Abstract
Human cancers frequently show a loss of heterozygosity on chromosome 7q31, which indicates the existence of broad-range tumour-suppressor gene(s) at this locus. Truncating mutations in the ST7 gene at this locus are seen frequently in primary colon cancer and breast cancer cell lines. Therefore, the ST7 gene represents a novel candidate gene for the tumour suppressor at this locus. However, more recent studies have reported that ST7 mutations are infrequent or absent in primary cancer and cell lines. To ascertain the frequency of mutations of the ST7 gene in cancer cells, we examined mutations in the ST7 coding sequence in 48 colorectal, 48 gastric, and 48 hepatocellular carcinomas using polymerase chain reaction–single-strand conformational polymorphism and direct sequencing. We detected somatic mutations, which were located near the exon–intron junction in intron 8, in only three out of 144 cases. We conclude that mutations in the ST7 gene are rare in primary colorectal, gastric, and hepatocellular carcinomas.
Keywords: ST7, RAY1, HELG, 7q31, mutation, tumour-suppressor gene, colorectal cancer, gastric cancer, hepatocellular carcinoma
Loss of heterozygosity (LOH) on human chromosome 7q31.1 is found frequently in different human neoplasms, which include cancers of the colon (Zenklusen et al, 1995), stomach (Nishizuka et al, 1997), pancreas (Achille et al, 1996), breast (Bieche et al, 1992), prostate (Latil et al, 1995), ovary (Edelson et al, 1997; Koike et al, 1997), head and neck (Zenklusen et al, 1995), kidney (Shridhar et al, 1997), myeloid system (Liang et al, 1998; Koike et al, 1999), and thyroid gland (Zhang et al, 1998). Previous studies on these cancers have suggested the existence of broad-range tumour-suppressor gene(s) in this chromosomal region.
To date, several genes, such as CAV1, CAV2 (Chang et al, 1994), MET (Vande Woude et al, 1997), CAPZ (Caldwell et al, 1989), WNT2 (Dale et al, 1996), ALP1 (Zenklusen et al, 2001), and CFTR (Seibert et al, 1997), have been located within this region. However, these genes are rarely inactivated by mutations or aberrant promoter methylation. The tumour-suppressor gene(s) responsible for this critical region have not yet been identified (Zenklusen et al, 1999).
The ST7 gene, which in other contexts is designated as RAY1 (Vincent et al, 2000) or HELG (Hughes et al, 2001), maps within this critical region. Recently, frameshift mutations in the ST7 gene have been observed frequently in primary colon cancer and breast cancer cell lines (Zenklusen et al, 2001). The introduction of ST7 cDNA suppressed the tumorigenicity of a prostate cancer cell line in vivo (Zenklusen et al, 2001). These results suggest that the ST7 gene is a candidate tumour-suppressor gene within this critical region. However, there have been reports that somatic mutation in the ST7 gene is extremely rare (Hughes et al, 2001; Thomas et al, 2001; Brown et al, 2002; Dong and Sidransky, 2002). Thus, the previous data on ST7 gene mutations show conflicting results.
In this study, we investigated the true frequency of ST7 gene mutations by examining 48 primary colorectal cancers, 48 primary gastric cancers that frequently show LOH on 7q31 (Nishizuka et al, 1997), and 48 primary hepatocellular carcinomas that show high-level expression of the ST7/RAY1 gene (Vincent et al, 2000; Zenklusen et al, 2001). We surveyed mutations in the entire ST7 coding sequence using polymerase chain reaction–single-strand conformational polymorphism (PCR–SSCP) analysis and direct DNA sequencing.
MATERIALS AND METHODS
Tissue specimens and DNA extraction
Specimens from 48 colorectal, 48 gastric, and 48 hepatocellular carcinomas and corresponding noncancerous tissues were obtained at surgery from Japanese patients. The samples were frozen immediately in liquid nitrogen and stored at −80°C until use. High-molecular-weight DNA was extracted using the standard phenol/chloroform procedure.
