread to target organs (3). Despite the potential importance of GPCRs in breast cancer metastasis, there have been few clinical trials of drugs directly inhibiting GPCRs in breast cancer treatment. The strategy of inhibiting a single GPCR is limited, in part, because breast cancer metastasis could be driven by several different GPCRs simultaneously, thereby having metastatic signal redundancy. Thus, identifying molecules responsible for regulating signals initiated by many GPCRs could be an effective, alternative strategy for breast cancer treatment.
The regulator of G-protein signaling (RGS) family of proteins modulates GPCR-mediated responses in cells (12). About 30 mammalian RGS proteins have been identified (13). All share a conserved RGS domain with GTPase-activating protein (GAP) activity that profoundly inhibits downstream consequences of GPCR activation by increasing the rate of signal-terminating GTP hydrolysis of G-proteins (13). Dysfunction of RGS proteins has been reported in various human diseases (14–16). There have been a few studies implicating RGS regulation of GPCR signaling in metastatic cancer cells (17–19) and we recently reported that down-regulation of RGS2 facilitates prostate cancer progression (20). However, the pathophysiologic roles of RGS proteins in cancer metastasis remain uncertain.
The initial purpose of this study was to determine whether RGS proteins play a role in regulating breast cancer migration and invasion, important steps of cancer metastasis. We found that RGS4, but not a RGS4 GAP-deficient mutant or other RGS proteins, inhibited breast cancer metastatic abilities in vitro and in an orthotopic xenograft mouse model, suggesting that RGS4 is a suppressor of breast cancer metastasis. We therefore investigated the underlying mechanisms by examining signaling pathways regulated by RGS4. RGS4 attenuated signaling initiated by multiple Gi-coupled receptors, which inhibited Rac1-dependent lamellipodia formation, a key structure for cancer cell migration and invasion (21). More importantly, our data suggest that proteasome degradation of RGS4 protein promotes breast cancer cell migration and invasion.
Materials and Methods
Cells and reagents
MDA-MB-231 and MDA-MB-436 were from the American Type Culture Collection (ATCC) and cultured in DMEM, 10% fetal bovine serum (FBS). MCF-7 cells (ATCC) were cultured in IMEM, 10% FBS, and 10 μ/mL insulin. MCF-10A cells (ATCC) were grown in MEBM with additives (ATCC).
Cell-permeable C3 transferase and F-actin visualization kit (Cytoskeleton); NSC23766, MG132, PSI, and chloroquine (Calbiochem); epidermal growth factor (EGF) and fibronectin (BD Biosciences); CXCL12 (R&D Systems); BMS-200261 and TFLLR (Peptides International); SCH79797 (Tocris); cycloheximide (Sigma); rabbit anti-PAR1 antibody (Santa Cruz Biotechnology); and monoclonal anti-hemagglutinin (HA) antibody (Covance). Rabbit anti-RGS4 and anti-Gαq antibodies were gifts from Susanne Mumby and Paul Sternweis (University of Texas Southwestern Medical Center), respectively. RGS4 GAP-deficient N128A mutant was generated from pcDNA3.1-RGS4 (gift from John Hepler, Emory University) with Stratagene QuikChange mutagenesis kit. Plasmids encoding other HA-tagged RGS proteins were described previously (20). Recombinant untagged RGS4 protein was purified from Escherichia coli cells (22).
Quantitative real-time PCR analysis
Primers and procedures for quantitative real-time PCR of the RGS4 gene were described previously (20). β-Actin was the internal control.
Immunohistochemistry analysis of human breast tissues
Formalin-fixed, paraffin-embedded blocks were collected from breast cancer patients by the Creighton University Department of Pathology, approved by the Creighton University Institutional Review Board. Immunohistochemistry was performed using standard techniques. Negative controls in each experiment used anti-RGS4 antibody preblocked by purified RGS4 protein. Levels of RGS4 protein were measured on immunohistochemically stained tissue sections using the Automated Cellular Imaging System (Chroma-Vision Medical Systems; ref. 23). A “histo-score” (H-score) was calculated by multiplying the percentage (P) of positive cells by the average intensity (I). Average H-scores from 10 randomly chosen 200× fields were used to grade RGS4 protein levels in each sample. The maximum H-score is 255 if all cells (P = 1.0) stained with maximal intensity (I = 255).
Rac and Rho activation assays
Active Rac or Rho was assayed using the Rac Activation Assay Kit or Rho Assay Reagent (Rhotekin RBD, agarose; Millipore). Western blotting of samples used anti-Rac1 (clone 23A8, Millipore) or anti-RhoA antibody (26C4, Santa Cruz Biotechnology).
Protein extraction, electrophoresis, and Western blot analysis
Protein was extracted from cells using 1× radioimmunoprecipitation assay lysis buffer (Santa Cruz Biotechnology), subjected to SDS-PAGE, and transferred to Immobilon-FL membrane (Millipore). Primary antibodies were used to detect specific protein and β-actin (see Cells and Reagents). Secondary antibodies were labeled with either IRDye700 or IRDye800 for detection with an Odyssey IR imaging system (LI-COR Biosciences).
RNA interference and stable cell lines
MDA-MB-231 cells were transfected with pRs plasmids encoding RGS4-specific shRNA (OriGene Technologies, TI339326) or control GFP-targeted shRNA. Transfected cells were selected with puromycin (1 μg/mL) for 3wk. Surviving transfected cells were verified for silencing of endogenous RGS4 by Western blot analysis. Stable MDA-MB-231 cell lines were established by a standard protocol using Lipofectamine 2000 (Invitrogen). Transfected cells were selected with G-418 (400 μg/mL) for 2 wk. Positive clones were selected and verified by Western blotting for COOH-terminal HA-tagged RGS4 or RGS4-N128A protein expression.
