Abstract
FUS1 is a tumor suppressor gene located on human chromosome 3p21, and expression of Fus1 protein is highly regulated at various levels, leading to lost or greatly diminished tumor suppressor function in many lung cancers. Here we show that selected microRNAs (miRNAs) interact with the 3’ untranslated region (3’ UTR) of FUS1, leading to down-regulation of protein expression. Using computational methods, we first predicted that FUS1 is a target of three miRNAs, miR-93, miR-98 and miR-197, and then showed that exogenous over-expression of these miRNAs inhibited Fus1 protein expression. We then confirmed that the three miRNAs target the 3’UTR region of the FUS1 transcript, and that individual deletion of the three miRNA target sites in the FUS1 3’UTR restores the expression level of Fus1 protein. We further found that miR-93 and miR-98 are expressed at higher levels in small cell lung cancer cell lines (SCLC) than in non-small cell lung cancer cell lines (NSCLC) and immortalized human bronchial epithelial cells (HBECs), and that miR-197 is expressed at higher levels in both SCLC and NSCLC than in HBECs. Finally, we found that elevated miR-93 and miR-197 expression is correlated with reduced Fus1 expression in NSCLC tumor specimens. These results suggest that the three miRNAs are negative regulators of Fus1 expression in lung cancers.
Keywords: microRNA, lung cancer, tumor suppressor gene
Introduction
FUS1, also known as TUSC2 (tumor suppressor candidate 2), is a tumor suppressor gene located on human chromosome 3p21.3, a region in which deficient gene expression is frequently seen in lung cancer (1). The tumor suppressor function of Fus1 has been demonstrated in studies showing that over-expression of Fus1 significantly inhibits tumor growth and progression in mouse models (2), and that FUS1 knockout mice show an increased frequency of spontaneous cancers (3). More recently, Prudkin et al. found a reduction or complete loss of Fus1 expression in 82% of NSCLCs and 100% of SCLCs studied, which was associated with significantly worse overall survival (4), further demonstrating that Fus1 plays an important role in the pathogenesis of lung cancer.
Loss or reduced FUS1 expression could be caused by various mechanisms. Allelic loss of the 3p21.3 chromosomal region containing FUS1 is the major cause of loss or reduction of Fus1 expression in lung cancer (1, 5). In other cases, however, the FUS1 gene and FUS1 mRNA expression level are normal, but the Fus1 protein is not expressed at detectable levels (1, 6), suggesting that Fus1 expression may be down-regulated at certain post-transcriptional stages. One mechanism regulating FUS1 expression and function has already been shown by Uno et al., who demonstrated that loss of myristoylation of Fus1 protein causes it to be rapidly degraded and removes its ability to suppress tumor growth (7). Recent work on non-coding RNAs suggests other mechanisms by which transcription and translation of FUS1 might also be effectively uncoupled, including translational repression and destabilization of the FUS1 mRNA, both of which can be mediated by miRNAs.
miRNAs are short, 21 to 23 nucleotide RNAs that regulate gene expression by binding to sequences in the 3’ untranslated region (3’ UTR) of an expressed mRNA, resulting in either modulation of translation efficiency or degradation of the mRNA (8–10). miRNAs have been shown to regulate expression of a variety of genes involved in embryonic development and in human disease (8, 11–21), including cancer (22–26). The 3’ UTR of FUS1 is highly conserved, strongly suggesting that it plays an important role in regulating FUS1 expression. Target prediction shows that at least 3 miRNAs — miR-93, miR-98 and miR-197 — potentially interact with the FUS1 3’UTR. We therefore examined the role of these miRNAs in regulating Fus1 protein expression.
Results
The 3’UTR of FUS1 plays a significant role in regulating FUS1 expression levels in NCI-H1299 cells
