Abstract
Prion diseases are a group of fatal neurodegenerative disorders characterized by the accumulation of a misfolded form (PrPSc) of the cellular prion protein (PrPC) in the brains of affected individuals. The conversion of PrPC to PrPSc is thought to involve a change in protein conformation from a normal, primarily α-helical structure into a β-sheet conformer. Few proteins have been identified that differentially interact with the two forms of PrP. It has been reported that plasminogen binds to PrPSc from a variety of prion phenotypes. We have examined potential motifs within the kringle region that may be responsible for binding to PrP. We synthesized 12–15-mer peptides that contain small, repetitive stretches of amino acid residues found within the kringle domains of plasminogen. These synthetic peptides were found to capture PrPSc from the brain homogenates of bovine spongiform encephalopathy affected cattle, chronic wasting disease affected elk, experimental scrapie of hamsters and that of subjects affected by Creutzfeldt-Jakob disease, without binding to PrPC in unaffected controls. Therefore, we have identified critical peptide motifs that may be important for protein-protein interactions in prion disease pathogenesis. The ability of these synthetic peptides to bind preferentially to PrPSc suggests a potential application in the diagnosis of prion diseases.
Keywords: Prion disease, prion protein, kringle domain, bovine spongiform encephalopathy, Creutzfeldt-Jakob disease, diagnosis
Introduction
Prion diseases, also called transmissible spongiform encephalopathies, are invariably fatal neurodegenerative diseases affecting both humans and animals. Prion diseases in animals include scrapie in sheep, chronic wasting disease in cervids, and bovine spongiform encephalopathy (BSE) in cattle [1]. In humans, the disease may occur spontaneously, as in sporadic Creutzfeldt-Jakob disease (CJD); have a genetic origin, in the form of familial CJD, fatal familial insomnia or Gerstmann-Sträussler-Scheinker disease (GSS); or it may be transmitted infectiously, as seen in iatrogenic CJD and variant CJD (vCJD) [2, 3]. Prion diseases are unique among infectious diseases in that its etiology involves no nucleic acid. It is thought to be caused when the normal cellular prion protein (PrPC), widely expressed in the central nervous system, undergoes a conformational change from a primarily α-helical structure in to a β-sheet structure. The disease causing β-sheet form of the protein is called PrPSc [1].
Plasminogen is a ubiquitous pro-protease which may be cleaved to form the serine protease plasmin, an important component of the fibrinolytic system [4]. The fibrinolytic pathway has been implicated in biological processes as diverse as cell migration [5], inflammation [6], neuronal plasticity [7] and embryonic implantation [8], and also excitotoxin-mediated neurotoxicity [9] or Alzheimer’s disease [10]. Plasminogen may be activated by one of two mechanisms, the urokinase plasminogen activator (uPA) and the tissue type plasminogen activator (tPA) [11]. The former is normally associated with pericellular proteolytic activity, while the latter is associated with the dissolution of intravascular clots [4]. It has previously been demonstrated that plasminogen forms a complex with PrPSc from mice, and a variety of other species, with no binding observed to PrPC [12, 13]. This interaction was thought to be mediated through the first three kringle domains of plasminogen [12]. Each kringle domain is composed of approximately 80 amino acids, and contains 3 disulfide bond [14]. They are joined to other kringle domains by varying lengths of linker amino acids. Kringle domains are not unique to plasminogen; they are also found in a variety of proteins associated with fibrinolysis, coagulation and angiogenesis[14]. The first three kringle domains in plasminogen contain a lysine binding domain, which may be responsible for the majority of its interactions with other proteins [15]. Structurally, plasminogen contains five kringle domains in total, as well as a C-terminal proteolytic domain. Although no interaction between plasminogen and PrPC was found using plasminogen to capture PrPC from brain homogenate [12, 13], later work indicated the possibility of an interaction with recombinant human PrPC [16–18]. While there has been no evidence of an interaction between plasminogen and PrPSc or PrPC in vivo, in vitro there have been several recent studies that indirectly suggest an interaction using both recombinant human PrPC, as well as investigations into the role of plasminogen in CJD [19].
Binding of recombinant human PrPC has been observed to both plasminogen and tPA [16]. Direct binding of full length recombinant PrP to the kringle domains of plasminogen was observed by ELISA, but was significantly reduced using the recombinant fragment PrP(89–231) [17]. Recombinant prion protein, bound to copper, was found to increase the rate of activation of plasminogen with tPA, but no effect was found in the absence of copper, or with the uPA activation system [18]. This activation of plasminogen using PrP was found to be conserved in the N-terminal region of the protein, which is a relatively unstructured region to which copper was known to bind[16, 20]. Further, plasmin was found to cleave PrPC, with the cleavage site located at lysine 110, and that the activation was further augmented in the presence of low molecular weight heparin [21]. Mutants of PrP were made in later work by the same group, and it was then discovered that both lysine clusters in the N terminal region of the prion protein are required for activation [22]. Again, the activation of the plasminogen was only found to be enhanced in the presence of PrPC in the tPA system.
The cortical neurons of individuals with CJD have been found to express the uPA receptor [19]. The expression of this receptor is thought to be associated with signaling cascades that lead to eventual neurodegeneration. Although neurons associated with the most uPA receptor had the most degenerative hallmarks, no interaction was demonstrated between the plasminogen itself and either form of the prion protein. When plasminogen concentrations and activities were examined in brains of patients with CJD, compared to those with other forms of dementia, it was found that the plasminogen concentrations were higher, but the activity was lower in CJD [23]. This suggests a relationship between plasminogen and the disease process of CJD, but it is not conclusive. However, other work, in which plasminogen knock out mice were used, found that the absence of plasminogen led to a faster disease course for scrapie infected animals, suggesting plasminogen has a neuroprotective effect [24].
With the discovery of a causal relationship between the consumption of beef contaminated with BSE and the development of vCJD, prion diseases for the first time presented a challenge for public health [25, 26]. At the present time, diagnosis of the disease is limited to the detection of proteinase K (PK) resistant PrPSc in a Western blot, or by immunohistochemistry, both of which are usually performed on brain tissue at autopsy [27]. We aimed to exploit the PrPSc binding properties of the plasminogen kringle domains through the development of short peptides, based on sequences that appear frequently within the plasminogen kringle domains. These peptides bind to PrPSc, but not PrPC, as detected by affinity-capture assay and immunoblotting. Of several peptides that were found to bind PrPSc in preliminary analysis, we selected two for further study, one 12 residues in length, designated Peptide 1 (P1), (sequence: YRGYRGYRGYRG), and the second, 15 residues in length, designated Peptide 2 (P2), (sequence: YRGRYGYKGKYGYRG).