Polymerase chain reaction–single-strand conformational polymorphism analysis
All samples were examined by PCR–SSCP analysis for mutations throughout the entire coding sequence of the ST7 gene (exons 1a–16b). The exon–intron boundaries were identified by comparing the cDNA sequences of ST7 (GenBank accession no. AY009152) and the genomic DNA sequence of chromosome 7q31 (AC002542). Using this information, we designed intronic primers for each genomic region, except for exons 1b and 16b (Table 1). The primers for exons 1b and 16b were prepared as described previously (Thomas et al, 2001).
Table 1. Primer sequences for PCR–SSCP analysis of the ST7gene.
| Primer sequences (5′–3′) |
||||
|---|---|---|---|---|
| Exon | Product size (bp) | Tm (°C) | Sense | Antisense |
| 1a | 273 | 62 | gaatcatcccggcagacac | gcgcgagttgcactaacttt |
| 1b | 234 | 58 | agcagagaggagcgctgaa | ttgcactaactttccggggc |
| 2 | 148 | 62 | ccttgttcttctccctttctc | ttaaatgagaaggactccacc |
| 3 | 230 | 58 | aacagtgaccataaacacgct | aaataatattgcaaactgaagg |
| 4 | 162 | 62 | gtagtgtcactgaacttacgc | ctgtctttgctctctgaacc |
| 5 | 369 | 58 | aggtcttgcttttctctctca | gaggggactcatttcaacata |
| 6 | 220 | 62 | ggattgacttggtgttttctc | atcctccagttcaaatgcagt |
| 7 | 176 | 62 | gtgactctctctgaatgttcc | tcatttggttagaagtagggc |
| 8 | 237 | 62 | ggctttgtaattgatggtggc | acaattctgatccccccaatgc |
| 9 | 342 | 58 | tcaacatcctcactcaaaagc | tctgtaagccactgatcccaa |
| 10 | 195 | 58 | attccttggtttcttctgccc | gggaaaatacatcaaaagagg |
| 11 | 191 | 58 | cctgcaaacttatgtgttcct | aacacatctcaattccggtca |
| 12 | 168 | 62 | ggatggtttttgtctttctgc | atcataacgagttcctgtggg |
| 13 | 237 | 62 | attaacacaagtgtgtcctgc | ttagcaccttttcatgctctt |
| 14 | 209 | 62 | cacaaacattggacatctctg | ctggctgaagagaggtgaga |
| 15 | 351 | 58 | gggtcagatgttggctatgg | cttggctttccccatccatt |
| 16a | 200 | 62 | ggtttctgctgacttctgtg | aaggagttggcacagaggag |
| 16b | 225 | 58 | aggcgagtgcaatcagaaag | gaggaggagcagttttggtg |
The genomic DNA template (50 ng) was incubated in a total volume of 10 μl PCR buffer that contained 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 100 nM of each primer, 200 μM of each deoxynucleotide triphosphate, 1.5 Ci of alpha-[32P]dCTP (Amersham Pharmacia), and 0.5 U of rTaq DNA polymerase (TaKaRa). The following PCR conditions were employed: 30 s at 95°C, 30 s at 58°C or 62°C, and 90 s at 72°C for 35 cycles, followed by 10 min at 72°C in a thermal cycler (GeneAmp 9700; Applied Biosystems). Single-strand conformational polymorphism analysis was performed with the low-pH buffer system, which allowed improved separation of fragments of up to 800 bp in length (Kukita et al, 1997). The 32P-labelled PCR products were denatured, loaded on nondenaturing polyacrylamide gels that contained 10% polyacrylamide (99 : 1 acrylamide to bisacrylamide) and TPE (30 mM Tris (pH 6.8), 20 mM PIPES, and 1 mM Na2EDTA), and electrophoresed in TPE buffer at 10°C. The gels were dried and analysed with the BAS 2000 system (Fuji Photo Films). To exclude potential PCR artefacts, all positive cases were tested independently at least three times.