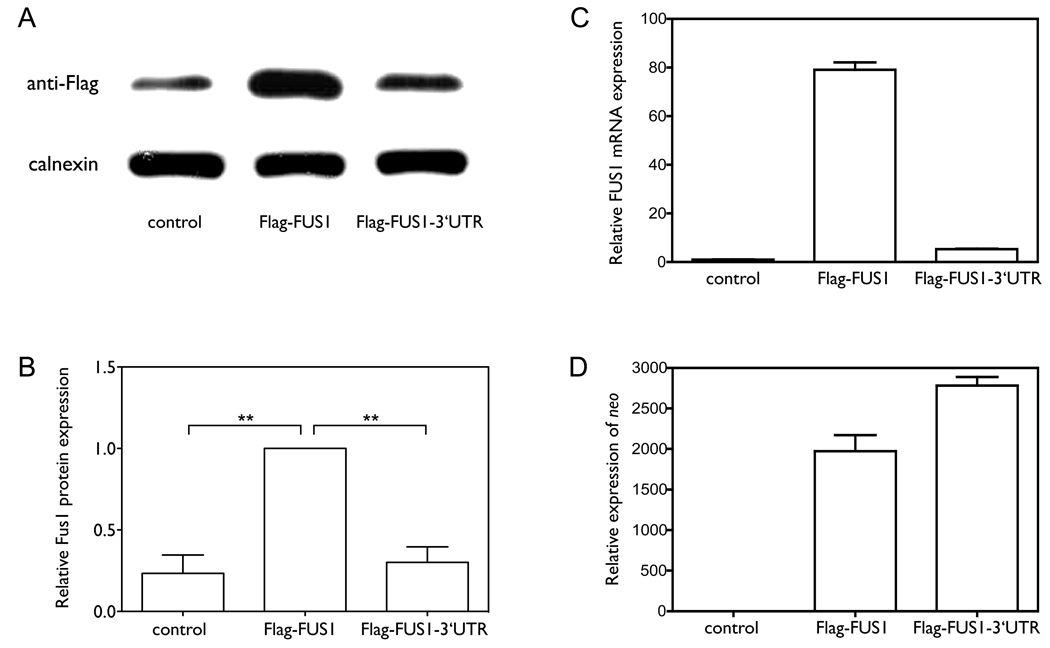
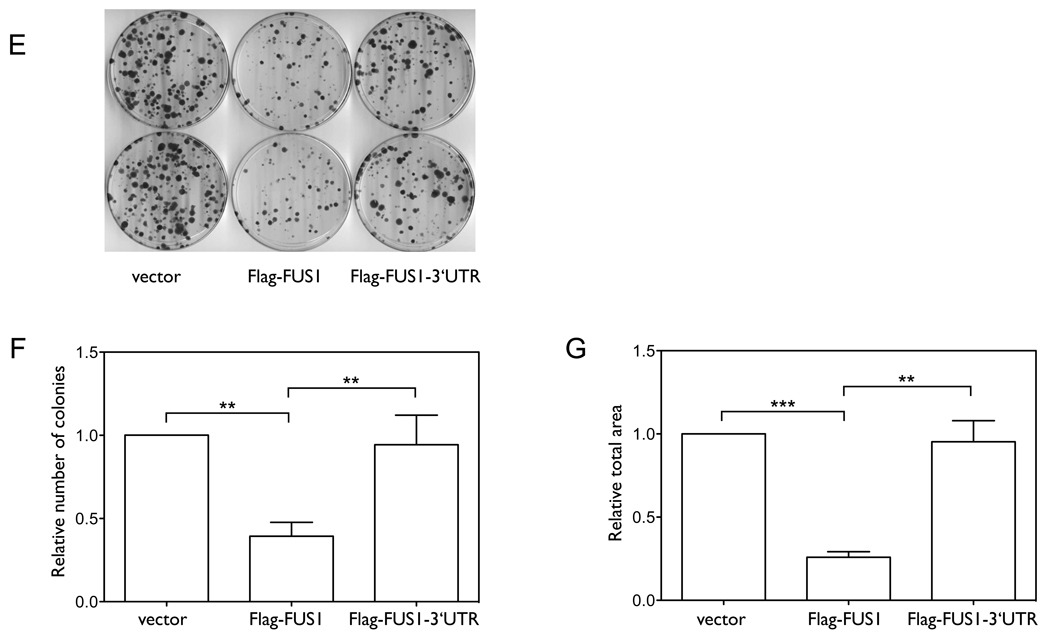
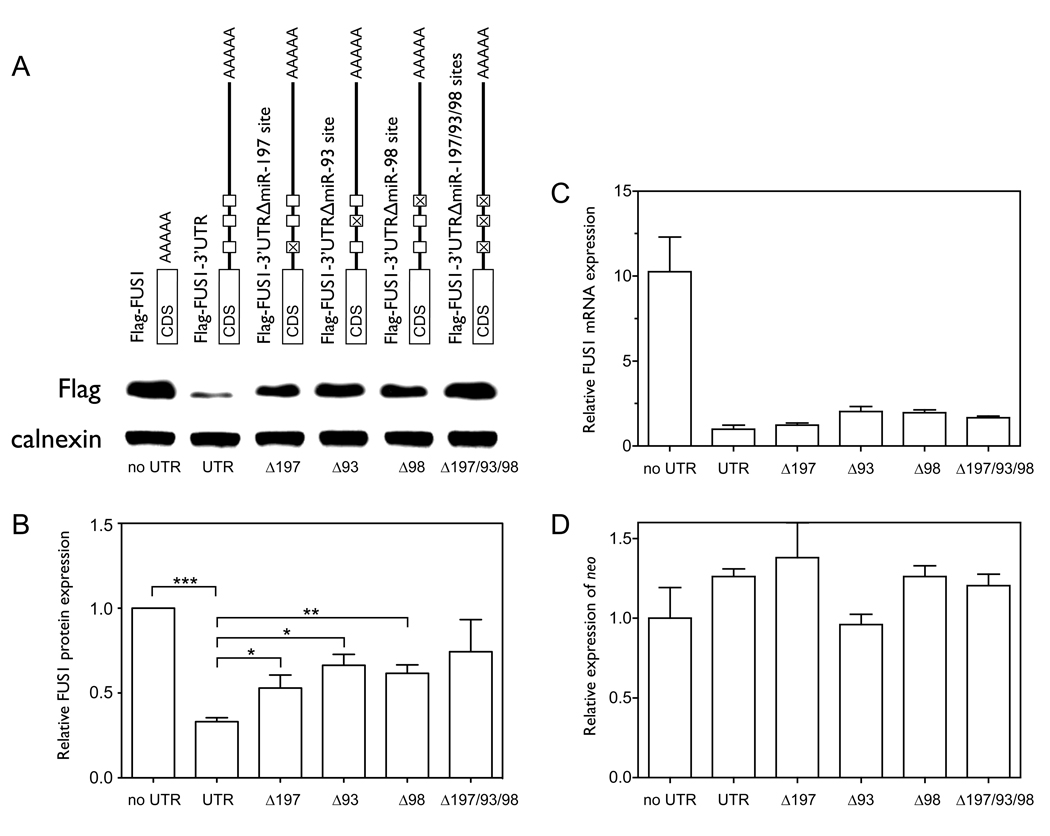
Previous studies have shown that Fus1 is expressed in normal lung epithelial cells, but the expression is frequently reduced or lost in lung cancer cell lines and in lung cancer specimens (4, 7), despite the FUS1 mRNA being expressed in these cells. To assess the role of the 3’UTR of FUS1 in regulating Fus1 protein expression, we compared the levels of expression of the recombinant proteins from Flag-FUS1 (Flag-tagged FUS1 without the 3’UTR), and Flag-FUS1-3’UTR (Flag-tagged FUS1 with the full-length 3’UTR) constructs. As shown in Figure 1A, 1B and 1C, both the mRNA and protein expression levels of Fus1 were significantly lower in cells expressing Flag-FUS1-3’UTR than in cells expressing Flag-FUS1 (p=0.0024 and p=0.0019, n=5). mRNA expression levels of neo (Figure 1D), which was also present in the expression vector and served as a control for expression efficiency of the vector (27), indicated that the higher expression level of Fus1 protein in cells expressing Flag-FUS1 was not due to higher expression efficiency of this construct relative to that of the Flag-FUS1-3’UTR. An inverse relationship between exogenously induced over-expression of FUS1 and the growth rate of H1299 cells has already been established (6). We therefore examined the contribution of the 3’UTR of FUS1 to cell proliferation by colony formation assay. As shown in Figure 1E–G, Flag-FUS1 expression significantly inhibited cell growth relative to Flag-FUS1-3’UTR and the control vector, with the number and size of colonies formed in the presence of Flag-FUS1 much smaller than in the presence of Flag-FUS1-3’UTR (p=0.0081 and p=0.0011) or the vector control (p=0.0048 and p=0.0004, by two-tailed paired t-test). Taken together, these results implicate the 3’UTR of FUS1 in reducing levels of Fus1 expression and tumor suppressor function.
Figure 1. The 3’UTR plays a significant role in regulating Fus1 protein and mRNA expression levels.
(A–D) We cloned constructs expressing either Flag-tagged FUS1 without the 3’UTR (Flag-FUS1) or Flag-tagged FUS1 with the full length 3’UTR (Flag-FUS1-3’UTR). NCI-H1299 cells were transfected for 48 h with either equal amounts (0.46 nM) of the above constructs, or were treated with only transfection reagents (control). (A) The protein expression level of Fus1 was detected by Western blot using an anti-Flag antibody, with calnexin as a loading control. (B) Quantification results of the Fus1 protein level were from three independent experiments. (C) The mRNA level of FUS1 as detected by quantitative PCR. (D) The mRNA level of neo as detected by quantitative PCR. (E–G) Colony formation s a function of the presence or absence of the FUS1 3’UTR. NCI-H1299 cells were transfected with equal amounts (0.46 nM) of either constructs expressing Flag-FUS1, Flag-FUS1-3’UTR or empty vector (control), respectively. Colonies were visualized by staining with 1% crystal violet (E). Quantification of (F) number of colonies and (G) total area of colonies was from three independent colony formation assays. (**, p<0.01; ***, p<0.001)
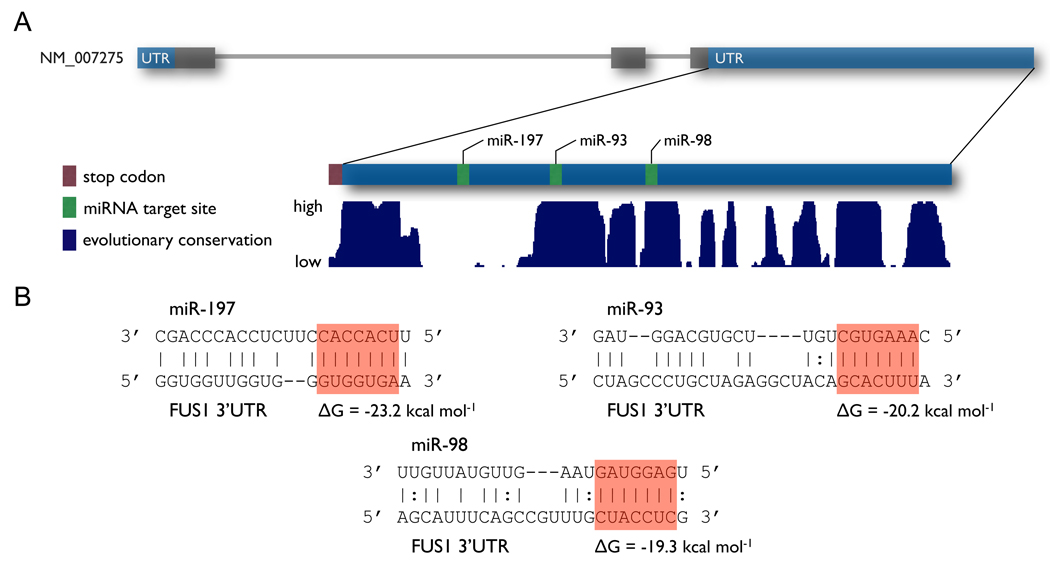
miR-93, miR-98 and miR-197 are predicted to target the 3’UTR of FUS1
Using a program we developed (miRmate), we identified three miRNAs — miR-93, miR-98 and miR-197 — that are highly likely to interact with the 3’ UTR of FUS1. As shown in Figure 2A, the predicted target sites of miR-93 and miR-98 are located in regions that are highly evolutionarily conserved, while the predicted target site of miR-197 does not fall in a highly conserved region. Figure 2B shows the structure of the miRNA:target interactions for miR-93, miR-98 and miR-197, with the conservation of the target sites across species. Both PicTar(28) and TargetScan(29) also predict several miRNA target sites in the FUS1 3’UTR, including those for miR-93 and miR-98.