Methods
Reagents and Peptides
Magnetic beads (M-280 streptavidin) were from Dynal (Oslo, Norway). Proteinase K (PK) was obtained from Sigma, and Pefabloc SC (4-(2-Aminoethyl)-benzenesulfonyl fluoride hydrochloride) protease inhibitor was obtained from Roche Applied Science (Indianapolis, IN). Antibodies used in this study were 3F4, recognizing an epitope of human and Syrian hamster PrP at residues 109–112 [28] and animal prion protein was detected using the monoclonal antibody 8H4 [29]. The horseradish peroxidase secondary antibody, and the chemiluminescence reagent were both purchased from Amersham Biosceinces (Piscataway, NJ). All other chemicals and reagents were purchased from Sigma, unless otherwise indicated. Peptides were synthesized using standard methods, obtained from Invitrogen (Carlsbad, CA). The sequence of P1 is NH2-YRGYRGYRGYRG[K-IcBiotin][CONH2], P2 is NH2-YRGRYGYKGKYGYRG[K-IcBiotin][CONH2], and the unrelated control peptide used was NH2-[biotin][AMCAP]-SEIKLLIS[CONH2].
Brain Tissues
Brain tissue was acquired at autopsy from individuals with and without prion diseases, and archived frozen at −80 °C at the National Prion Disease Pathology Surveillance Center. The tissue was then homogenized in lysis buffer (100 mM NaCl, 10 mM EDTA, 0.5 % Nonidet P-40, 0.5 % sodium deoxycholate, 10 mM Tris HCl, pH 7.5) containing a mixture of protease inhibitors (Roche Applied Science) to a concentration of 10 % (w/v). The brain homogenate was stored at −80 °C. The diagnosis of the various phenotypes of prion diseases was confirmed using standard methods, including immunohistochemistry, immunoblotting, and DNA typing. Animal brain tissues were homogenized and stored in the same manner, with the diagnosis confirmed by immunoblotting and immunohistochemistry.
Peptide Capture Assay
The biotinylated P1 or P2 (70 µl of a 1 mg ml−1 preparation) was conjugated to 2.3×108 streptavidin superparamagnetic beads in 1 ml of phosphate-buffered saline, pH 7.5 (PBS) at 37 °C for 20 h. The conjugated beads were then incubated in 0.1 % BSA in PBS, at 37 °C for 4 h, to block nonspecific binding. Once sodium azide is added (0.1 % final concentration), the conjugated beads are stable at 4 °C for at least 8 weeks. The angiostatin (K(1+2+3)) were obtained from Sigma, and conjugated to 2.3×108 tosyl-activated paramagnetic beads in 1 ml of PBS for 20 h at 37°C. The conjugated beads were incubated in 0.1 % BSA 0.2 M Tris-HCl, pH 8.6, for 4 h at 37 °C, to deactivate any unbound tosyl groups, and to block non-specific binding. The beads are stored at 4 °C, in 0.1 % BSA in PBS. The peptide capture assay was performed using 100 µl of the conjugated beads in 900 µl of capture buffer (PBS, 3 % Tween 20, 3 % Nonidet P-40), to which the 10 % brain homogenate is added (6 µl of human brain homogenate, or 3 µl of animal brain homogenate). The mixture was incubated with constant rotation at room temperature for 3 h. Following the incubation, the beads were washed 3 times with wash buffer (PBS, 2 % Tween-20, 2 % Nonidet P-40), to remove unbound materials. The beads were then resuspended in SDS Sample buffer (3 % SDS, 2 mM EDTA, 10 % glycerol, 2.5 % β mercaptoethanol and 50 mM Tris HCl, pH 6.8), and heated to 100 °C for 10 min.
Immunoblotting
The samples were loaded on to an SDS-PAGE gel (15 % Tris-glycine pre-cast gel, BioRad), separated, and electrotransferred onto a polyvinylidene difluoride membrane (Millipore). PrP was detected using the anti-PrP monoclonal antibody (mAb) 3F4 (1:50,000) for hamster and human forms of the protein, and 8H4 mAb (1:3,000) for other animal forms. Following the addition of a horseradish peroxidase conjugated secondary antibody, the ECL Plus kit (Amersham Biosciences) was used to visualize immunoreactivity.
PK Digestion
PK was added to samples of total brain homogenate, or conjugated beads resuspended in 20 µl of lysis buffer, at 50 µg ml−1. Incubation, with shaking, was at 37°C for 1 h. Pefabloc SC (Roche Applied Science, Indianapolis, IN) was added, at a final concentration of 5 mM, to stop the reaction. 2X SDS sample buffer was then added, before immunoblotting.
Conformational Stability Immunoassay
The conformational stability immunoassay was performed according to published methods [30, 31]. After incubation of 10 % brain homogenate from a Syrian hamster infected with experimental scrapie (strain 263K) in 0–3.0 M guanidine hydrochloride, and removal of the guanidine HCl with the addition of a 5-fold volume of methanol, the samples were evaluated by either PK digestion followed by immunoblotting, as described above, or by a peptide capture assay using peptide 1.