Cell invasion and migration assays
Matrigel invasion assays were performed at 37°C for 5 h using 24-well Transwell inserts (Corning) coated with 30 μg of Matrigel (BD Biosciences; ref. 9). Cells (50,000) suspended in 200 μL serum-free medium were seeded into the upper chamber and 600 μL chemoattractants in the lower chamber. Cells that migrated and invaded through the membrane were counted and normalized relative to 10,000 seeded cells. Transwell cell migration assays were performed similarly without Matrigel. Conditioned medium (CM) from NIH-3T3 mouse embryonic fibroblasts (ATCC) was collected (9) and used as a chemoattractant. To study the effect of RGS4 on the PAR1, CXCR4, or EGF receptor–dependent cell migration, the insert membrane was precoated with fibronectin (2 μg/mL). PAR1 agonist TFLLR, CXCR4 agonist CXCL12, or EGF was used as the chemoattractant. Cell migration was also examined by an in vitro wound-healing assay (24).
In vitro cell growth assay and orthotopic mammary fat pad model in nude mice
MDA-MB-231 cells were seeded into 24-well plates at a density of 1.5 × 104 per well in DMEM plus 10% FBS for 8 d until they reached confluence, and counted every 24 h using a Coulter Counter ZM (Coulter Electronics). Results represent an average of four independent experiments in triplicate. The in vivo experiments were performed using 6-wk-old athymic nude female mice (Charles River) following a protocol approved by the Creighton University Institutional Animal Care and Use Committee. MDA-MB-231 cells stably transfected with empty vector, RGS4, or RGS4-N128A mutant were injected into an inguinal mammary fat pad (5 × 106 cells in 100 μL Matrigel, 12 mg/mL). Tumor dimensions were measured twice per week using a digital caliper and tumor volume was calculated using the ellipsoid formula. Primary tumors with adherent tissue were excised, fixed in 10% formalin/PBS, and embedded in paraffin. Tissue sections (4 μm) were prepared and stained with H&E. Tumor sections (at least three per mouse) were examined by Drs. Bo Wang and Caishu Deng, certified clinical pathologists, in a blinded manner to assess tumor invasiveness.
Fluorescence microscopy
F-actin was visualized with rhodamine-labeled phalloidin. Cortactin was visualized by anticortactin antibody (Millipore) followed by FITC-conjugated secondary antibody. Lamellipodia were identified as smoothly protruding regions along the edge of the cell with perpendicular phalloidin-stained F-actin. The total cell periphery and cell periphery occupied by lamellipodia were outlined and measured in length using Image-Pro Plus. The summed length of lamellipodia was expressed as a percentage of total cell circumference (25).
Measurement of intracellular Ca2+
Fura-2–loaded cells on glass coverslips were excited with light from a DeltaRAM X monochronometer (Photon Technology International). Paired fluorescent images of cells were captured by a CoolSNAP HQ2 camera on a Nikon microscope with the intracellular Ca2+ concentration shown as the ratio of fluorescence intensities (500-nm emission) at the excitation of 340 nm compared with 380 nm. Data are averages of 50 cells.
Statistical analysis
Results are the mean ± SE of at least three determinations and statistical comparisons used a Student's t test or the two-way ANOVA with a Bonferroni correction where there were multiple comparisons. A probability (P) value of <0.05 was considered significant.
Results
RGS4 selectively inhibits breast cancer cell migration
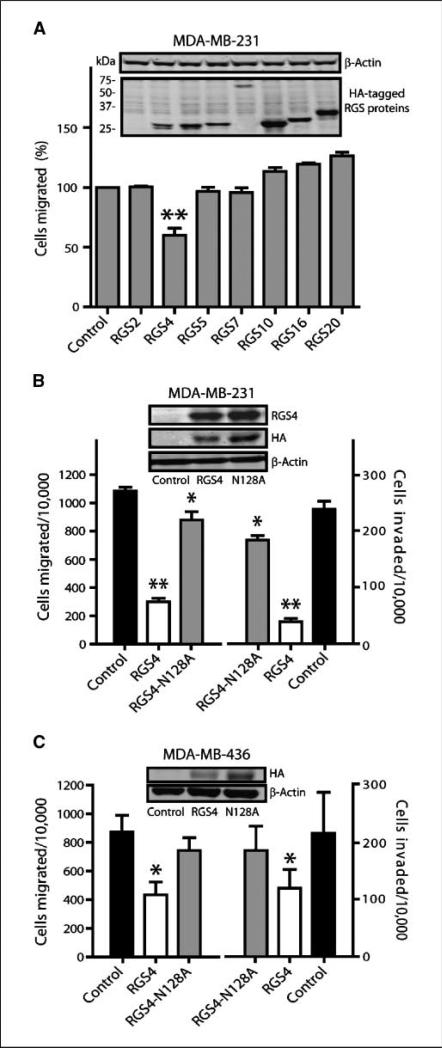
We first examined transient expression (∼50% transfection efficiency) of seven HA-tagged RGS proteins, representing four subgroups of the mammalian RGS gene family (13), on migration of metastatic breast cancer MDA-MB-231 cells. The chemoattractant was NIH-3T3 fibroblast conditioned medium (CM), widely used in in vitro cancer metastasis assays (26). Although expression of RGS4 protein (lane 3 of Fig. 1A, inset) was similar to other RGS proteins, only RGS4 caused significant inhibition of MDA-MB-231 cell migration (40%, Fig. 1A).
Figure 1.
Exogenously expressed RGS4 protein inhibits breast cancer cell migration and invasion. Columns, mean; bars, SE with *, P < 0.05 and **, P < 0.01 compared with control cells. A, comparison of Transwell migration of MDA-MB-231 cells transfected with HA-tagged RGS proteins normalized to vector controls (n = 6, 100% = 755 ± 98 per 10,000 seeded cells). Transwell migration and invasion of MDA-MB-231 cells stably expressing RGS4 or RGS4-N128A (B, n = 5) or MDA-MB-436 cells transiently expressing RGS4 or RGS4-N128A (C, n = 3). Insets, Western blot of HA-tagged RGS proteins and β-actin.