Sequencing analysis
PCR fragments that showed different mobilities were purified using the QIAquick PCR Purification Kit (QIAGEN), and directly sequenced in both directions using the BigDye Terminator Kit and the ABI 3100 DNA Sequencing System (Applied Biosystems).
Analysis of microsatellite instability
We assessed microsatellite instability using five reference markers (D2S123, BAT25, BAT26, D5S346, and D17S250) and the criteria recommended by the National Cancer Institute workshop (Boland et al, 1998; Yamada et al, 2002).
Statistical analysis
Statistical analysis was performed using the StatView 5.0. program (SAS Institute Inc.). The χ2, Fisher's exact, and Mann–Whitney U-tests were used for background and clinicopathological data. A P-value of less than 0.05 was considered to be statistically significant.
Ethics
This study was carried out with the approval of the ethical committee of Gunma University Faculty of Medicine.
RESULTS
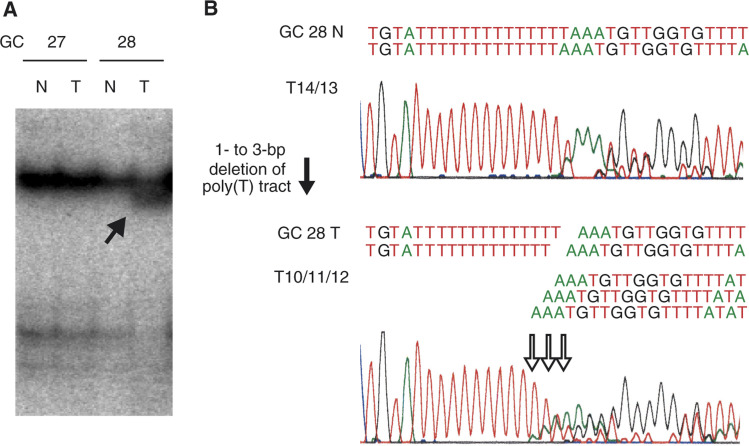
We detected a somatic mutation in the polypyrimidine tract within the splice-acceptor site of the intron 8–exon 9 junction. Deletions in intron 8 (−32 nucleotides from exon 9) were found in one out of 48 (2.1%) of the colorectal cancers, and in two out of 48 (4.1%) of the gastric cancers (Table 2). The number of deleted nucleotides in one tumour sample ranged from one to three bases, which demonstrates the highly heterogeneous nature of the tumour. A representative case is shown in Figure 1. All the three patient groups exhibited high-frequency microsatellite instability (microsatellite instability-high; MSI-H).
Table 2. Mutations in the ST7 gene.
| Case | Type of tumour | Locations | Mutations | Amino-acid substitution | Status | Microsatellite instability |
|---|---|---|---|---|---|---|
| CRC39 | Colorectal | Intron 8 (−32 nt from exon 9) | 1 to 3 nt deletion (heterogeneous) | None | Somatic mutation | MSI-H |
| GC18 | Gastric | Intron 8 (−32 nt from exon 9) | 1 to 3 nt deletion (heterogeneous) | None | Somatic mutation | MSI-H |
| GC28 | Gastric | Intron 8 (−32 nt from exon 9) | 1 to 3 nt deletion (heterogeneous) | None | Somatic mutation | MSI-H |
| CRC18 | Colorectal | Exon 5 | 427 G to A (heterozygous) | 143 Ala to Thr | Germline mutation or rare polymorphism | MSS |
nt=nucleotide; CRC=colorectal cancer; GC=gastric cancer; MSI-H=microsatellite instability-high; MSS=microsatellite stable.
Figure 1.