Competition Assay
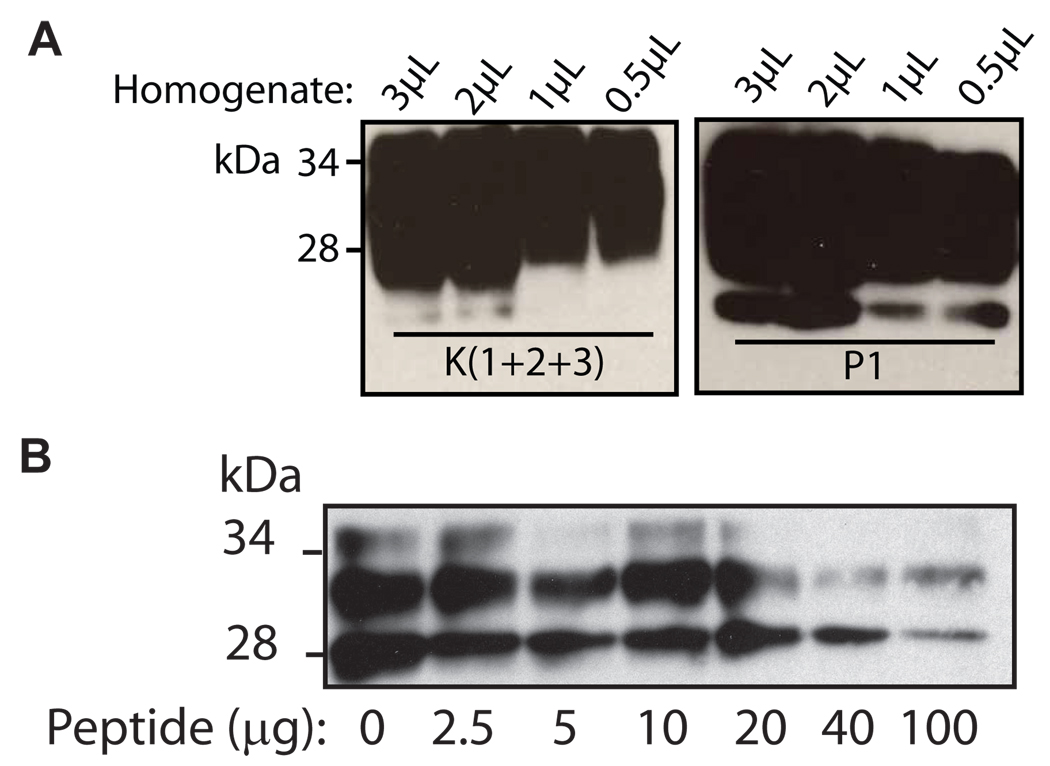
The competition assay was performed in a similar manner to the peptide capture assay described above. Increasing amounts of free peptide (P1, 0–100 µg) were added to each peptide capture reaction, and incubated at room temperature for 3 h, before immunoblotting as described above.
Plasma spiking assay
Plasma samples containing 80 mM EDTA from individuals unaffected by CJD were obtained from the National Prion Disease Pathology Surveillance Center. The peptide capture assay was performed as described previously, with the capture buffer replaced with a solution of 900 µl plasma and 100µl capture buffer, followed by immunoblotting as described above.
Results
The mini-kringle Peptide 1 and 2 specifically capture PrPSc, but not PrPC
The mini-kringle peptides, P1 (YRGYRGYRGYRG) and P2 (YRGRYGYKGKYGYRG), were derived from the YRG sequence that appears frequently through out the sequence of the kringle domains found in plasminogen (Fig. 1). Biotinylated P1 and P2 were conjugated to streptavidin magnetic beads, and then incubated with homogenates from normal or diseased brains for 3 hours at room temperature. To avoid non-specific protein precipitation, the reaction was performed in the presence of high concentrations of detergents (3 % Tween 20 and 3 % Nonidet P-40), and beads were washed and recovered by magnetic force applied to the side of the reaction tubes. As shown in Fig. 2, PrP was successfully captured by P1 and P2 in large quantities from brains of humans affected by prion diseases. No PrP was captured from human brains unaffected by prion diseases. To ensure that the binding of PrP to the peptides was specific, streptavidin beads without conjugated peptide and beads conjugated with an unrelated peptide served as negative controls.
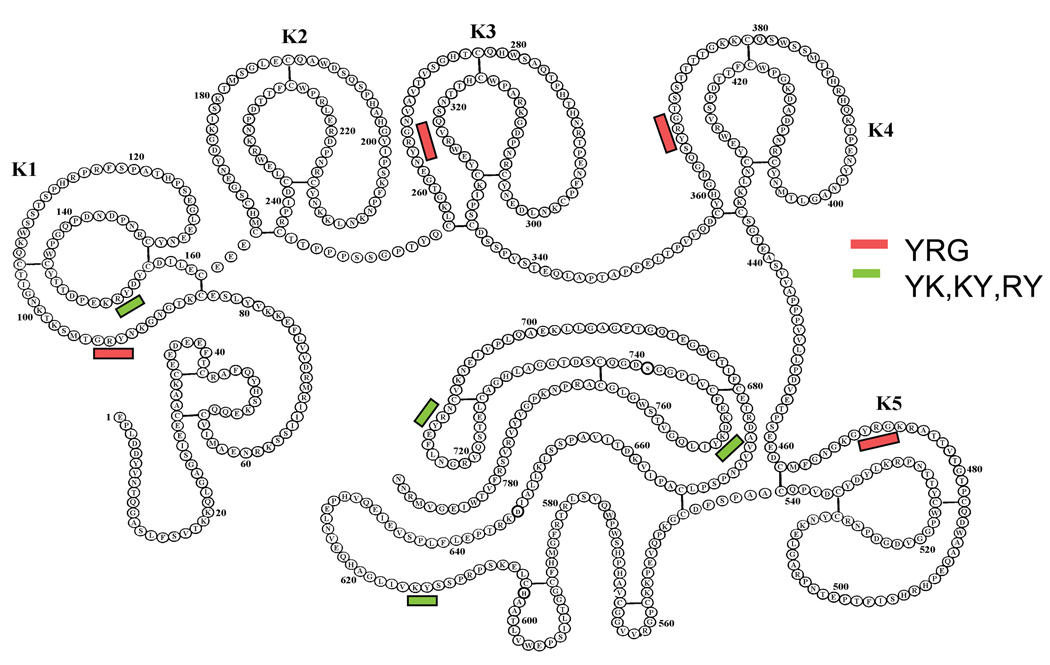
Fig. 1. The presence of the YRG motif in the kringle domains of plasminogen.
Plasminogen molecule consists of a total of five kringle domains labeled as K1–K5, and a C-terminal region. The YRG sequence appears in four out of the five kringle domains. The shorter YK, KY and RY sequences appear also in different regions of the plasminogen molecule.