Figure 2. miR-93, miR-98 and miR-197 are predicted to target FUS1.
(A) The FUS1 gene structure, showing the predicted target sites of each miRNA and the conservation of the 3’UTR across species. (B) The structure of the miRNA:target interactions for miR-197, miR-93, and miR-98, with the sequence of the miRNA, the sequence of the target site and the predicted free energy of hybridization. The seed sequence is highlighted in red.
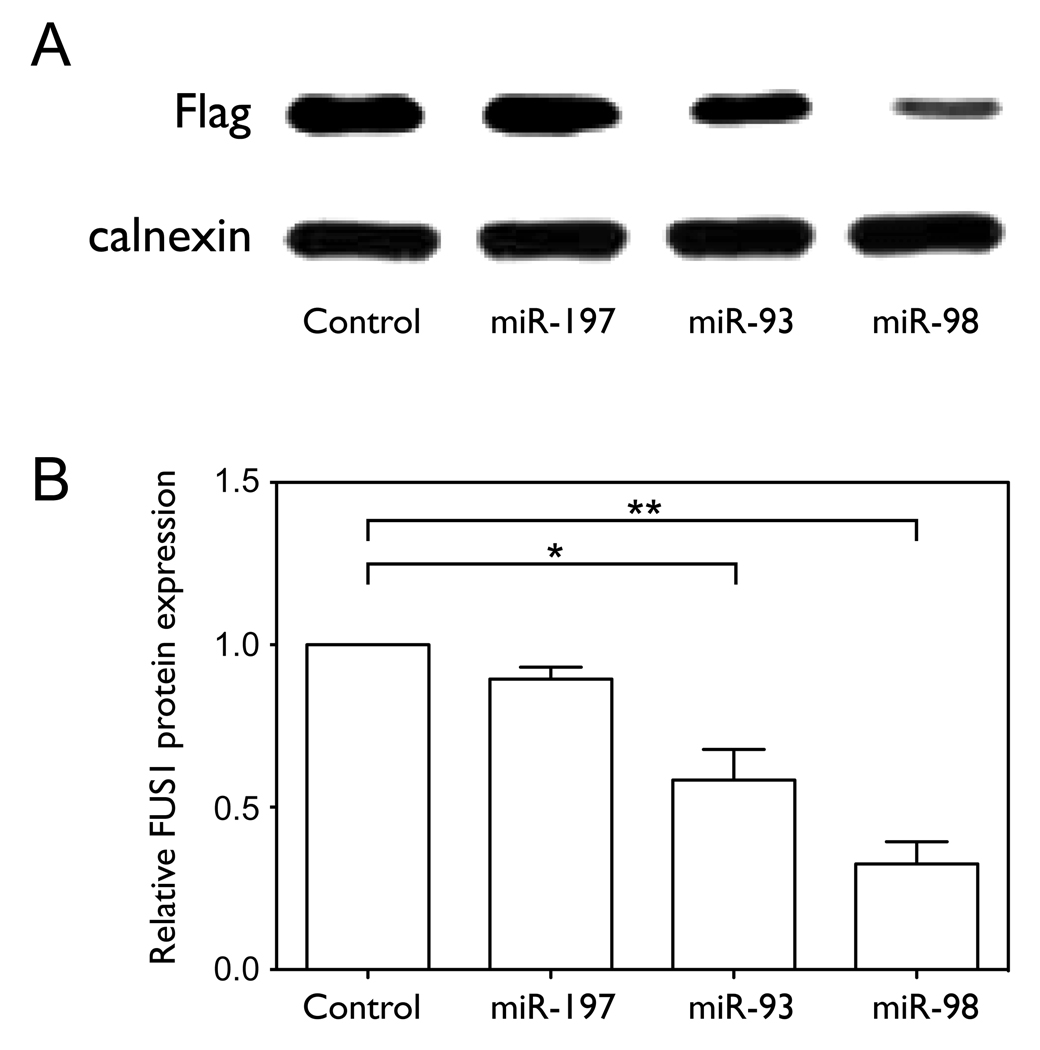
miR-93, miR-98 and miR-197 translationally repress Fus1 by targeting specific sequences in the 3’UTR of FUS1
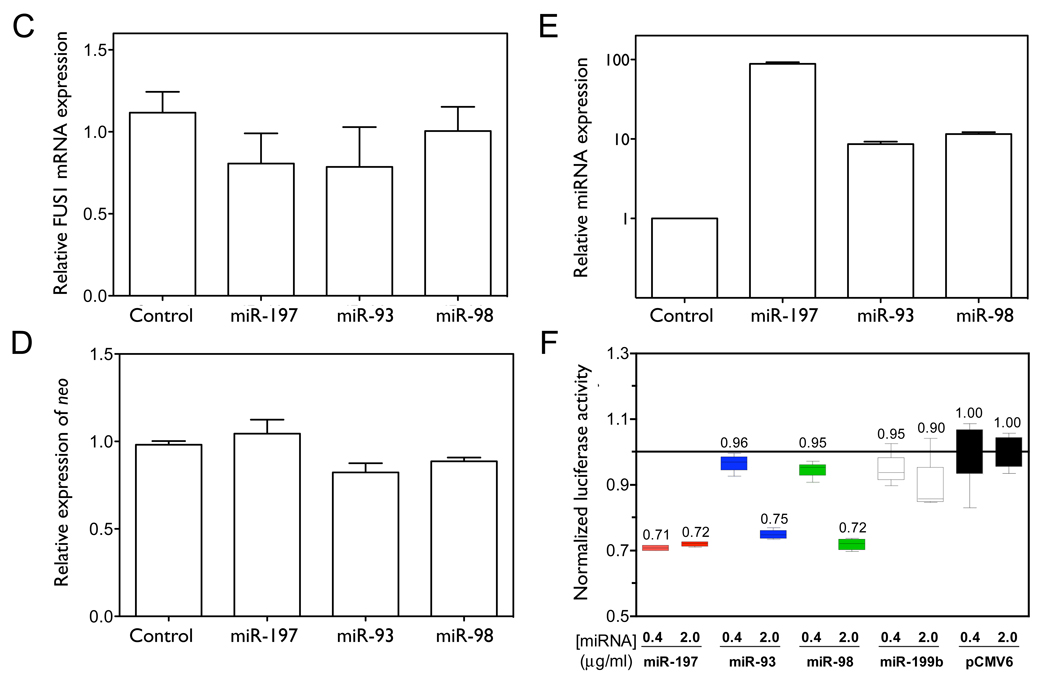
In order to test the effect of the three miRNAs on Fus1 protein expression, we co-transfected NCI-H1299 cells with the Flag-FUS1-3’UTR construct and with either control or miRNA expression constructs. As shown in Figure 3A and 3B, over-expression of miR-93 or miR-98 significantly inhibited Fus1 protein expression (0.58±0.16 and 0.32±0.12 relative to control, with p=0.0239 and p=0.0050, by one-tailed paired t-test, n=3), with miR-98 showing a greater effect than miR-93. miR-197 does not significantly decrease Fus1 protein expression (0.89±0.06 relative to control, with p=0.0515, n=3), although the over-expressed level of miR-197 relative to endogenous control is much higher than that of miR-93 and miR-98, as shown in Figure 3E. FUS1 mRNA levels are not significantly affected by miRNA over-expression (Figure 3C), and neo expression levels measured as a control for transfection efficiency are not significantly different between samples (Figure 3D).