GAP activity is required for RGS4 to inhibit breast cancer cell migration and invasion
We next determined whether this inhibitory effect of RGS4 was due to deactivation of G-proteins through its GAP activity. Stable MDA-MB-231 cell clones expressing either RGS4 or a GAP-deficient RGS4-N128A mutant (27) were generated. All three of the RGS4-expressing clones had reduced cell migration compared with control cells or cells expressing RGS4-N128A (Supplementary Fig. S1). We chose control (vector) clone 1, RGS4-N128A clone 36, and RGS4 clone 8 for further study. Although the expression level of RGS4-N128A was similar to that of RGS4 (Fig. 1B, inset), expression of RGS4 decreased migration and invasion by 75% and 85%, respectively, whereas expression of RGS4-N128A caused only a 14% reduction (Fig. 1B). Those results suggest that RGS4 is more potent than its GAP-deficient mutant in MDAMB-231 cells and were further supported by data from a wound-healing assay (Supplementary Fig. S1C). A similar dependence on GAP activity was seen in metastatic breast cancer MDA-MB-436 cells when RGS4 or RGS4-N128A was transiently expressed (∼50% transfection efficiency; Fig. 1C).
RGS4 inhibits tumor growth and suppresses tumor invasiveness in nude mice
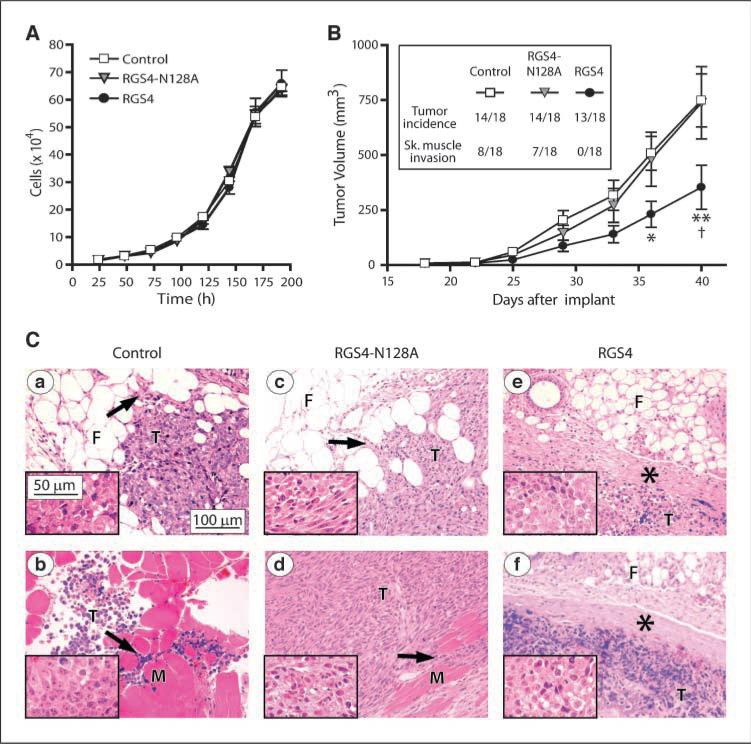
We compared the growth characteristics of MDA-MB-231 control cells and cells stably expressing RGS4 or its N128A mutant in culture and in vivo. Under identical culture conditions, these cell variants exhibited similar growth rates, with doubling times of 30.1 ± 2.1, 29.6 ± 2.4, and 29.2 ± 2.6 hours for control, RGS4, and RGS4-N128A–expressing cells, respectively (Fig. 2A). However, whereas control MDA-MB-231 cells and RGS4-N128A cells implanted in the mammary fat pad of nude mice tended to produce tumors of similar sizes over time, tumors derived from RGS4 cells grew more slowly (Fig. 2B) but with similar tumor incidence (Fig. 2B, inset). Approximately half of the tumors derived from control cells and RGS4-N128A cells, but none from RGS4 cells, also had grossly apparent skeletal muscle invasion when harvested (Fig. 2B, inset). Figure 2C shows representative images of orthotopic tumors. The tumors were masses composed of pleomorphic neoplastic cells with patchy areas of tumor necrosis. In control cell tumors (Fig. 2Ca, b), the borders of the tumor (T) had infiltrated into the surrounding mammary fibroadipose tissue (F) and skeletal muscle (M; see arrows in Fig. 2Ca, b, respectively). Tumors derived from RGS4-N128A cells exhibit the same morphology and histopathology as the control tumors (Fig. 2Cc, d) with both mammary fat pad and skeletal muscle invasion. However, tumors derived from cells expressing RGS4 were quite different. In all instances, they formed a well-demarcated tumor mass (see asterisks,Fig.2Ce, f) that was easily dissected free of the abdominal skeletal muscle.
Figure 2.
RGS4 inhibits breast tumor growth and suppresses tumor invasion in nude mice. A, in vitro growth of MDA-MB-231 control cells or cells stably expressing RGS4 or RGS4-N128A. Points, mean (n = 4 triplicate experiments); bars, SE. B, in vivo tumor growth derived from MDA-MB-231 cell variants injected into mouse mammary fat pads (n = 18 mice per group; inset, tumor incidence and skeletal muscle invasion). Points, mean for formed tumors; bars, SE. *, P < 0.05 and **, P < 0.001 compared with control, †, P < 0.001 compared with RGS4-N128A using two-way ANOVA with Bonferroni comparison of means. C, representative H&E-stained micrographs of tumor tissue from mice injected with MDA-MB-231 control cells showing (a) mammary fat pad and (b) skeletal muscle invasion, RGS4-N128A–expressing cells showing (c) mammary fat pad and (d) skeletal muscle invasion, and RGS4-expressing cells showing well demarcated tumor boundaries (e and f, asterisks). Arrows, infiltration of the tumor cells (T) into the fibroadipose tissue (F) or skeletal muscle (M). Insets, higher magnification images of the tumor cells.