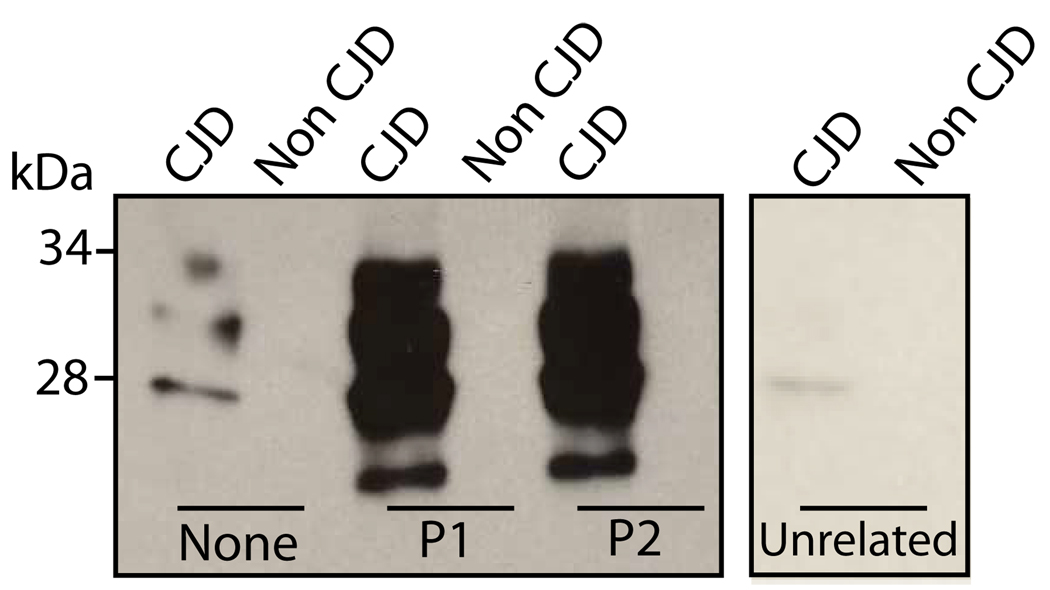
Fig. 2. Capture of disease-associated PrP by P1 and P2.
A. P1 and P2, biotinylated and coupled to streptavidin magnetic beads, were used to capture PrP from the brain homogenates of CJD patients, as well as unaffected controls (Non-CJD). Beads with no conjugated peptide, but blocked with 0.1 % BSA, were used as a negative control (None). An unrelated peptide was biotinylated and conjugated to streptavidin magnetic beads, as a second negative control (Unrelated).
The mini-kringle peptides capture PrP from a variety of human and animal prion diseases
The peptide capture assay using P1 and P2 was used to capture PrP from a wide variety of human and animal prion diseases, including the animal prion diseases with the greatest significance for human health, chronic wasting disease and BSE (Fig. 3). P1 and P2 successfully captured PrP from these diseases, as well as from Syrian hamsters affected with the experimental scrapie strain 263K, but not from the brain of any animal unaffected by prion diseases (Fig. 3). In addition, P1 and P2 successfully captured PrP from brain homogenate of patients affected by type 1 and type 2, as well as a case of mixed type 1/2 of sporadic CJD[2] (Fig. 4A). Also tested with similar findings were a familial CJD case (E200K mutation), a case of GSS (P102L mutation), and CJD cases with an infectious etiology, iatrogenic and variant CJD (Fig. 4). In all cases, brains from individuals who were unaffected by CJD were included. PrP was never captured from these non-CJD control brains, although it is abundantly present prior to capture by P1 and P2 (Fig. 4).
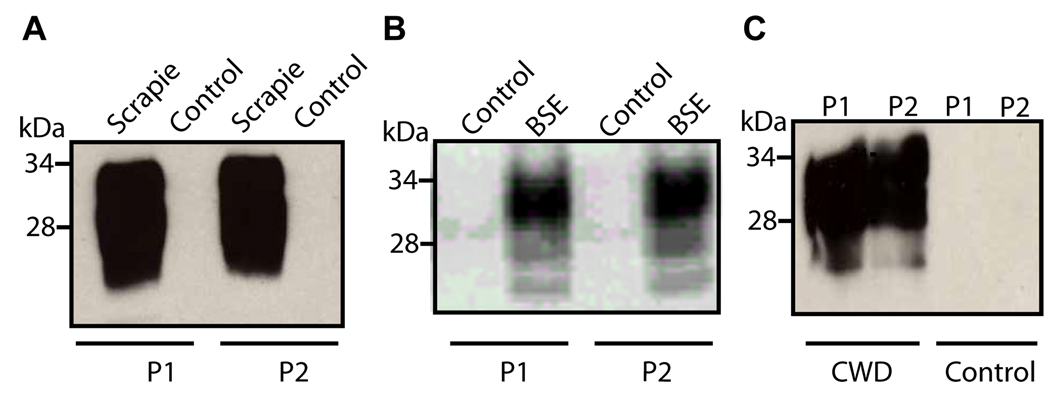
Fig. 3. Capture of PrPSc from animal forms of prion disease by Peptides 1 and 2.
P1 and P2, coupled to the magnetic beads, were used to pull down PrPSc in brain homogenates from Syrian hamsters infected with scrapie strain 263K (left panel), cattle affected by BSE (middle panel) and elk affected by chronic wasting disease (right panel). For the capture assay, peptide conjugated beads were incubated with 6 µl of 10 % brain homogenate in 1 ml PBS containing 3 % Tween-20 and 3 % NP-40, for three h at room temperature. Beads were recovered by applying magnetic field and proteins bound to beads were subjected to immunoblotting using either 3F4 mAb (in A) or 8H4 mAb (in B).