Figure 3. miR-93, miR-98 and miR-197 translationally repress FUS1.
(A–D) NCI-H1299 cells were plated in 100 mm dishes and co-transfected with Flag-FUS1-3’UTR expression vector (2 µg) and either pCMV6 empty vector (control) or pCMV6/miRNA expression vectors (10 µg), respectively, using Lipofectamine 2000 transfection reagent. After 48 h cell lysates were harvested and total RNA was isolated. Fus1 expression levels (A, B) were measured by Western blot using an anti-Flag antibody. Calnexin levels were used as a loading control. Quantification results were from three independent experiments. Relative levels of FUS1 (C) and neo (D) mRNAs, as well as miRNA levels (E) were measured by qRT-PCR. (F) The 3’UTR of FUS1 was cloned into a reporter construct containing a luciferase cDNA under the control of a mammalian promoter/terminator system. HEK-293 cells were plated in 24-well plates and co-transfected with pMIR-REPORT/luciferase-FUS1 3’UTR (0.4 µg), pCMV6/miRNAs (0.4 µg or 2 µg) and pMIR-REPORT/β gal control expression constructs (0.8 µg) using FuGENE 6 Transfection Reagent. After 48 h of transfection, cells were lysed and luciferase and β-galactosidase activity were measured. Shown are normalized luciferase activity for miR-93, miR-98 and miR-197, with miR-199b and an empty vector (pCMV6) as negative controls. The concentrations are of the transfected miRNA expression vector, and are 1X and 5X relative to that of the transfected luciferase reporter construct. (*, p<0.05; **, p<0.01).
To test the specificity of the interaction between the three miRNAs and the 3’UTR of FUS1, we placed the 3’UTR of FUS1 downstream of the luciferase coding region in an expression construct. We co-expressed the reporter construct with either a vector expressing a candidate miRNA, a control vector expressing miR-199b, which is not predicted to target the 3’UTR of FUS1, or an empty vector (pCMV6), and monitored luciferase activity. As shown Figure 3F, each miRNA repressed luciferase activity by 25–30%, as compared with the control. This magnitude of repression is consistent with that observed in previous studies of miRNA regulation using luciferase reporter vectors (28, 30).
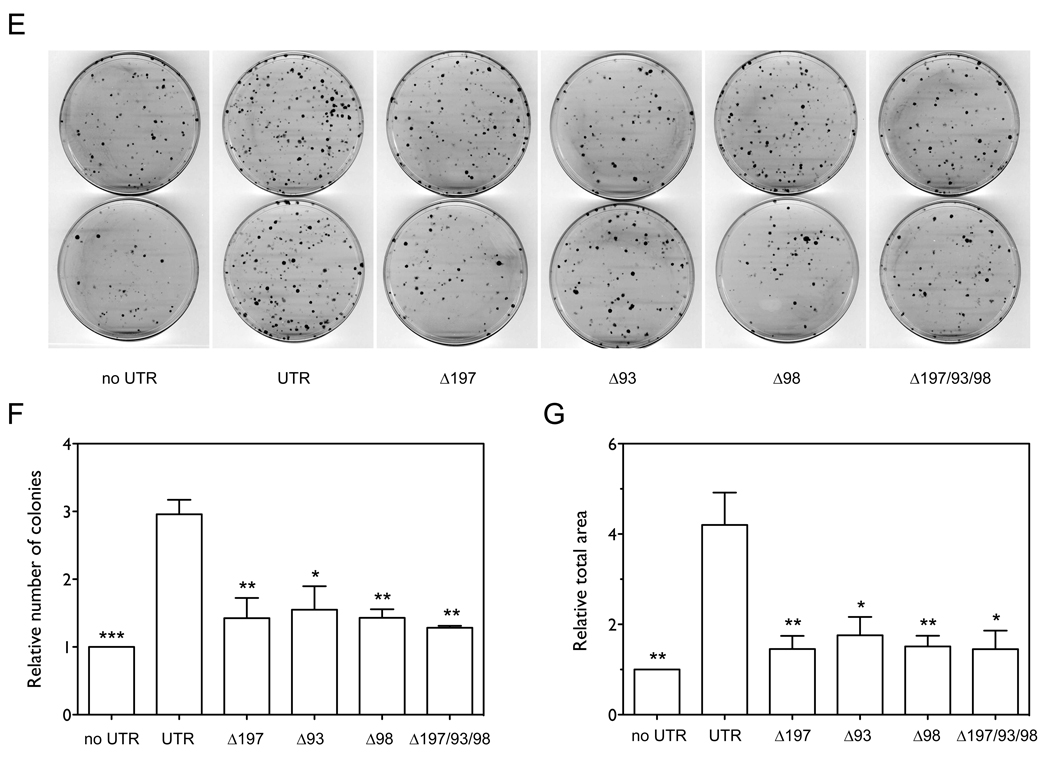
To further confirm the specific target sites of the three miRNAs in the 3’UTR of FUS1, we deleted the seed sequences of the miR-93, miR-98 and miR-197 target sites (Figure 2B) in the 3’UTR of the Flag-FUS1-3’UTR construct, and compared protein expression and tumor suppressor function of FUS1 with that of the wildtype Flag-FUS1 and Flag-FUS1-3’UTR constructs in transfected NCI-H1299 cells. As shown in Figure 4A and 4B, individual deletions of the seed sequences of the miR-197, miR-93, and miR-98 target sites significantly increased the expression level of Fus1 protein relative to wildtype Flag-FUS1-3’UTR (1.6 fold with p=0.0477, 2 fold with p=0.0259, and 1.85 fold with p=0.0067, respectively, by one-tailed paired t-test, n=4) with the combinatorial deletion showing higher levels of protein expression than individual deletions (2.24 fold). Relative levels of FUS1 (Figure 4C) and neo mRNA (Figure 4D) measured by qRT-PCR were not altered by the deletions. As shown in Figure 4E–G, over-expression of the Flag-FUS1-3’UTR expression constructs with individual deletions of the miR-197, miR-93 and miR-98 seed target sequences as wells as combined deletion of the three target sites significantly decreased the number (p=0.0012, p=0.0328, p=0.0036 and p=0.0024, respectively, by one-tailed paired t-test, n=3) and size (p=0.0044, p=0.0500, p=0.0083 and p=0.0101, respectively, by one-tailed paired t-test, n=3) of the colonies formed as compared with wildtype Flag-FUS1-3’UTR construct. The above results indicate that removal of the three miRNAs target sites in the 3’UTR of FUS1 significantly restores Fus1 protein expression, and correspondingly, restores its function as a tumor suppressor.
Figure 4. Deletion of the target sites of the three miRNAs restores Fus1 protein expression levels in NCI-H1299 cells.
NCI-H1299 cells were transfected with equal amounts (0.46 nM) of vectors expressing Flag-FUS1, wildtype Flag-FUS1-3’UTR and several mutated versions of Flag-FUS1-3’UTR, in which the seed sequences of the target sites in the FUS1 3’UTR are deleted, using Lipofectamine 2000 transfection reagent. After 48 h cell lysates were harvested and Fus1 expression levels (A,B) were measured by Western blot using an anti-Flag antibody. Calnexin levels were used as a loading control. The quantitation was generated from two independent transfections, each with two repeated Western blots. Relative levels of FUS1 mRNA (C) and neo mRNA (D) were measured by qRT-PCR. (E–G) Colony formation as a function of the Fus1 protein. NCI-H1299 cells were transfected with equal amounts (0.46 nM) of the above constructs. Colonies were visualized and quantified as described previously. (*, p<0.05; **, p<0.01; ***, p<0.001).