RGS4 blocks lamellipodia formation by inhibiting Rac1 activation in breast cancer cells
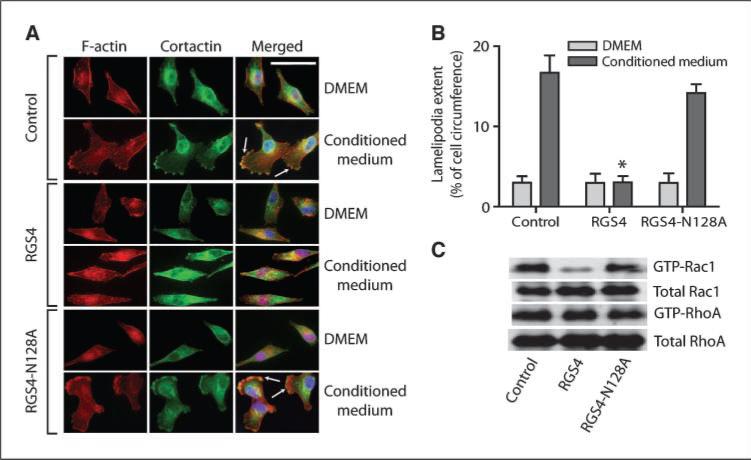
GPCR activation triggers actin cytoskeleton reorganization to form lamellipodia (28), the flattened F-actin–rich leading edge of migrating cells (21). Interestingly, RGS4, but not RGS4-N128A, inhibited NIH-3T3 CM-induced lamellipodia formation (Fig. 3A, B) and translocation of cortactin from cytosol to lamellipodia in MDA-MB-231 cells (Fig. 3A). These processes are regulated by active Rac, a member of the Rho family of GTPases (29), previously implicated in breast cancer metastasis (30). Therefore, we examined effects of RGS4 on Rac activities in breast cancer cells.
Figure 3.
RGS4 selectively inhibits Rac-activated lamellipodia formation in breast cancer cells. A and B, RGS4 blocks lamellipodia formation and the translocation of cortactin to lamellipodia by inhibiting Rac activity in MDA-MB-231 cells. Arrows, lamellipodia at cell edges (A). Scale bar, 50 μm. B, lamellipodial extent at cell leading edges was quantified as fraction of cell circumference on 20 randomly selected cells in each group. Columns, means; bars, SE (n = 3), *, P < 0.001 compared with control. C, representative image of Western blot of total (10%) and active GTP-bound Rac1 and RhoA (n = 3) in MDA-MB-231 control cells and cells expressing RGS4 or RGS4-N128A.
MDA-MB-231 cells stably expressing RGS4 had significantly lower levels of activated GTP-bound Rac1 when compared with control cells or cells expressing RGS4-N128A (Fig. 3C). In contrast, little difference was observed in the activity of RhoA, another member of the Rho family (Fig. 3C). Interestingly, 100 μmol/L NSC23766, a widely used concentration to block Rac1 activation (31), also reduced MDA-MB-231 cell migration and invasion by more than 80% and 70%, respectively, whereas cell-permeable C3 transferase, a Rho inhibitor (32), only caused 25% inhibition (Supplementary Fig. S2).
RGS4 inhibits migration of breast cancer cells by attenuating Gi-coupled receptor–mediated signal transduction
Figure 4A shows that pretreatment of MDA-MB-231 cells with PAR1 peptide antagonist BMS-200261 (50 μmol/L) inhibited NIH-3T3 CM-induced cell migration and invasion by more than 70%. A PAR1 nonpeptide antagonist, SCH79797, also dose-dependently blocked CM-induced cell migration with maximum inhibition of 80% (IC50 = 185 ± 45 nmol/L, inset). These results are consistent with a previous report showing the importance of PAR1 activation in breast cancer metastasis (9). Activated PAR1 couples to Gi, Gq, and/or G12/13 families of G-proteins (33). We found that the Gi inhibitor pertussis toxin blocked CM-stimulated MDA-MB-231 cell migration and invasion by 45% and 60%, respectively. Our data suggest that NIH-3T3 CM induces breast cancer cell migration and invasion through activation of PAR1 in a Gi-dependent manner. Therefore, we investigated RGS4-mediated regulation of a PAR1-dependent increase in intracellular Ca2+ in MDA-MB-231 cells.
Figure 4.
RGS4 inhibits PAR1 and CXCR4 signal transduction and cell migration. A, NIH-3T3 CM stimulates migration and invasion of MDA-MB-231 cells through Gi-coupled receptor PAR1. Cells were pretreated with or without pertussis toxin (PTX; 100 ng/mL, 18 h) or PAR1 antagonists BMS-200261 (BMS,50 μmol/L) or SCH79797 (inset) for 10min. Columns, means; bars, SE (n = 3), *, P <0.05. B, PAR1 agonist TFLLR increases intracellular Ca2+ in a Gi-dependent manner (top), which is attenuated by RGS4 GAP activity (bottom). Inset, Western blot of PAR1 in MDA-MB-231 cells. Cells pretreated with or without pertussis toxin and subjected to F-actin staining (scale bar, 25 μm; C) or Transwell migration assays (D) in response to TFLLR (25 μmol/L), CXCL12 (30ng/mL), or EGF (5 ng/mL). Columns, means; bars, SE (n = 4).
Figure 4B (top) shows that the PAR1 agonist TFLLR (25 μmol/L) caused a transient increase in intracellular Ca2+ in MDA-MB-231 cells that was inhibited by 1 μmol/L SCH79797 or pertussis toxin, indicating a Gi-coupled PAR1-dependent response. Stable expression of RGS4 reduced the PAR1-stimulated Ca2+ peak by more than 50%, whereas RGS4-N128A only attenuated the Ca2+ response by 10% (Fig. 4B, bottom) when PAR1 protein levels were similar (Fig. 4B, inset).