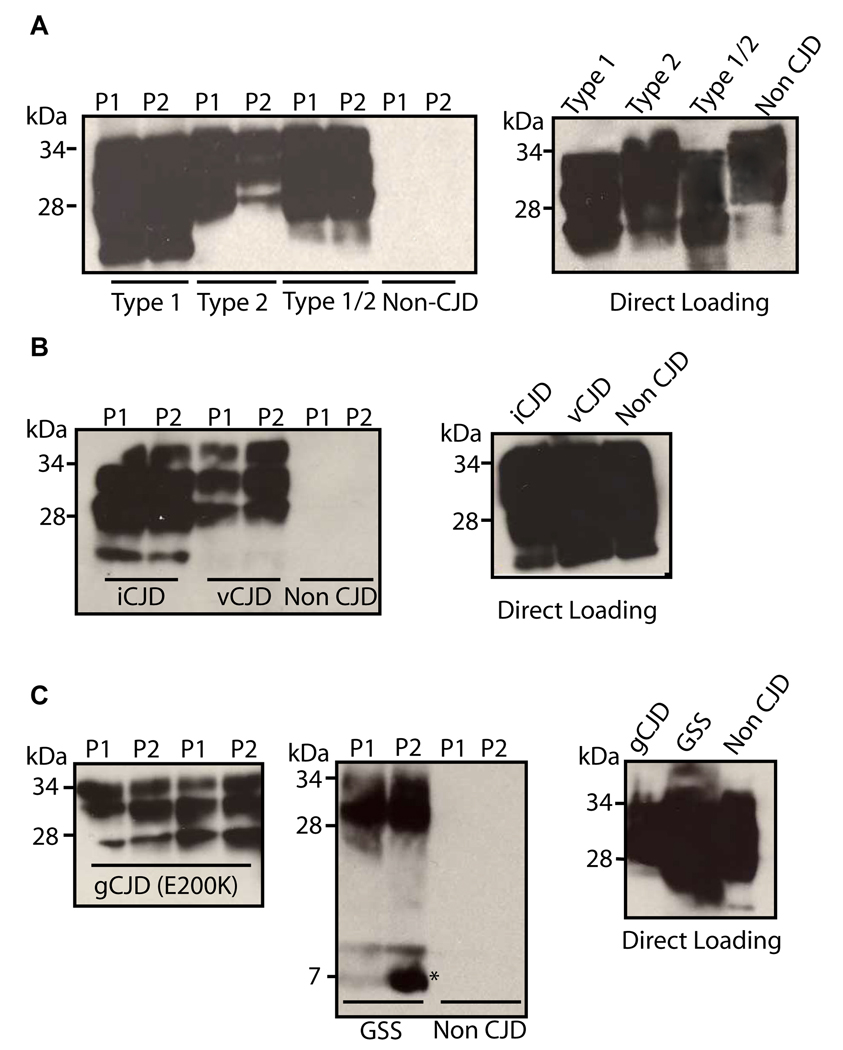
Fig. 4. P1 and P2 capture PrPSc from different subtypes of human prion diseases.
A. P1 and P2, coupled to streptavidin magnetic beads, were used to pull down PrPSc from 10% brain homogenate of individuals affected by Type 1 sCJD (Lanes 1 and 2), Type 2 sCJD (lanes 3 and 4), Type 1/2 sCJD (Lanes 5 and 6), as well as a CJD unaffected control (lanes 7 and 8), accompanied by a direct loading control of the same cases (Direct Loading). B. P1 and P2 were used to capture PrPSc from 10% brain homogenates of individuals affected by iatrogenic (lanes 1 and 2) and variant (lanes 3 and 4) CJD, as well as a CJD unaffected control (lanes 5 and 6), accompanied by a direct loading control of the same cases (Direct Loading). C. P1 and P2 were used to capture PrPSc from 10% brain homogenate from cases of familial CJD (E200K) and GSS. The 7kDa internal fragment of GSS (in C) is indicated by an asterisk. Immunoblotting was performed using 3F4 mAb.
PrPSc from humans and animals captured by P1 and P2 is PK resistant
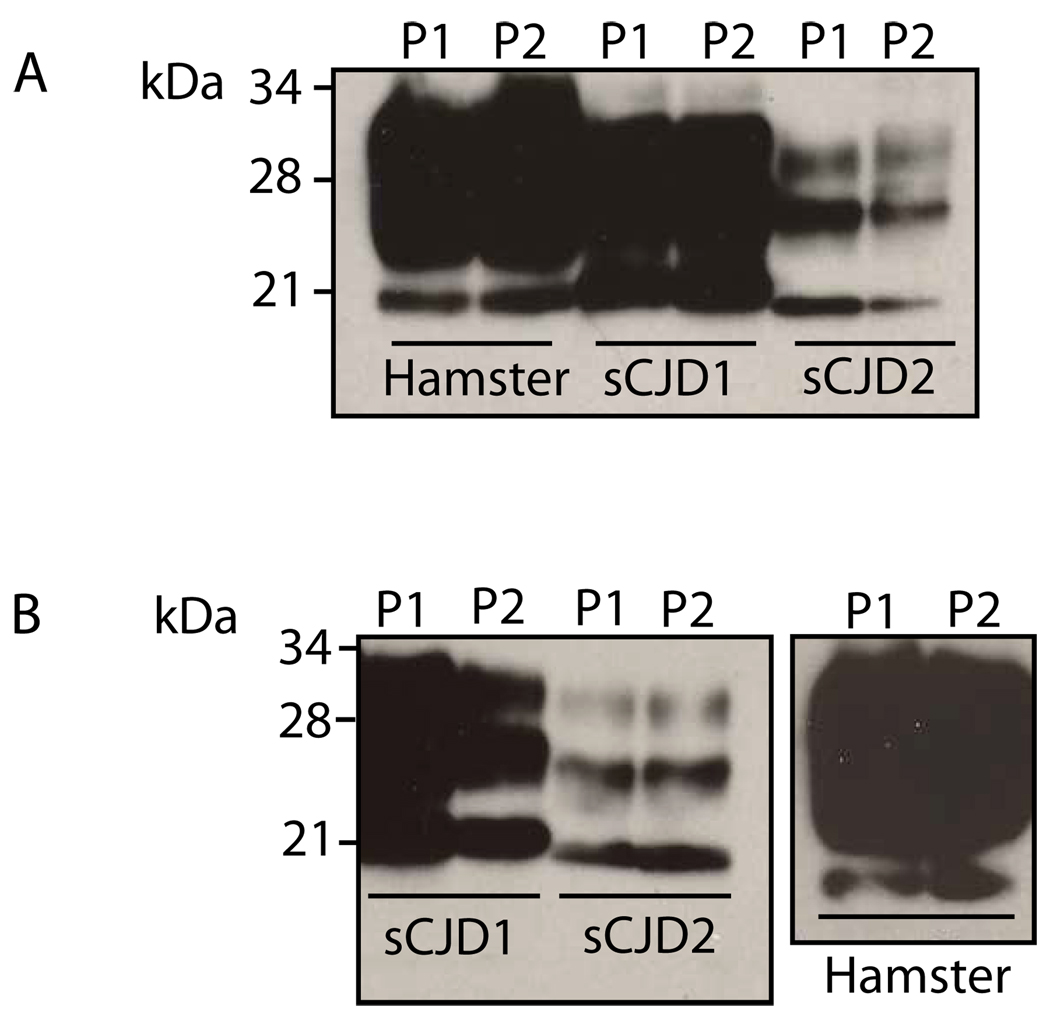
One of the most important biochemical characteristics of PrPSc is its resistance to PK digestion, and this feature is the basis for distinguishing PrPSc from PrPC in current diagnostic tests. In order to determine whether the PrP captured by the beads was PK resistant, it was incubated in the presence of PK (50 µg ml−1). Samples used included beads incubated with homogenate from a Syrian hamster infected with experimental scrapie strain 263K, as well as homogenate from human cases of both type 1 and type 2 sporadic CJD. Previous work has shown that on an SDS-PAGE gel, the migration of the core PK resistant fragment of PrPSc differs between type 1 and type 2 cases of sporadic CJD [2]. The same difference in migration is seen in the two cases of sporadic CJD following capture by peptides 1 and 2 followed by PK digestion (Fig. 5A). This indicates that the PrP captured by P1 and P2 is PK resistant, with similar characteristics to those expected of different types of prion diseases.