Overall, these results indicate that miR-93, miR-98 and miR-197 negatively regulate FUS1 tumor suppressor function by inhibiting Fus1 protein expression through targeting specific sites in the FUS1 3’UTR.
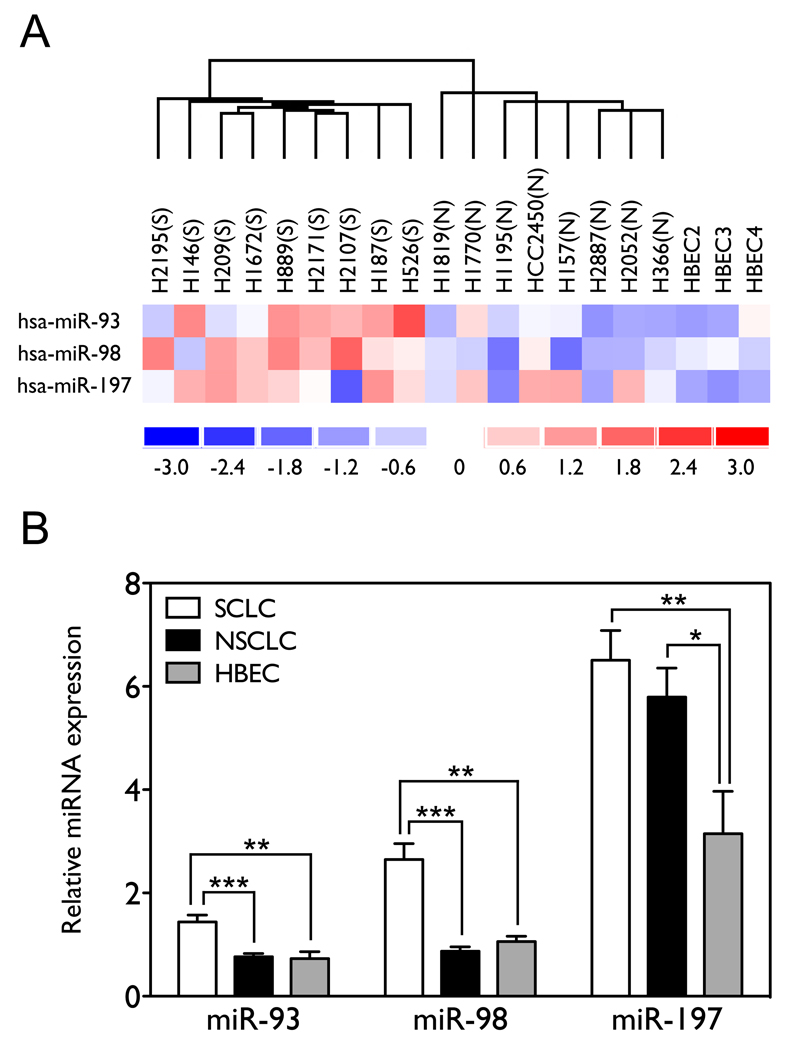
miR-93 and miR-98 are expressed at higher levels in SCLC than in NSCLC and HBEC cell lines, and miR-197 is expressed at higher levels in both SCLC and NSCLC relative to HBEC cell lines
To evaluate the physiological relevance of these miRNAs in lung cancer pathogenesis, we analyzed endogenous expression levels of these three miRNAs in 9 SCLC cell lines, 8 NSCLC cell lines, and 3 immortalized normal human bronchial epithelial cell lines (HBECs). We profiled expression levels of 136 miRNAs in these cell lines in duplicate using miRNA microarrays. The normalized expression profiles allowed us to identify miRNAs with statistically significant differential expression between SCLC and NSCLC cell lines and between lung cancer cell lines and HBECs. We found that miR-93 and miR-98 are over-expressed in SCLCs relative to NSCLC and HBECs, and miR-197 is over-expressed in both SCLCs and NSCLC relative to HBECs. As shown in Figure 5A and 5B, the expression levels of miR-93, miR-98 and miR-197 differ significantly among the different cell lines, and generally miR-93 and miR-98 are expressed at higher levels in SCLC (n=9) as compared with NSCLC (1.88 fold with p=0.0001 by two-tailed t-test, and 3.04 fold with p<0.0001, n=8) and HBECs (1.98 fold with p=0.0100, and 2.50 fold with p=0.0077, n=3), whereas miR-197 is expressed at higher levels in both SCLCs and NSCLCs as compared with HBECs (2.07 fold with p=0.0060, and 1.84 fold with p=0.0206). This suggests that aberrant expression of miR-197 might contribute to both SCLC and NSCLC tumorigenesis in general, while miR-93 and miR-98 may play a more important role in the development of SCLCs than NSCLCs.
Figure 5. miR-93, miR-98 and miR-197 are differentially expressed among SCLC, NSCLC, and HBEC cell lines.
(A) Expression levels of miR-93, miR-98 and miR-197 across the panel of cell lines. The expression levels have been normalized by subtracting the mean expression and dividing by the standard deviation. Red represents miRNAs that are over-expressed relative to the reference; blue represents miRNAs that are under-expressed. (B) The mean expression levels of miR-93, miR-98 and miR-197 by group. Significance was assessed by two-tailed t-test. SCLC and (S), small-cell lung cancer; NSCLC and (N), non-small-cell lung cancer. (*, p<0.05; **, p<0.01; ***, p<0.001).
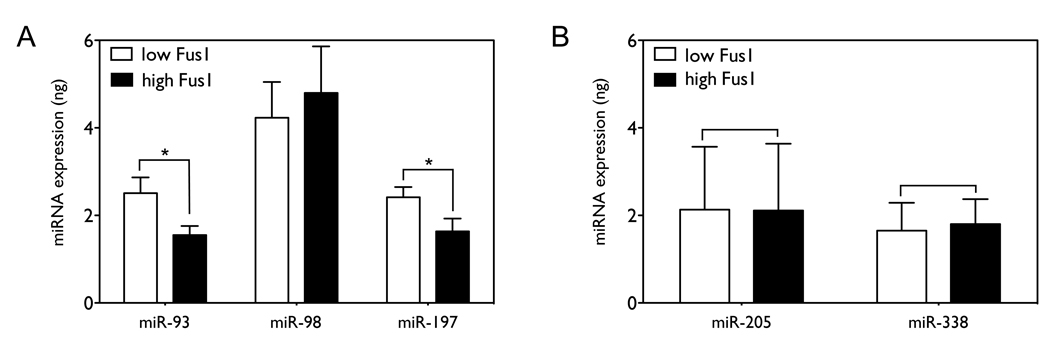
Expression of miR-93 and miR-197 is inversely correlated with Fus1 expression in lung cancer tumor specimens
Levels of all three miRNAs differ dramatically between the NSCLC cell lines, suggesting that aberrant expression of the miRNAs may contribute to the decrease of Fus1 expression in specific NSCLCs. In order to examine the correlation of the three miRNAs with Fus1 level, we measured the expression levels of miR-93, miR-98 and miR-197 in 20 NSCLC tumor specimens (14 adenocarcinoma and 6 squamous cell carcinoma) samples by qRT-PCR. These samples had been previously characterized as positive for Fus1 expression by immunohistochemical staining (4) and were classified into two groups, “low” and “high”, based on Fus1 expression. As shown in Figure 6A, expression of miR-93 and miR-197 is significantly higher in samples in the “low” Fus1 group than in the “high” Fus1 group (1.62 fold with p=0.0164, and 1.48 fold with p=0.0265 by one-tailed t-test with n=10), while expression of miR-98 is not significantly different between the two groups (p=0.3390, n=10). In contrast, expression levels of miR-205 and miR-338, which are not predicted to target FUS1 mRNA, are not significantly different between the two groups (p=0.4961 and p=0.4318), as shown in Figure 6B.