TFLLR also induced lamellipodia formation (Fig. 4C) and caused a 3-fold increase in MDA-MB-231 cell migration (Fig. 4D), which was largely blocked by pertussis toxin. Stable expression of RGS4, but not RGS4-N128A, significantly attenuated TFLLR-induced lamellipodia formation and migration of MDA-MB-231 cells (Fig. 4C, D).
CXCR4 plays an important role in breast cancer metastasis (34, 35) and initiates signaling through Gi proteins when its ligand, CXCL12, is bound (36). Indeed, CXCL12 induced lamellipodia formation (Fig. 4C) and increased MDA-MB-231 cell migration by 2.3-fold (Fig. 4D) in a Gi-dependent manner. As expected, CXCL12-stimulated responses were blocked by RGS4 but not RGS4-N128A. In contrast, EGF-stimulated lamellipodia formation and migration of MDA-MB-231 cells, processes not dependent on G-proteins, were not affected by pertussis toxin and RGS4 (Fig. 4C and D). Thus, RGS4 functions as a specific regulator of signals initiated by at least two metastasis-associated Gi-coupled receptors.
Reduced RGS4 protein levels in human breast carcinomas
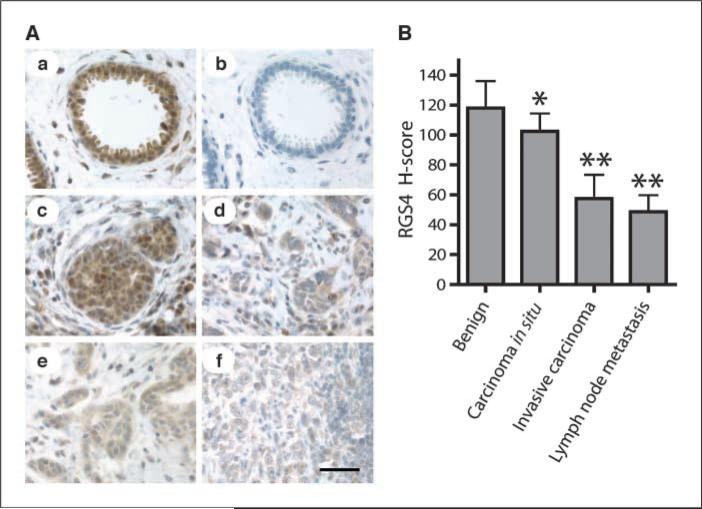
We performed immunohistochemical analysis of RGS4 protein levels in human breast cancer specimens using a well-characterized RGS4-specific antibody (37). In benign breast tissue, RGS4 staining was dense and localized predominantly in luminal epithelial cells (Fig. 5Aa), which was completely blocked by preincubation of the antibody with purified recombinant RGS4 (Fig. 5Ab). There was a small reduction in RGS4 staining in noninvasive ductal carcinoma in situ (Fig. 5Ac). In contrast, immunostaining of RGS4 protein was markedly decreased in invasive breast carcinoma (Fig. 5Ad, e) and metastases to regional lymph nodes (Fig. 5Af). Average H-scores (23), an index of RGS4 protein levels in tissues from nine breast cancer patients, are shown in Fig. 5B. The H-score of invasive breast carcinoma and lymph node metastases was statistically significantly reduced by 50% to 60% compared with nearby cells of benign breast tissue, but was only 13% lower in noninvasive ductal carcinoma in situ. These data show down-regulation of RGS4 protein in human breast cancer, which is proportional to the degree of invasiveness of these cancers.
Figure 5.
Reduction of RGS4 protein in human breast carcinomas. A, sections of breast tissue stained for RGS4 protein with anti-RGS4 antibody. Scale bar, 50 μm. a, benign breast tissue; b, benign breast tissue with antibody preblocked by RGS4 protein; c, ductal carcinoma in situ; d and e, invasive carcinoma; f, breast cancer metastasis in lymph nodes. B, RGS4 protein levels in human breast tissues (n = 9 patients) determined by H-score analysis of immunohistochemically stained tissue sections. Columns, mean; bars, SD. *, P < 0.05 and **, P < 0.001 compared with nearby benign breast tissue by paired t test.
Loss of RGS4 in metastatic breast cancer cells is due to proteasome degradation
RGS4 protein levels were much lower in metastatic MDA-MB-231 cells compared with immortalized human breast epithelial MCF-10A cells and nonmetastatic MCF-7 cells (Fig. 6A, inset). However, this reduction was not due to silencing of the RGS4 gene because RGS4 mRNA levels were increased by 1,900-fold in MDA-MB-231 cells (Fig. 6A). RGS4 is susceptible to degradation by the proteasome pathway (38). Accordingly, as shown in Supplementary Fig. S3A, RGS4 protein levels were significantly increased when MDA-MB-231 cells were exposed to proteasomal inhibitors MG132 or PSI, but not to the lysosomal inhibitor chloroquine (left). In contrast, RGS4 protein levels in MCF-10A cells were not influenced by these inhibitors (right). MG132 had no effect on endogenous levels of Gαq protein and inhibition of protein synthesis by cycloheximide abolished MG132-induced accumulation of RGS4 (Fig. 6B, inset). These results show that RGS4 levels are regulated by proteasome degradation. Interestingly, we found that COOH-terminal HA-tagged RGS4 is more stable than untagged or NH2-terminal HA-tagged protein when expressed in metastatic breast cancer cells (data not shown).
Figure 6.
Endogenous RGS4 protein inhibits breast cancer cell migration and invasion. Columns, mean; bars, SE with *, P < 0.01 and **, P < 0.001. A, quantitative real-time PCR analysis of RGS4 mRNA levels (n = 3). Inset, representative Western blot (n = 5) of RGS4 protein levels. MDA-MB-231 cell lysate plus 0.25 ng purified RGS4 protein as positive control. MDA-MB-231 cells (B) and MDA-MB-436 cells (C) treated for 4 h with or without MG132 (20 μmol/L) ± cycloheximide (CHX;0.1 mmol/L; n = 5) before Western blot analysis (insets) or Transwell migration and invasion assays. D, silencing endogenous RGS4 expression reverses the proteasome blockade-induced inhibition of MDA-MB-231 cell migration and invasion. Cells stably expressing RGS4 shRNA or GFP shRNA were treated with or without MG132 (n = 3) and then subjected to Western blot analysis (top) or Transwell migration and invasion assays (bottom). Untreated cells expressing GFP shRNA were set as 100%.