Fig. 5. Binding of P1 and P2 to the PK-resistant core fragment of PrPSc.
A. Total brain homogenate from a scrapie-infected hamster (Hamster) or sporadic CJD type MM1 (sCJD1) and VV2 (sCJD2), was captured by P1 and P2 coupled to streptavidin magnetic beads, followed by digestion with 50 µg/ml PK for 1 h at 37°C and immunoblotting with 3F4. B. Total brain homogenate of sporadic CJD type MM1 (sCJD1) and VV2 (sCJD2), as well as scrapie-adapted hamster (Hamster) was digested with 50 µg ml−1 PK for 1 h at 37 °C, prior to capture by P1 and P2. Immunoblotting was performed using 3F4 mAb.
The P1 and P2 bind to the PK-resistant core fragment of PrPSc
The P1 and P2 were also used to capture PrP that had already been digested with PK. The fragment of PrPSc produced by PK digestion corresponds to a C terminal core fragment of the protein, commonly referred to as PrP(27–30) [32]. After incubation with P1 and P2, the C-terminal fragment of PrPSc, PrP(27–30), was captured (Fig. 5B). This suggests that the peptides interact with PrPSc in the core C-terminal PK resistant fragment, PrP(27–30). This fragment in PrPSc type 1 spans residues 82–231, and in type 2, 97–231 [27, 33]. Again, when cases of type 1 and type 2 sporadic CJD were used, the expected difference in migration was observed.
Mini-kringle peptide binding to PrPSc is conformation dependent
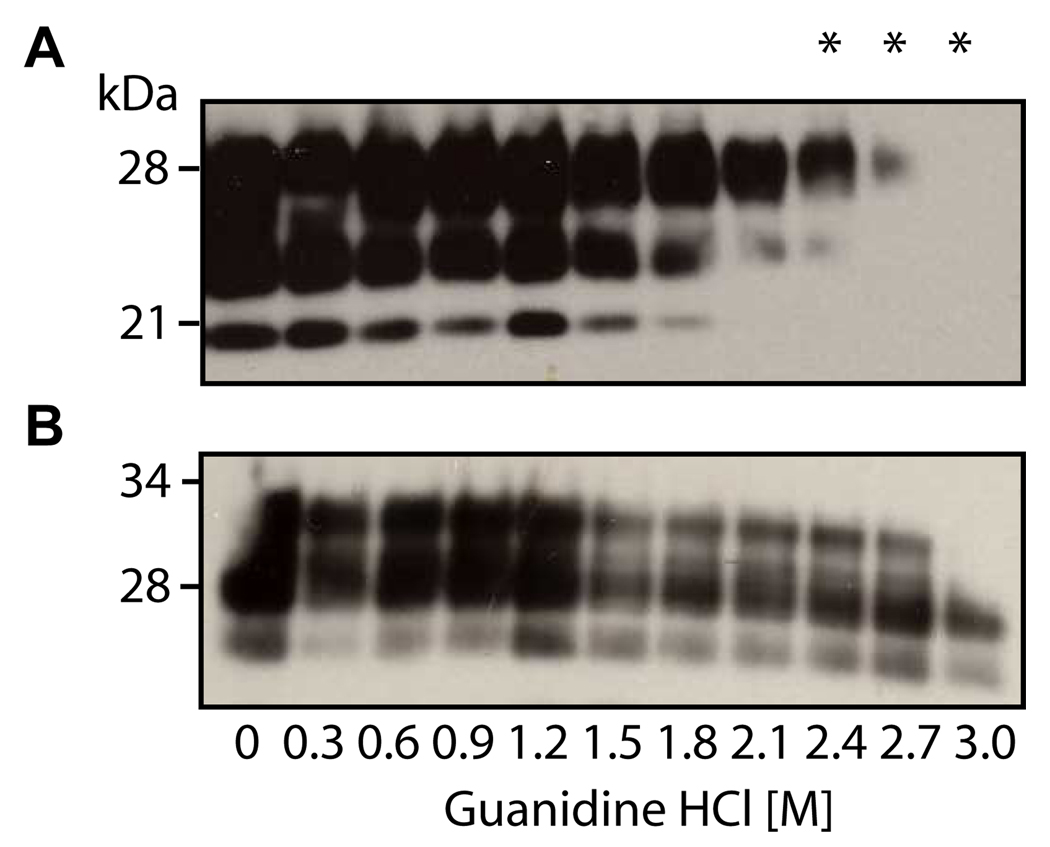
Since P1 and P2 have similar capacity in capture PrPSc from diseased brains, detailed characterization of the binding between the mini-kringle peptides and PrPSc was carried out using P1. The conformational dependence of the P1 binding to PrPSc was investigated in the following assays. When PrPSc is denatured using guanidine HCl, as the concentration of the denaturant increases, PrPSc undergoes a conformational change to a form resembling PrPC. This property can be exploited to determine the conformational dependence of the interaction between PrPSc and another molecule (Safar et al. 1998, Zou et al., 2000). Aliquots of brain homogenate from a hamster affected by experimental scrapie were incubated with 0–3 M guanidine HCl. As expected, denaturation by guanidine hydrochloride resulted in a gradual loss of PK-resistant PrPSc (Fig. 6A). PrPSc experienced an almost complete loss of its PK resistance at a concentration of 2.7M guanidine HCl. The amount of PrP captured by P1 also showed a gradual decrease as the native conformation of PrPSc was lost (Fig. 6B). The observation that P1 binding to PrPSc decreased in the presence of increasing concentrations of guanidine HCl suggest that such binding is dependent on the conformation of the PrPSc. However, approximately 20 % of the original amount of PrPSc was still bound to P 1 at a guanidine concentration of 2.7 M that rendered PrPSc sensitive to PK. This indicates that P1 binds a small amount of PK sensitive PrPSc. This finding is consistent with other studies using conformation-dependent analyses, that have previously indicated that PrPSc may contain both PK-resistant and PK-sensitive species [34].