Figure 6. Expression of miR-93 and miR-197 is inversely correlated with Fus1 expression in lung cancer tumor specimens.
miR-93, miR-98 and miR-197 levels were measured in 6 squamous cell carcinoma and 14 adenocarcinoma tumor specimens. The 20 samples were divided into two groups of 10 specimens, the low Fus1 group and the high Fus1 group, based on Fus1 protein expression levels measured by immunohistochemical staining. Significance was assessed by one-sided t-test. (*, p<0.05).
Discussion
The 3’UTRs of many protein-coding mRNAs have been shown to play an important role in regulating protein expression. Regulatory elements typically located in 3’UTRs include AU-rich elements (ARE) and iron-response elements (IREs) (31). Binding of the trans-acting factors ARE binding proteins (ARE-BP) and IRE binding proteins (IRE-BP) to AREs and IREs, respectively, leads to either translational repression/enhancement or mRNA stabilization/destabilization. In this study, we show that the 3’UTR significantly represses the expression of FUS1 at both the mRNA and protein levels. Analysis of the FUS1 3’UTR shows, however, that it lacks typical ARE and IRE elements, suggesting a different regulatory mechanism. Using computational methods, we predicted that the 3’UTR of FUS1 contains target sites of at least three miRNAs, including miR-93, miR-98 and miR-197. We showed that deletion of the seed sequences of the miRNA target sites in the 3’UTR significantly restores protein expression levels (Figure 4A) and, correspondingly, the tumor suppressor function of FUS1 (Figure 4E–G), suggesting that Fus1 expression is under significant repression by endogenous expression of the three miRNAs.
In order to confirm the direct interaction of the miRNAs with the 3’UTR of FUS1, we conducted luciferase reporter assays in HEK 293 cells. Our results (Figure 3F) show that for miR-197, both a 1:1 ratio and a 5:1 ratio of miRNA vector to luciferase reporter vector result in the maximum reduction of the luciferase activity. For miR-93 and miR-98, however, a 1:1 ratio of miRNA expression vector to luciferase reporter vector is not enough to significantly inhibit luciferase activity, and a 5:1 ratio is needed to reach the maximum effect. These data indicate that the 3’UTR of FUS1 is more sensitive to lower doses of miR-197 than of miR-93 and miR-98, which suggests that miR-197 might have a more substantial direct effect on the 3’UTR of FUS1 than do miR-93 and miR-98. However, when over-expressing the three miRNAs in NCI-H1299 cells, miR-93 and miR-98 significantly inhibited Fus1 protein expression, whereas unexpectedly, miR-197 did not have a significant inhibitory effect (Figure 3A). The following possibilities could lead to the above outcome: 1) miR-197 regulates Fus1 expression through both direct and indirect mechanisms. In H1299 cells, miR-197 might inhibit the expression of another gene that down-regulates Fus1 expression. The resulting down-regulation of this gene will compensate for the direct inhibitory effect of miR-197 on the 3’UTR of FUS1. If both pathways are active in H1299 cells, then over-expression of miR-197 in H1299 would not significantly decrease Fus1 expression due to the opposing directions of these two regulatory pathways. 2) In H1299 cells, but not in HEK293 cells, the 3’UTR of FUS1 is maximally repressed by endogenous miR-197, so that exogenous introduction of miR-197 cannot lead to further significant inhibition of Fus1 expression (Figure 3A). In this case, removal of the miR-197 target site would release FUS1 from repression by endogenous miR-197, leading to the observed significant increase in Fus1 protein (Figure 4A and 4B).
As for the role of the 3’UTR in regulating FUS1 mRNA levels, our results indicate that the 3’UTR is responsible not only for decreasing expression of the Fus1 protein product, but also for significantly down-regulating mRNA expression (Figure 1A and 1C). On the other hand, our results (Figure 3C and Figure 4C) also show that the three miRNAs do not significantly affect FUS1 mRNA levels. The above results clearly indicate that there are additional undefined site(s) in the 3’UTR of FUS1 that influence mRNA stability. However, combined removal of the three miRNA target sites restores protein expression nearly completely (Figure 4A). This result not only suggests that the low levels of FUS1-3’UTR mRNA can be translated very efficiently upon removal of the miRNA target sites, but also seems to imply that mRNA stability (as reflected in mRNA levels) does not play a significant role in controlling Fus1 protein levels. We speculate that the disruption of only the mRNA de-stabilizing site(s) would significantly increase both the FUS1 mRNA and protein level. It is also reasonable to infer, however, that the removal of both the miRNA regulatory sites and the mRNA de-stabilizing sites altogether (by removing the 3’UTR as shown in Figure 4A) could not bring the Fus1 protein to a significantly higher level than what would be observed after removing either the miRNA target sites or the mRNA de-stabilizing site(s), since the significantly elevated Fus1 protein level might induce more intensive down-regulation of Fus1 through a mechanism such as protein degradation, thereby leading to lower than “predicted” Fus1 levels. These possibilities suggest attractive future directions for studying the regulation of FUS1 mRNA stability and Fus1 protein regulation at the post-translational level.
Of the three miRNAs that translationally repress Fus1, miR-93 and miR-98 are overexpressed in SCLC relative to NSCLC and HBECs. This finding is of particular interest given the early dissemination and rapid clinical evolution of SCLC relative to the slow clinical progression of NSCLC. Coupled with the finding that average Fus1 expression is significantly lower in SCLC tissue specimens than in NSCLC tissue specimens and normal lung epithelia (4), it is reasonable to speculate that the overexpression of these two miRNAs in SCLC is one of the factors contributing to the loss or reduction of Fus1 expression. miR-93 and miR-197 are expressed at higher levels in specimens where Fus1 expression is low, and at lower levels in specimens where Fus1 expression is high, suggesting that miR-93 and miR-197 are responsible, at least partially, for the reduced Fus1 expression in the former. The expression of miR-98, however, does not differ significantly between the two groups, possibly because the endogenous level of miR-98 is not high enough to affect the Fus1 level in NSCLC tissue specimens, especially in the presence of high levels of miR-93 and miR-197 in the specimens with low Fus1 expression. Overall, our results indicate that the expression of the three miRNAs is related to Fus1 expression in lung cancer, suggesting that the aberrant expression of these miRNAs might play a role during the tumorigenesis of lung cancer by regulating Fus1 expression.
miR-93 and miR-98 have been linked to cancer in previous studies. For example, Blenkiron et al. recently showed that miR-93 expression is upregulated in primary breast cancer as compared with normal breast cells (32). Yanaihara, et al. reported that over-expression of miR-93 correlated with worse prognosis of lung adenocarcinoma patients (33). These studies, however, did not investigate whether miR-93 has an oncogenic function. Our results thus provide the first evidence that miR-93 acts as an oncogene by down-regulating the expression of tumor suppressor gene FUS1. Hebert et al. showed that miR-98 targets Hmga2 in head and neck squamous cell carcinoma (HNSCC) cells (34). Hmga2 expression has been linked to several other types of cancers and has been shown to be associated with enhanced selective chemosensitivity of cancer cells to the topoisomerase II inhibitor doxorubicin (35–37). Interestingly, translocations frequently append the 3’UTR of HMGA2 to the 3’ end of tumor suppressor genes such as FHIT, RAD51L1 and HEI10, suggesting that translational repression of these tumor suppressor genes by miR-98 may contribute to tumorigenesis. Coupling these results, we speculate that miR-98 may play a critical role in the development of cancer by regulating expression of multiple cancer-related proteins, including Fus1. In contrast to miR-93 and miR-98, little is known about the function of miR-197. Our results therefore implicate miR-197 in carcinogenesis through its down-regulation of Fus1 expression. Unlike miR-93 and miR-98, which are overexpressed in SCLC, miR-197 is overexpressed in both NSCLC and SCLC relative to normal HBECs, suggesting that miR-197 plays a more important role in down-regulating Fus1 expression in NSCLC than either miR-93 or miR-98.