Proteasome blockade inhibits breast cancer cell migration and invasion
The MG132-induced increase of endogenous RGS4 protein was associated with inhibition of cell migration and invasion by more than 85% and 70% (Fig. 6B), respectively, without any significant effect on cell viability as assessed by trypan blue exclusion (data not shown). Cycloheximide alone inhibited cell migration and invasion by 50%, but adding MG132 did not cause additional inhibition when RGS4 protein synthesis was blocked by cycloheximide. Similarly, MG132 increased RGS4 levels 6-fold in MDA-MB-436 cells (Fig. 6C, inset), and blocked cell migration and invasion by 70% (Fig. 6C). Cycloheximide markedly attenuated the MG132 effects on RGS4 degradation, cell migration, and invasion (Fig. 6C, inset). However, MG132 had little effect on RGS4 protein level and migration of MCF-10A cells (Supplementary Fig. S3).
Silencing endogenous RGS4 expression prevents protea-some blockade–induced inhibition of breast cancer cell migration and invasion
Expression of RGS4-targeted shRNA reduced endogenous RGS4 protein to undetectable levels in the absence of MG132 and to ∼30% of that in cells expressing a control GFP shRNA in the presence of MG132 (Fig. 6D, top). In the absence of MG132, migration and invasion of cells expressing RGS4 shRNA were 30% higher than control cells (Fig. 6D, bottom), suggesting that, despite rapid degradation, residual endogenous RGS4 still plays a regulatory role. Silencing RGS4 reduced the MG132-induced inhibition of cell migration and invasion from more than 85% to 35%. Thus, our results show that the inhibitory effect of MG132 on cell migration and invasion is largely dependent on increased endogenous RGS4 protein.
Discussion
RGS4 can markedly inhibit Gi- and Gq-coupled GPCR signaling through its GAP activity (39) and has regulatory roles in the cardiovascular (16) and central nervous systems (14). Our data show that increases or decreases in RGS4 protein with GAP activity cause corresponding decreases or increases in migration and invasion of breast cancer cells. Orthotopic tumors in nude mice from breast cancer cells expressing RGS4, but not its GAP-deficient mutant, had slower growth and a noninvasive phenotype. Moreover, our data show that the molecular basis of the RGS4 action on breast cancer invasiveness involves blocking Rac1 activation, which disrupts lamellipodia formation. Our results are consistent with a previous study showing that RGS4 overexpression blocked migration of endothelial cells to form new blood vessels (40), another important step during cancer metastasis. Furthermore, the GAP-deficient RGS4 mutant failed to inhibit metastatic abilities of breast cancer cells in vitro and in vivo, suggesting that attenuating GPCR signaling by deactivating G-proteins accounts for the action of RGS4 to suppress breast cancer metastasis. Many GPCRs, including PAR1 and CXCR4, are excessively activated in breast cancer cells (3). We found that activation of PAR1 or CXCR4 can stimulate lamellipodia formation, thus promoting breast cancer cell migration in a Gi-dependent manner. RGS4 specifically attenuates PAR1- and CXCR4-dependent signaling by deactivating Gi-protein through its GAP activity. In contrast, EGF-dependent lamellipodia formation and cell migration were not affected by RGS4. Interestingly, activation of other Gi-coupled GPCRs, including the type 1 lysophosphatidic acid receptor, also promotes breast cancer metastasis (41). Therefore, our data may provide a unifying molecular mechanism by which RGS4 suppresses breast cancer metastasis and a therapeutic rationale for increasing levels of RGS4 in breast cancer cells.
Indeed, we found an inverse correlation between expression levels of RGS4 protein in human breast cancer cell lines and their generally accepted metastatic capability. More importantly, our studies using human breast cancer specimens showed a progressive loss of RGS4 protein from benign breast tissues to noninvasive ductal carcinoma in situ, to invasive breast carcinoma. In contrast, there was no significant difference in RGS4 protein levels between invasive breast carcinoma and breast cancer that had metastasized to a lymph node. These results suggest that decreased RGS4 protein may be an early event in development of an invasive phenotype of breast cancer. Currently, a larger study of breast cancer specimens with known clinical outcomes is in progress to determine if RGS4 protein down-regulation correlates with specific clinicopathologic features.
Holland and colleagues (35) recently showed that increased expression of GPCRs such as CXCR4 may not be adequate for breast cancer cells to acquire metastatic potential. Instead, they propose that loss of the tight regulation of GPCR signaling may play a more important role in the transition from nonmetastatic to malignant tumors. Our finding that RGS4 protein, an inhibitor of many GPCRs, is down-regulated in metastatic breast cancer cells provides a mechanism for those previous observations. Thus, identifying the mechanism underlying RGS4 down-regulation should provide an effective, alternative strategy for invasive breast cancer treatment.
Alterations in RGS4 gene transcription have been previously identified in other cancers, including gliomas (42), ovarian cancer (43), and pancreatic cancer (44); however, protein levels and/or functions of endogenous RGS4 were not examined. In fact, our data show a complete lack of correlation between RGS4 mRNA and protein in metastatic breast cancer cells due to rapid degradation through proteasomes. RGS4 is not intrinsically unstable, but is made unstable by N-arginylation at the oxidized Cys-2 residue through a nonribosomal arginine-transferase and then degraded by the proteasome pathway (45). Our findings point to the existence of this posttranslational regulation of RGS4 function in breast cancer cells, which may have important implications for acquisition of a metastatic phenotype. Interestingly, proteasome activity is often elevated in breast cancers (46). Proteasome inhibitors have some efficacy in breast cancer treatment (47, 48), but they may require combination with other antitumor therapies (49). Preventing RGS4 degradation by inhibiting proteasome activity should retard the spread of migrating and invading breast cancer cells, making them better targets for surgery, radiation, and immune treatments. Thus, our discovery provides a rationale for designing new combinations of drug therapies for breast cancer.