Representative example of an ST7 frameshift mutation. (A) SSCP analysis of the intron 8–exon 9 junction of the ST7 gene. The solid arrow indicates a shifted band in the tumour sample. T: tumour samples; N: corresponding normal tissue samples. (B) Sequence analysis. The open arrow indicates deletions in the polypyrimidine tract within the splice-acceptor site of intron 8 (−3 nucleotides from exon 9). The number of nucleotides deleted ranged from one to three.
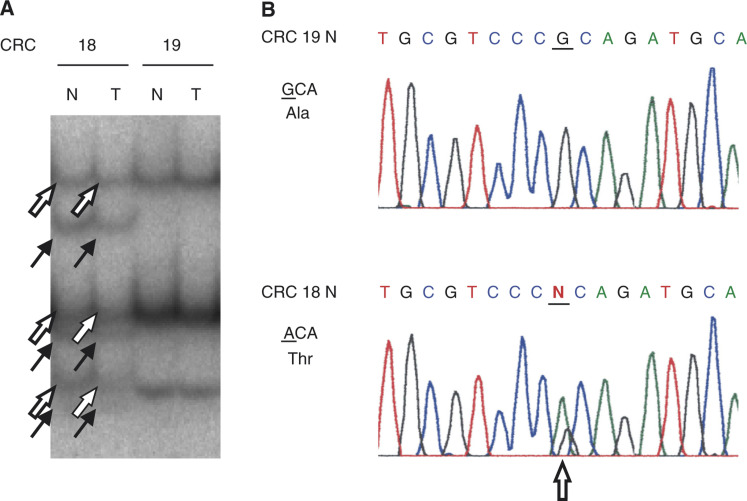
We also detected a G to A substitution at the first nucleotide of codon 143 (GenBank accession no. AY009152) in exon 5 of one of the colorectal cancer cases (Figure 2, Table 2). This substitution resulted in an amino-acid change from Ala to Thr. The same substitution was found in the corresponding normal tissue from the same patient. Thus, the change represents a germline mutation or rare polymorphism.
Figure 2.
Representative example of a 1-bp substitution in the coding sequence of the ST7 gene. (A) SSCP analysis of the intron 8–exon 9 junction of the ST7 gene. The solid arrow indicates the shifted band that was predicted to carry substitutions, in both the tumour and corresponding normal tissue sample. The open arrow indicates another allele without a substitution. T: tumour samples; N: corresponding normal tissue samples; CRC=colorectal cancer. (B) Sequence analysis. The open arrow indicates a one-nucleotide substitution in exon 9 of the ST7 gene.
In addition, we identified four single-nucleotide polymorphisms (SNPs) in introns 8, 10, 11, and 15 of the ST7 gene (Table 3). There were no correlations between these SNPs and the clinicopathological data.
Table 3. Polymorphisms in the ST7 gene locus detected in this study.
| Allele frequency |
||||
|---|---|---|---|---|
| Location | Nt | CRC | GC | HCC |
| Intron 8 (−31 nt from exon 9) | A/T | 0.05/0.95 | 0.02/0.98 | 0.03/0.97 |
| Intron 10 (+8 nt from exon 9) | T/C | 0.20/0.80 | 0.20/0.80 | 0.18/0.82 |
| Intron 11 (+17 nt from exon 10) | C/T | 0.20/0.80 | 0.20/0.80 | 0.16/0.84 |
| Intron 15 (+28 nt from exon 14) | T/G | 0.00/1.00 | 0.01/0.99 | 0.00/1.00 |
nt=nucleotide; CRC=colorectal cancer; GC=gastric cancer; HCC=hepatocellular carcinoma. Allele frequency was determined in each of the 48 cancers.