Overall, our results are the first demonstration that three miRNAs – miR-197, miR-93 and miR-98 – down-regulate Fus1 expression by targeting the 3’UTR of FUS1, and also clearly indicate that there are additional undefined site(s) in the 3’UTR of FUS1 that influence mRNA stability. In addition, given the rapid growth of the miRNA family, it is reasonable to expect that there are unknown miRNAs that regulate Fus1 expression. For example, recent work by Lee et al. has shown that miR-378 can down-regulate Fus1 expression in U87 cells (38). Further study is needed to define the full set of regulatory sites in the 3’UTR of FUS1 and of regulators of Fus1 expression. From a clinical perspective, while our preliminary investigation in lung cancer cell lines and clinical samples suggests that aberrant expression of these miRNAs plays a role in down-regulating Fus1 expression, further investigation is needed to fully define their role in the development and progression of lung cancer in vivo.
Materials and Methods
Cell lines
Lung cancer cell lines (NCI-H146, NCI-H157, NCI-H187, NCI-H209, NCI-H526, NCI-H889, NCI-H1299, NCI-H1648, NCI-H1672, NCI-H1770, NCI-H1819, NCI-H2052, NCIH2107, NCI-H2171, NCI-H2195, NCI-H2122, NCI-H2887, HCC366, HCC970, HCC1195, and HCC2450) were from the Hamon Center collection. HEK-293 cells were obtained from ATCC (Manassas, VA). Lung cancer cell lines and HBECs were grown in RPMI 1640 supplemented with 5% FBS (Atlanta Biologicals, Lawrenceville, GA). HEK-293 cells were grown in Dulbecco’s MEM (Mediatech, Manassas, VA) supplemented with 10% FBS.
Construction of expression vectors
miR-197, miR-93, and miR-98 and miR-199b were amplified by PCR from genomic human DNA using the following primers:
miR-197 forward, 5’- ATTACTTTGCCCATATTCATTTTGA-3’;
miR-197 reverse, 5’-GCAACAGTGTGAATGTACTTAATGC-3’;
miR-93 forward, 5’-AGTCTCTGGCTGACTACATCACAG-3’;
miR-93 reverse, 5’-CTACTCACAAAACAGGAGTGGAATC-3’;
miR-98 forward, 5’-TGTATGACTGCCGTATGTTTCCTATT-3’;
miR-98 reverse, 5’-AATTCTTAAAGTATGCTTTCCATTCC-3’;
miR-199b forward, 5’-CTTCGGGGTTGGACACTAGGTAGGAG-3’;
miR-199b reverse, 5’-GTGTATGTCTTTGGGATGTGAGGATGG-3’.
The amplified sequences were inserted into the EcoRI and XmaI restriction sites of the multiple cloning site of expression vector pCMV6 and verified by sequencing. To investigate the regulation of Fus1 by miRNAs, we added a Flag tag to the N-terminus of Fus1 and made three derivative constructs: Flag-FUS1, Flag-FUS1-3'UTR and Flag (as a negative control) based on the pcDNA3.1 expression vector. We also made mutated constructs based on Flag-FUS1-3'UTR with the miR-93, miR-98 and miR-197 seed sequences of the target sites deleted individually and in combination. In order to test the interaction between specific miRNAs and the 3’UTR of FUS1, the 1.2 kb 3’UTR was cloned into pMIR-REPORT (Ambion, Austin, TX), a reporter construct containing a luciferase cDNA under the control of a mammalian promoter/terminator system.
Transfection
Cells were plated and cultured overnight. Cells were then transfected with specified expression vectors using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) or FuGENE 6 Transfection Reagent (Roche, Basel, Switzerland). After 48 hours, cells were harvested and specified assays were conducted.
Luciferase Assay
Luciferase activity and β-galactosidase activity were measured using the Luciferase Assay System and β-galactosidase Assay System (Promega, Madison, WI), respectively. Luciferase activity was adjusted for transfection efficiency by normalizing by the β-galactosidase activity of each sample.
Quantitative RT-PCR
Total RNA was prepared using the mirVana™ miRNA Isolation Kit (Ambion, Austin, TX) followed by treatment with DNase. Quantitative RT-PCR (qRT-PCR) of miRNAs was conducted on an ABI PRISM 7900 Sequence Detection System using TaqMan® Universal PCR Master Mix and miRNA Expression Assay primer and probe sets (Applied Biosystems, Foster City, CA). RNU19 RNA was used as an internal control for normalization of cDNA loading. FUS1 and neo expression were measured using TaqMan® reverse transcription kits with qRT-PCR conducted using TaqMan® Universal PCR Master Mix and Gene Expression Assay primer and probe sets (Applied Biosystems). GAPDH mRNA was used as an internal control for normalization of cDNA loading. Threshold cycle times (Ct) were obtained and relative gene expression was calculated using the comparative cycle time method (39).
qRT-PCR in lung tumor samples
Four 20 µm sections were collected from each of 20 formalin-fixed paraffin-embedded (FFPE) lung tumor tissue specimens (14 adenocarcinoma, 6 squamous cell carcinoma) previously characterized for Fus1 expression by immunohistochemical staining (4). Deparaffinization, digestion, and RNA isolation were performed using the RecoverAll™ Total Nucleic Acid Isolation kit (Ambion, Austin, TX), according to manufacturer instructions. RNA concentration and quality were assessed by spectrophotometer. miRNA expression was measured using 10 ng of total RNA from each sample with TaqMan® microRNA Assays on an ABI 7900HT Real Time PCR Instrument (Applied Biosystems). We used RNU19 as a loading control and normalized all measurements to RNU19 levels. We determined the miRNA expression levels using the absolute quantification method.
Western blots
Cells were washed with ice-cold phosphate buffered saline (PBS) and cell lysates prepared by incubating cells in NP-40 buffer for 1 h on ice. Cell lysates were centrifuged at 13,000 rpm for 15 minutes at 4°C and the pellets were discarded. Protein concentration of the supernatant was determined using the PIERCE BCA assay (PIERCE, Rockford, IL). For electrophoresis, equal amounts of cell lysate protein (30 µg) were resolved by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA). Flag-tagged Fus1 protein was detected by immunoblotting with mouse anti-Flag primary antibody (Sigma-Aldrich, St. Louis, MO) followed by incubation with goat anti-mouse HRP-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Calnexin protein was detected by immunoblotting with rabbit anti-calnexin primary antibody Stressgen Biotechnologies, San Diego, CA) followed by incubation with goat anti-rabbit HRP-conjugated secondary antibody (Santa Cruz Biotechnology). Bands were detected by exposing the membrane to Kodak™ X-OMAT™ Blue XB-1 film (Eastman Kodak, Rochester, NY) for 1–2 minutes. Band intensities were quantitated using a Kodak™ Image Station 2000.
miRNA:target prediction method
Interactions between miR-197, miR-93 and miR-98 and the 3’UTR of FUS1 were predicted by miRmate, a microRNA target prediction algorithm developed in our lab. The determinants of the interaction between microRNAs and their target sites are believed to be (1) perfect complementarity in the seed sequence (bases 2–8 of the miRNA), (2) a favorable free energy of hybridization (ΔG ≈ −20–25 kcal/mol), and (3) accessibility of the target site in the 3’UTR. Most target prediction methods include one or more of these, and often include evolutionary conservation of the target site. miRmate considers complementarity (including the G·U basepairing found in RNA duplexes), based on variable weighting of positions across the miRNA, so as to encourage (but not require) complete complementarity at positions 2–8 of the miRNA (the seed region) and mismatches and insertions in the central bulge at positions 9–11 of the miRNA. This allows strong complementarity at the 3’ end to compensate for suboptimal complementarity at the 5’ end. This analysis is coupled with an estimate of the hybridization free energy of the predicted mRNA-miRNA duplexes. Evolutionary conservation across species is used post hoc to prioritize candidate target sites. Of the relatively small number of experimentally validated miRNA:target pairs, miRmate correctly identifies more with lower (better) rank fractions and misses fewer than other commonly used methods, including TargetScan, miRanda and PicTar.