Fig. 6. The binding of PrPSc to P1 is conformation dependent.
A. Brain homogenate from scrapie-adapted hamsters was incubated for 1 h with guanidine HCl (0–3 M), followed by digestion with 50 µg ml−1 PK for 1 h at 37 °C and immunoblotting with 3F4.. B. Brain homogenate from scrapie-adapted hamsters was incubated for 1 h with guanidine HCl (0–3 M), followed by incubation with Peptide 1 conjugated beads and immunoblotting with 3F4. The asterisks indicate where there is a difference between PK-resistant PrP and the PrP captured directly by P1.
P1 competes PrPSc binding by the full-length kringle domains
We compared the capture of PrPSc by P1 with that by angiostatin containing three intact kringle domains, K(1+2+3), previously documented to bind to PrPSc [12, 13]. Each individual capture reaction contained approximately 7 µg of P1 and 50 µg of K(1+2+3), each conjugated to 2.3×107 beads.. The peptide capture assay was performed with decreasing amounts of brain homogenate (3-0.5 µl) from an experimentally infected Syrian hamster. An equimolar comparison was not made, due to the difference in molecular weight between K(1+2+3) and P1, and because any differences between the stoichiometry of PrPSc binding to either K(1+2+3) or P1 are not known. Nevertheless, there was negligible difference in the amount of PrPSc captured by K(1+2+3) and P1, even using the smallest amount of brain homogenate (Fig. 7A). Therefore, the affinity of PrPSc for peptide 1 and K(1+2+3) appears to be comparable. Because the sequence of P1 is derived from recurring residues in the kringle domains, a competition assay was used to assess whether the binding site was shared. In a peptide capture assay, in which increasing amounts of free P1 was added to the reaction, the amount of PrPSc bound to the K(1+2+3) conjugated beads decreased as increasing amounts of free P1 were added (Fig. 7B). This indicates that free P1 is able to bind to PrPSc, and that such binding is sufficient to inhibit the pre-existing association between K(1+2+3) and PrPSc. While the binding site for this interaction has not been determined, the ability of P1 to successfully compete with K(1+2+3) for PrPSc binding suggests that P1 may be used as a capture reagent for PrPSc already bound to other molecules.
Fig. 7. Binding of P1 to PrPSc compared to binding of angiostatin (K(1+2+3)).
A. Decreasing quantities of 10 % brain homogenate from scrapie-adapted hamsters (3 µl to 0.5 µl) were incubated with 50 µg of K(1+2+3) conjugated to 2.3×107 tosyl-activated beads or 7 µg of P1 conjugated 2.3×107 streptavidin beads. The mixtures were incubated for 3 hours at room temperature, followed by immunoblotting with 3F4. B. Angiostatin (K(1+2+3), 1.7 mM) was conjugated to tosyl-activated magnetic beads and incubated with 4 µl of 10 % brain homogenate from a case of sporadic CJD. The binding of (K(1+2+3) to PrP was competed with increasing amounts (2–100 µg) of P1, followed by immunoblotting with 3F4.
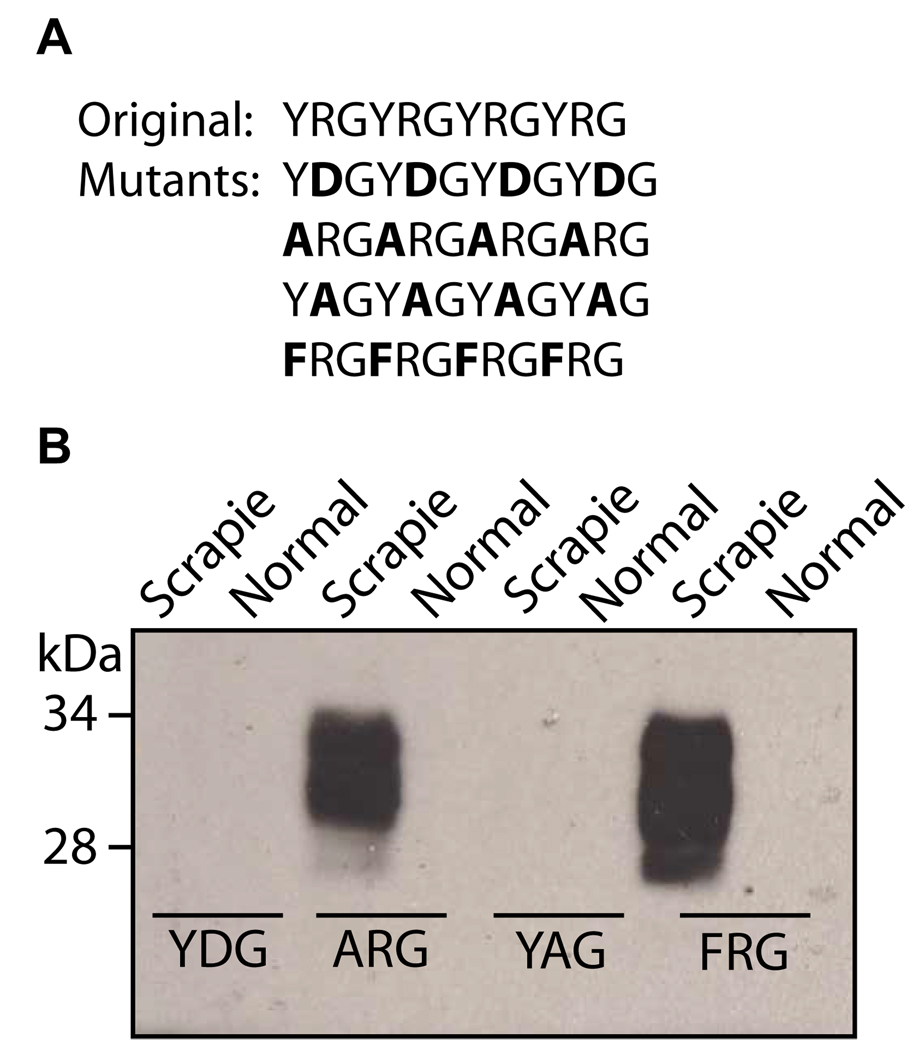
Substitution of the arginine residue of P1 eliminates PrPSc binding
In order to determine which residues of P1 were important for its interaction with PrPSc, several new peptides were synthesized, with substitutions made for the tyrosine or arginine residues (Fig. 8A). When the peptide capture assay was performed using the substituted peptides, those peptides in which the arginine residue had been substituted was no longer able to bind PrPSc. This was true in both a peptide in which the arginine had been substituted for a negatively charged residue (aspartic acid) and a peptide in which it had been substituted for the neutral residue alanine (Fig. 8B). However, when the arginine was retained, and the tyrosine was substituted, the peptide maintained its ability to capture PrP. This was true regardless of whether tyrosine was substituted for another aromatic residue, or whether the aromatic character was eliminated (Fig. 8B). This indicates that the most important residues in the interaction with PrP are the positively charged residues such as arginine and lysine.