DISCUSSION
We detected somatic mutations in the polypyrimidine tract within the splice-acceptor site of intron 8, although the frequency of mutation was low. The polypyrimidine tract is essential for efficient branch-point utilisation and splice-site recognition, and deletions within this region affect splicing efficiency (Roscigno et al, 1993). We were unable to examine whether the mutation at the polypyrimidine tract induced insufficient splicing, because the appropriate RNA samples were not available. Therefore, we could not confirm the involvement of this mutation in carcinogenesis and the progression of colorectal and gastric cancers. However, considering the fact that mutations were found only in the tumour samples and not in the corresponding noncancerous samples, we cannot exclude the possibility that this mutation confers advantages upon these cancer cells under selective pressure.
All of the cases with the polypyrimidine-tract mutation showed high-frequency microsatellite instability. It is known that a simple mononucleotide repeat can act as a mutational target in tumours that show high-frequency MSI (Perucho, 1999; Yamada et al, 2002). Therefore, the polypyrimidine tract, in which we found mutations, may be a mutational target in MSI-positive tumours, and mutations therein may be involved in carcinogenesis and the progression of MSI-positive tumours. Microsatellite instability-high is found rarely in hepatocellular carcinomas, and we could not detect any mutations in the polypyrimidine tract in the 48 cases of hepatocellular carcinoma (Saeki et al, 2000; Yamamoto et al, 2000; Wang et al, 2001).
We also detected a single-nucleotide substitution with amino-acid change in one patient with colon cancer. This substitution was identical to that identified previously in the breast cancer cell line MDA-MB435 (Thomas et al, 2001). Since the corresponding noncancerous cell line was not available, these investigators could not determine whether the change was somatic or germline specific. In contrast, we found the same substitution in the corresponding normal tissue from the same patient. Therefore, the change is not somatic, but represents a germline mutation or rare polymorphism. Further functional studies are needed to clarify the ramifications of this amino-acid substitution.
In addition, we detected four SNPs in the ST7 gene locus. There were no correlations between these SNPs and the clinicopathological data. The consequences of these SNPs for colorectal, gastric, and hepatocellular carcinomas are unclear.
Contrary to the result of Zenklusen et al, we rarely detected mutations in the ST7 gene of patients with colorectal, gastric, or hepatocellular carcinoma, a finding that has been corroborated by other groups (Hughes et al, 2001; Thomas et al, 2001; Brown et al, 2002; Dong and Sidransky, 2002). In our study, there were no technical problems in detecting mutations; the frequent detection of SNPs demonstrates the high sensitivity of our procedure. Although the reason for the discrepancy between our results and those of Zenklusen et al is unclear, we (and the aforementioned groups) propose the following possible explanations: the use of selected specimens, the presence of PCR artefacts, and the effects of culture passages (Hughes et al, 2001; Thomas et al, 2001; Brown et al, 2002).
We conclude that ST7 gene mutations are rare in colorectal, gastric, and hepatocellular carcinomas. Our results do not exclude the possibility that the ST7 gene is inactivated by other molecular mechanisms, such as aberrant hypermethylation or haplo-insufficiency (Merlo et al, 1995; Largaespada, 2001). Since there have been no reports on the expression of the ST7 gene in cancer cells, further studies are needed to understand the role of this gene in carcinogenesis and the progression of these cancers.
Acknowledgments
This work was supported in part by the Harnasou Foundation Cancer Research Subsidizing Fund, the Maebashi Norte Hospital Fund, the Kan-etsu Chuo Hospital Fund and the Kato Surgical Hospital Fund.