Colony formation assay
NCI-H1299 cells were plated in 100 mm dishes, grown for 24 h, and transfected with plasmids pcDNA3.1/Flag, pcDNA3.1/Flag-FUS1, and pcDNA3.1/Flag-FUS1-3'UTR. After 48 h of transfection, the cells were trypsinized, and 5,000 cells were replated in 10 cm dishes and cultured in G418 (800 µg/ml) supplemented medium (RPMI 1640, 5% fetal bovine serum) for 2 weeks. Colonies were then stained with 1% crystal violet in ethanol/PBS (15%/85%).
miRNA expression profiling
Oligonucleotide probes for 136 nonredundant, conserved human and mouse miRNAs, antisense to the published mature sequences, were synthesized and spotted in duplicate on Corning GAPS-2 coated slides using a robotic spotter (40). RNA species smaller than 200 nt were isolated from total RNA samples and labeled with a fluorescent modified dinucleotide (5’-phosphate-cytidyl-uridyl-Cy3-3’) using T4 RNA ligase (40). Samples were hybridized to the array along with an equimolar reference oligonucleotide set corresponding to all mature miRNAs, which had been labeled with Cy5. The reference set allows normalization of different datasets to a common standard and in principle allows measurement of absolute expression levels. Array images were analyzed with GenePix Pro 4.1 (Molecular Devices, Sunnyvale, CA). We measured raw Cy3/Cy5 ratios, which we log transformed, normalized by median centering and clustered using average linkage hierarchical clustering based on Pearson correlation.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the technical assistance of J. Michael Thomson and Summer Goodson and thank David Shames, Michael Peyton and Cheng Hui Lee for thoughtful insights and discussions and critical reading of the manuscript. This work was supported in part by Public Health Service grant number P50 CA75907 from the UT Southwestern/MD Anderson Cancer Center Lung Specialized Program of Research Excellence (UTSW/MDACC Lung SPORE) and the National Cancer Institute.
References
- 1.Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res. 2000;60(21):6116–6133. [PubMed] [Google Scholar]
- 2.Ji L, Nishizaki M, Gao B, et al. Expression of several genes in the human chromosome 3p21.3 homozygous deletion region by an adenovirus vector results in tumor suppressor activities in vitro and in vivo. Cancer Res. 2000;62(9):2715–2720. [PMC free article] [PubMed] [Google Scholar]
- 3.Ivanova AV, Ivanov SV, Pascal V, et al. Autoimmunity, spontaneous tumourigenesis, and IL-15 insufficiency in mice with a targeted disruption of the tumour suppressor gene Fus1. The Journal of pathology. 2007;211(5):591–601. doi: 10.1002/path.2146. [DOI] [PubMed] [Google Scholar]
- 4.Prudkin L, Behrens C, Liu DD, et al. Loss and reduction of FUS1 protein expression is a frequent phenomenon in the pathogenesis of lung cancer. Clin Cancer Res. 2008;14(1):41–47. doi: 10.1158/1078-0432.CCR-07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene. 2002;21(45):6915–6935. doi: 10.1038/sj.onc.1205835. [DOI] [PubMed] [Google Scholar]
- 6.Kondo M, Ji L, Kamibayashi C, et al. Overexpression of candidate tumor suppressor gene FUS1 isolated from the 3p21.3 homozygous deletion region leads to G1 arrest and growth inhibition of lung cancer cells. Oncogene. 2001;20(43):6258–6262. doi: 10.1038/sj.onc.1204832. [DOI] [PubMed] [Google Scholar]
- 7.Uno F, Sasaki J, Nishizaki M, et al. Myristoylation of the fus1 protein is required for tumor suppression in human lung cancer cells. Cancer Res. 2004;64(9):2969–2976. doi: 10.1158/0008-5472.can-03-3702. [DOI] [PubMed] [Google Scholar]
- 8.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science (New York, NY. 2004;304(5670):594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 9.Vasudevan S, Tong Y, Steitz JA. Science (New York, NY. 5858. Vol. 318. 2007. Switching from repression to activation: microRNAs can up-regulate translation; pp. 1931–1934. [DOI] [PubMed] [Google Scholar]
- 10.Novotny GW, Sonne SB, Nielsen JE, et al. Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17-92 cluster. Cell death and differentiation. 2007;14(4):879–882. doi: 10.1038/sj.cdd.4402090. [DOI] [PubMed] [Google Scholar]
- 11.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16(19):2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G. RNA (New York, NY. 2. Vol. 9. 2003. Numerous microRNPs in neuronal cells containing novel microRNAs; pp. 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu LL, Shi Y, Petrovics G, et al. PMEPA1, an androgen-regulated NEDD4-binding protein, exhibits cell growth inhibitory function and decreased expression during prostate cancer progression. Cancer Res. 2003;63(15):4299–4304. [PubMed] [Google Scholar]
- 15.Calin GA, Liu CG, Sevignani C, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101(32):11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2009;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang J, Nicolas E, Marks D, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biology. 2004;1(2):106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 18.Chen CZ, Li L, Lodish HF, Bartel DP. Science (New York, NY. 5654. Vol. 303. 2004. MicroRNAs modulate hematopoietic lineage differentiation; pp. 83–86. [DOI] [PubMed] [Google Scholar]
- 19.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279(50):52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 20.Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6(11):1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 21.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 22.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008 doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18(5):549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 24.Boyerinas B, Park SM, Shomron N, et al. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68(8):2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 25.Takakura S, Mitsutake N, Nakashima M, et al. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer science. 2008 doi: 10.1111/j.1349-7006.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, Lee UJ, Kim MN, et al. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283(26):18158–18166. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 27.Jeyaseelan K, Ma D, Armugam A. Real-time detection of gene promoter activity: quantitation of toxin gene transcription. Nucleic Acids Res. 2001;29(12):E58–E58. doi: 10.1093/nar/29.12.e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nature genetics. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 29.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282(19):14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 31.Lopez de Silanes I, Quesada MP, Esteller M. Aberrant regulation of messenger RNA 3'-untranslated region in human cancer. Cell Oncol. 2007;29(1):1–17. doi: 10.1155/2007/586139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumour subtype. Genome Biol. 2007;8(10):R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 34.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarhadi VK, Wikman H, Salmenkivi K, et al. Increased expression of high mobility group A proteins in lung cancer. The Journal of pathology. 2006;209(2):206–212. doi: 10.1002/path.1960. [DOI] [PubMed] [Google Scholar]
- 36.Sgarra R, Rustighi A, Tessari MA, et al. Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer. FEBS letters. 2004;574(1–3):1–8. doi: 10.1016/j.febslet.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Boo LM, Lin HH, Chung V, et al. High mobility group A2 potentiates genotoxic stress in part through the modulation of basal and DNA damage-dependent phosphatidylinositol 3-kinase-related protein kinase activation. Cancer Res. 2005;65(15):6622–6630. doi: 10.1158/0008-5472.CAN-05-0086. [DOI] [PubMed] [Google Scholar]
- 38.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007;104(51):20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bookout AL, Mangelsdorf DJ. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nuclear receptor signaling. 2003;1:e012. doi: 10.1621/nrs.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1(1):47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.