Supplementary Material
Acknowledgments
Grant support: NIH grant R011CA125661 and Nebraska State grant LB595 and L506 awards (Y. Tu), and the National Basic Research Program of China (2004DB720000 and 2006CB911001; T. Wei).
We thank Dr. Ming-Fong Lin for helpful discussions, and Lyudmila Batalkina and Lisa Linder-Stephenson for expert technical assistance
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Condeelis J, Singer RH, Segall JE. The great escape: when cancer cells hijack the genes for chemotaxis and motility. Annu Rev Cell Dev Biol. 2005;21:695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- 3.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 4.Ma PC, Maulik G, Christensen J, et al. c-Met: structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev. 2003;22:309–25. doi: 10.1023/a:1023768811842. [DOI] [PubMed] [Google Scholar]
- 5.Tan M, Li P, Klos KS, et al. ErbB2 promotes Src synthesis and stability: novel mechanisms of Src activation that confer breast cancer metastasis. Cancer Res. 2005;65:1858–67. doi: 10.1158/0008-5472.CAN-04-2353. [DOI] [PubMed] [Google Scholar]
- 6.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–74. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 7.Gururaj AE, Rayala SK, Kumar R. p21-activated kinase signaling in breast cancer. Breast Cancer Res. 2005;7:5–12. doi: 10.1186/bcr961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moy B, Goss PE. Lapatinib: current status and future directions in breast cancer. Oncologist. 2006;11:1047–57. doi: 10.1634/theoncologist.11-10-1047. [DOI] [PubMed] [Google Scholar]
- 9.Boire A, Covic L, Agarwal A, et al. PAR1 is a matrix metalloprotease-1 receptor that promotes invasion and tumorigenesis of breast cancer cells. Cell. 2005;120:303–13. doi: 10.1016/j.cell.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 10.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–6. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 11.Guise TA, Yin JJ, Mohammad KS. Role of endothelin-1 in osteoblastic bone metastases. Cancer. 2003;97:779–84. doi: 10.1002/cncr.11129. [DOI] [PubMed] [Google Scholar]
- 12.Druey KM, Blumer KJ, Kang VH, et al. Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature. 1996;379:742–6. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- 13.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 14.Levitt P, Ebert P, Mirnics K, et al. Making the case for a candidate vulnerability gene in schizophrenia: convergent evidence for regulator of G-protein signaling 4 (RGS4). Biol Psychiatry. 2006;60:534–7. doi: 10.1016/j.biopsych.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 15.Tekumalla PK, Calon F, Rahman Z, et al. Elevated levels of DeltaFosB and RGS9 in striatum in Parkinson's disease. Biol Psychiatry. 2001;50:813–6. doi: 10.1016/s0006-3223(01)01234-3. [DOI] [PubMed] [Google Scholar]
- 16.Rogers JH, Tamirisa P, Kovacs A, et al. RGS4 causes increased mortality and reduced cardiac hypertrophy in response to pressure overload. J Clin Invest. 1999;104:567–76. doi: 10.1172/JCI6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurst JH, Henkel PA, Brown AL, et al. Endogenous RGS proteins attenuate Gα(i)-mediated lysophosphatidic acid signaling pathways in ovarian cancer cells. Cell Signal. 2008;20:381–9. doi: 10.1016/j.cellsig.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Kelly P, Moeller BJ, Juneja J, et al. The G12 family of heterotrimeric G proteins promotes breast cancer invasion and metastasis. Proc Natl Acad Sci U S A. 2006;103:8173–8. doi: 10.1073/pnas.0510254103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Liu M, Kozasa T, et al. Thrombin and lysophosphatidic acid receptors utilize distinct rhoGEFs in prostate cancer cells. J Biol Chem. 2004;279:28831–4. doi: 10.1074/jbc.C400105200. [DOI] [PubMed] [Google Scholar]
- 20.Cao X, Qin J, Xie Y, et al. Regulator of G-protein signaling 2 (RGS2) inhibits androgen-independent activation of androgen receptor in prostate cancer cells. Oncogene. 2006;25:3719–34. doi: 10.1038/sj.onc.1209408. [DOI] [PubMed] [Google Scholar]
- 21.Condeelis JS, Wyckoff JB, Bailly M, et al. Lamellipodia in invasion. Semin Cancer Biol. 2001;11:119–28. doi: 10.1006/scbi.2000.0363. [DOI] [PubMed] [Google Scholar]
- 22.Tu Y, Popov S, Slaughter C, et al. Palmitoylation of a conserved cysteine in the regulator of G protein signaling (RGS) domain modulates the GTPase-activating activity of RGS4 and RGS10. J Biol Chem. 1999;274:38260–7. doi: 10.1074/jbc.274.53.38260. [DOI] [PubMed] [Google Scholar]
- 23.Gao X, Mohsin SK, Gatalica Z, et al. Decreased expression of e6-associated protein in breast and prostate carcinomas. Endocrinology. 2005;146:1707–12. doi: 10.1210/en.2004-1198. [DOI] [PubMed] [Google Scholar]
- 24.Shan D, Chen L, Njardarson JT, et al. Synthetic analogues of migrastatin that inhibit mammary tumor metastasis in mice. Proc Natl Acad Sci U S A. 2005;102:3772–6. doi: 10.1073/pnas.0500658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlunck G, Damke H, Kiosses WB, et al. Modulation of Rac localization and function by dynamin. Mol Biol Cell. 2004;15:256–67. doi: 10.1091/mbc.E03-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albini A, Benelli R. The chemoinvasion assay: a method to assess tumor and endothelial cell invasion and its modulation. Nat Protoc. 2007;2:504–11. doi: 10.1038/nprot.2006.466. [DOI] [PubMed] [Google Scholar]