References
- Achille A, Biasi MO, Zamboni G, Bogina G, Magalini AR, Pederzoli P, Perucho M, Scarpa A (1996) Chromosome 7q allelic losses in pancreatic carcinoma. Cancer Res 56: 3808–3813 [PubMed] [Google Scholar]
- Bieche I, Champeme MH, Matifas F, Hacene K, Callahan R, Lidereau R (1992) Loss of heterozygosity on chromosome 7q and aggressive primary breast cancer. Lancet 339: 139–143 [DOI] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S (1998) A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predis-position: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58: 5248–5257 [PubMed] [Google Scholar]
- Brown VL, Proby CM, Barnes DM, Kelsell DP (2002) Lack of mutations within ST7 gene in tumour-derived cell lines and primary epithelial tumours. Br J Cancer 87: 208–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell JE, Heiss SG, Mermall V, Cooper JA (1989) Effects of CapZ, an actin capping protein of muscle, on the polymerization of actin. Biochemistry 28: 8506–8514 [DOI] [PubMed] [Google Scholar]
- Chang WJ, Ying YS, Rothberg KG, Hooper NM, Turner AJ, Gambliel HA, De Gunzburg J, Mumby SM, Gilman AG, Anderson RG (1994) Purification and characterization of smooth muscle cell caveolae. J Cell Biol 126: 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale TC, Weber-Hall SJ, Smith K, Huguet EL, Jayatilake H, Gusterson BA, Shuttleworth G, O'Hare M, Harris AL (1996) Compartment switching of WNT-2 expression in human breast tumors. Cancer Res 56: 4320–4323 [PubMed] [Google Scholar]
- Dong SM, Sidransky D (2002) Absence of ST7 gene alterations in human cancer. Clin Cancer Res 8: 2939–2941 [PubMed] [Google Scholar]
- Edelson MI, Scherer SW, Tsui LC, Welch WR, Bell DA, Berkowitz RS, Mok SC (1997) Identification of a 1300 kilobase deletion unit on chromosome 7q31.3 in invasive epithelial ovarian carcinomas. Oncogene 14: 2979–2984 [DOI] [PubMed] [Google Scholar]
- Hughes KA, Hurlstone AF, Tobias ES, McFarlane R, Black DM (2001) Absence of ST7 mutations in tumor-derived cell lines and tumors. Nat Genet 29: 380–381 [DOI] [PubMed] [Google Scholar]
- Koike M, Takeuchi S, Yokota J, Park S, Hatta Y, Miller CW, Tsuruoka N, Koeffler HP (1997) Frequent loss of heterozygosity in the region of the D7S523 locus in advanced ovarian cancer. Genes Chromosomes Cancer 19: 1–5 [DOI] [PubMed] [Google Scholar]
- Koike M, Tasaka T, Spira S, Tsuruoka N, Koeffler HP (1999) Allelotyping of acute myelogenous leukemia: loss of heterozygosity at 7q31.1 (D7S486) and q33–34 (D7S498, D7S505). Leuk Res 23: 307–310 [DOI] [PubMed] [Google Scholar]
- Kukita Y, Tahira T, Sommer SS, Hayashi K (1997) SSCP analysis of long DNA fragments in low pH gel. Hum Mutat 10: 400–407 [DOI] [PubMed] [Google Scholar]
- Largaespada DA (2001) Haploinsufficiency for tumor suppression: the hazards of being single and living a long time. J Exp Med 193: F15–F18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latil A, Cussenot O, Fournier G, Baron JC, Lidereau R (1995) Loss of heterozygosity at 7q31 is a frequent and early event in prostate cancer. Clin Cancer Res 1: 1385–1389 [PubMed] [Google Scholar]
- Liang H, Fairman J, Claxton DF, Nowell PC, Green ED, Nagarajan L (1998) Molecular anatomy of chromosome 7q deletions in myeloid neoplasms: evidence for multiple critical loci. Proc Natl Acad Sci USA 95: 3781–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D (1995) 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 1: 686–692 [DOI] [PubMed] [Google Scholar]