- 27.Tamirisa P, Blumer KJ, Muslin AJ. RGS4 inhibits G-protein signaling in cardiomyocytes. Circulation. 1999;99:441–7. doi: 10.1161/01.cir.99.3.441. [DOI] [PubMed] [Google Scholar]
- 28.Hobson JP, Rosenfeldt HM, Barak LS, et al. Role of the sphingosine-1-phosphate receptor EDG-1 in PDGF-induced cell motility. Science. 2001;291:1800–3. doi: 10.1126/science.1057559. [DOI] [PubMed] [Google Scholar]
- 29.Weed SA, Parsons JT. Cortactin: coupling membrane dynamics to cortical actin assembly. Oncogene. 2001;20:6418–34. doi: 10.1038/sj.onc.1204783. [DOI] [PubMed] [Google Scholar]
- 30.Baugher PJ, Krishnamoorthy L, Price JE, et al. Rac1 and Rac3 isoform activation is involved in the invasive and metastatic phenotype of human breast cancer cells. Breast Cancer Res. 2005;7:R965–74. doi: 10.1186/bcr1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, Dickerson JB, Guo F, et al. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci U S A. 2004;101:7618–23. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Just I, Selzer J, Jung M, et al. Rho-ADP-ribosylating exoenzyme from Bacillus cereus. Purification, characterization, and identification of the NAD-binding site. Biochemistry. 1995;34:334–40. doi: 10.1021/bi00001a041. [DOI] [PubMed] [Google Scholar]
- 33.Coughlin SR. How the protease thrombin talks to cells. Proc Natl Acad Sci U S A. 1999;96:11023–7. doi: 10.1073/pnas.96.20.11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li YM, Pan Y, Wei Y, et al. Upregulation of CXCR4 is essential for HER2-mediated tumor metastasis. Cancer Cell. 2004;6:459–69. doi: 10.1016/j.ccr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 35.Holland JD, Kochetkova M, Akekawatchai C, et al. Differential functional activation of chemokine receptor CXCR4 is mediated by G proteins in breast cancer cells. Cancer Res. 2006;66:4117–24. doi: 10.1158/0008-5472.CAN-05-1631. [DOI] [PubMed] [Google Scholar]
- 36.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–7. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 37.Krumins AM, Barker SA, Huang C, et al. Differentially regulated expression of endogenous RGS4 and RGS7. J Biol Chem. 2004;279:2593–9. doi: 10.1074/jbc.M311600200. [DOI] [PubMed] [Google Scholar]
- 38.Lee MJ, Tasaki T, Moroi K, et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci U S A. 2005;102:15030–5. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang C, Hepler JR, Gilman AG, et al. Attenuation of Gi- and Gq-mediated signaling by expression of RGS4 or GAIP in mammalian cells. Proc Natl Acad Sci U S A. 1997;94:6159–63. doi: 10.1073/pnas.94.12.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albig AR, Schiemann WP. Identification and characterization of regulator of G protein signaling 4 (RGS4) as a novel inhibitor of tubulogenesis: RGS4 inhibits mitogen-activated protein kinases and vascular endothelial growth factor signaling. Mol Biol Cell. 2005;16:609–25. doi: 10.1091/mbc.E04-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boucharaba A, Serre CM, Guglielmi J, et al. The type 1 lysophosphatidic acid receptor is a target for therapy in bone metastases. Proc Natl Acad Sci U S A. 2006;103:9643–8. doi: 10.1073/pnas.0600979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatenhorst L, Senner V, Puttmann S, et al. Regulators of G-protein signaling 3 and 4 (RGS3, RGS4) are associated with glioma cell motility. J Neuropathol Exp Neurol. 2004;63:210–22. doi: 10.1093/jnen/63.3.210. [DOI] [PubMed] [Google Scholar]
- 43.Puiffe ML, Le PC, Filali-Mouhim A, et al. Characterization of ovarian cancer ascites on cell invasion, proliferation, spheroid formation, and gene expression in an in vitro model of epithelial ovarian cancer. Neoplasia. 2007;9:820–9. doi: 10.1593/neo.07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niedergethmann M, Alves F, Neff JK, et al. Gene expression profiling of liver metastases and tumour invasion in pancreatic cancer using an orthotopic SCID mouse model. Br J Cancer. 2007;97:1432–40. doi: 10.1038/sj.bjc.6604031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davydov IV, Varshavsky A. RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J Biol Chem. 2000;275:22931–41. doi: 10.1074/jbc.M001605200. [DOI] [PubMed] [Google Scholar]
- 46.Chen L, Madura K. Increased proteasome activity, ubiquitin-conjugating enzymes, and eEF1A translation factor detected in breast cancer tissue. Cancer Res. 2005;65:5599–606. doi: 10.1158/0008-5472.CAN-05-0201. [DOI] [PubMed] [Google Scholar]
- 47.Cardoso F, Ross JS, Picart MJ, et al. Targeting the ubiquitin-proteasome pathway in breast cancer. Clin Breast Cancer. 2004;5:148–57. doi: 10.3816/cbc.2004.n.020. [DOI] [PubMed] [Google Scholar]
- 48.Orlowski RZ, Dees EC. The role of the ubiquitination-proteasome pathway in breast cancer: applying drugs that affect the ubiquitin-proteasome pathway to the therapy of breast cancer. Breast Cancer Res. 2003;5:1–7. doi: 10.1186/bcr460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang CH, Gonzalez-Angulo AM, Reuben JM, et al. Bortezomib (VELCADE) in metastatic breast cancer: pharmacodynamics, biological effects, and prediction of clinical benefits. Ann Oncol. 2006;17:813–7. doi: 10.1093/annonc/mdj131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.