- Nishizuka S, Tamura G, Terashima M, Satodate R (1997) Commonly deleted region on the long arm of chromosome 7 in differentiated adenocarcinoma of the stomach. Br J Cancer 76: 1567–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perucho M (1999) Correspondence re: C.R. Boland et al, A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 59: 249–256 [PubMed] [Google Scholar]
- Roscigno RF, Weiner M, Garcia-Blanco MA (1993) A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. J Biol Chem 268: 11222–11229 [PubMed] [Google Scholar]
- Saeki A, Tamura S, Ito N, Kiso S, Matsuda Y, Yabuuchi I, Kawata S, Matsuzawa Y (2000) Lack of frameshift mutations at coding mononucleotide repeats in hepatocellular carcinoma in Japanese patients. Cancer 88: 1025–1029 [PubMed] [Google Scholar]
- Seibert FS, Loo TW, Clarke DM, Riordan JR (1997) Cystic fibrosis: channel, catalytic, and folding properties of the CFTR protein. J Bioenerg Biomembr 29: 429–442 [DOI] [PubMed] [Google Scholar]
- Shridhar V, Sun QC, Miller OJ, Kalemkerian GP, Petros J, Smith DI (1997) Loss of heterozygosity on the long arm of human chromosome 7 in sporadic renal cell carcinomas. Oncogene 15: 2727–2733 [DOI] [PubMed] [Google Scholar]
- Thomas NA, Choong DY, Jokubaitis VJ, Neville PJ, Campbell IG (2001) Mutation of the ST7 tumor suppressor gene on 7q31.1 is rare in breast, ovarian and colorectal cancers. Nat Genet 29: 379–380 [DOI] [PubMed] [Google Scholar]
- Vande Woude GF, Jeffers M, Cortner J, Alvord G, Tsarfaty I, Resau J (1997) Met-HGF/SF: tumorigenesis, invasion and metastasis. Ciba Found Symp 212: 119–130 [DOI] [PubMed] [Google Scholar]
- Vincent JB, Herbrick JA, Gurling HM, Bolton PF, Roberts W, Scherer SW (2000) Identification of a novel gene on chromosome 7q31 that is interrupted by a translocation breakpoint in an autistic individual. Am J Hum Genet 67: 510–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Bani-Hani A, Montoya DP, Roche PC, Thibodeau SN, Burgart LJ, Roberts LR (2001) hMLH1 and hMSH2 expression in human hepatocellular carcinoma. Int J Oncol 19: 567–570 [PubMed] [Google Scholar]
- Yamada T, Koyama T, Ohwada S, Tago K, Sakamoto I, Yoshimura S, Hamada K, Takeyoshi I, Morishita Y (2002) Frameshift mutations in the MBD4/MED1 gene in primary gastric cancer with high-frequency microsatellite instability. Cancer Lett 181: 115–120 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Itoh F, Fukushima H, Kaneto H, Sasaki S, Ohmura T, Satoh T, Karino Y, Endo T, Toyota J, Imai K (2000) Infrequent widespread microsatellite instability in hepatocellular carcinomas. Int J Oncol 16: 543–547 [DOI] [PubMed] [Google Scholar]
- Zenklusen JC, Conti CJ, Green ED (2001) Mutational and functional analyses reveal that ST7 is a highly conserved tumor-suppressor gene on human chromosome 7q31. Nat Genet 27: 392–398 [DOI] [PubMed] [Google Scholar]
- Zenklusen JC, Thompson JC, Klein-Szanto AJ, Conti CJ (1995) Frequent loss of heterozygosity in human primary squamous cell and colon carcinomas at 7q31.1: evidence for a broad range tumor suppressor gene. Cancer Res 55: 1347–1350 [PubMed] [Google Scholar]
- Zenklusen JC, Weintraub LA, Green ED (1999) Construction of a high-resolution physical map of the approximate 1-Mb region of human chromosome 7q31.1–q31.2 harboring a putative tumor suppressor gene. Neoplasia 1: 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JS, Nelson M, McIver B, Hay ID, Goellner JR, Grant CS, Eberhardt NL, Smith DI (1998) Differential loss of heterozygosity at 7q31.2 in follicular and papillary thyroid tumors. Oncogene 17: 789–793 [DOI] [PubMed] [Google Scholar]