Fig. 8. Effect of substitution of arginine and tyrosine residues on the binding of P1 to PrPSc.
A. The sequences of P1 and the four substituted peptides. B. The substituted peptides were conjugated to streptavidin magnetic beads and were incubated at room temperature for 3 h with 3 µl of brain homogenate from scrapie-adapted hamster, or 6 µl of brain homogenate from a normal hamster. Immunoblotting was performed using 3F4 mAb.
P1 captures spiked PrPSc from human plasma
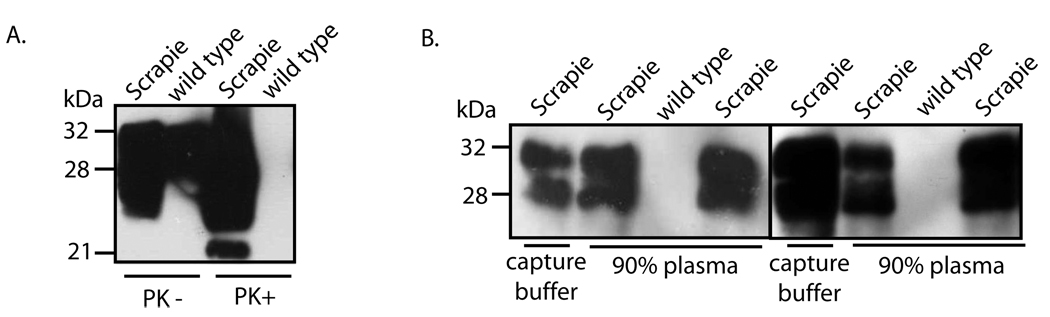
Because all of our experiments described above are based on capture of PrPSc from brain homogenates, it is yet to know if such assay will work for PrPSc in blood in which kringle domain containing proteins such as plasminogen are present in high concentrations. This issue will likely affect the potential for a blood-based assay for PrPSc using kringle based peptides. As a first step, we tested the ability of P1 to capture spiked PrPSc from human plasma. Plasma was obtained from six different individuals unaffected by prion diseases, and was spiked with either 3 µl of 10% brain homogenate from hamsters infected with the 263K strain of scrapie, or from wild type hamsters. Following addition of P1 conjugated beads, capture reaction was carried out in either 1 ml of capture buffer, or in 1 ml of 90% of human plasma (900 µl of plasma and 100 µl capture buffer). Results from experiments in two different plasma samples showed that P1 successfully captured spiked PrPSc from the plasma (Fig 9B). This indicates that P1 readily captures PrPSc and other proteins present in plasma do not appear to significantly inhibit the P1 interaction with PrPSc, at least in vitro. No prion protein was captured from the plasma spiked with wild type brain homogenate (Fig 9A). The presence of input PrPSc and PrPC in spiking sources of scrapie-infected and normal animals, respectively, was demonstrated by immunoblotting and PK digestion (Fig.9A).
Fig. 9. Capture of spiked PrPSc from human plasma by P1.
A. Aliquots (900 µl) of human plasma obtained from two different individuals was spiked with 3 µl of 10 % total brain homogenate from hamsters infected with the 263K scrapie or from normal hamster in P1 was conjugated to streptavidin magnetic beads and were incubated at room temperature with (Lanes 1, 3, 4, and 6), followed by immunoblotting with 3F4. The conjugated beads were also incubated with 6 µl of 10 % total brain homogenate from wild type hamsters in two separate samples of human plasma (Lanes 2 and 5). Immunoblotting was performed using 3F4 mAb. B. A 10 % total brain homogenate from a hamster infected with the 263K strain of scrapie and brain homogenate from a wild type hamster were incubated in the absence (PK−) or presence (PK+) of 50 µg ml−1 PK for 1 h at 37 °C, followed by immunoblotting with 3F4.
Discussion
We have discovered that the PrPSc-binding properties of the kringle domains of plasminogen may be recapitulated in short, synthetic peptides based on the sequence of the kringle domains. While the in vivo role, if any, of plasminogen in prion diseases is not clear, we have shown that the PrPSc binding properties of plasminogen may be exploited for diagnostic purposes, using very short peptides based on the sequence of plasminogen’s kringle domains. In our studies, these peptides, P1 and P2, were able to bind to PrPSc from prion diseases from a variety of sources, with different etiologies, including BSE and vCJD. P1 and P2 were found to bind to the core C-terminal fragment of PrPSc, PrP(27–30), as well as to the 6–7 kDa internal PrP fragment found in GSS. The internal fragment of GSS spans residues, migrating at 7–8 kDa spans residues 74–90 to 146–153 [35–37] Because P1 and P2 bind to PrP(27–30), the sequence of which is represented by recombinant PrP 90–231, it would appear that if the interaction is lysine based, as has previously been suggested for PrP interactions with plasminogen [22], that the N terminal lysine cluster (residues 23, 24 and 27) is not involved. The lysine cluster present at residues 101, 104, 106, and 110 remains in PrP 90–231, as well as in the internal fragment of GSS. However, the involvement of this cluster in binding is not clear because, in each peptide, 33% of the residues are positively charged. It seems unlikely that the lysine cluster is interacting with a peptide carrying an overall positive charge. Our results indicate that binding is retained when the positive charge of the peptide is present suggest that the lysine clusters may not be responsible for the binding to the peptides. The binding site for the peptides, and whether the interaction with the peptides is direct, or through other elements in a complex with PrPSc, is a subject for further study.
PrPC was not detected by P1 and P2 at any time using the peptide capture assay. Previous work, as mentioned earlier, has revealed a binding site for the lysine binding region of the plasminogen kringle domains in the lysine clusters of PrPC, and PrPC also contains a binding site for tPA. However, in our study, and in a similarly designed previous study using full length plasminogen, PrPC was not detected in the brain homogenates of animal or human cases of prion diseases [12, 13]. Although there is a degree of proteolytic activity even in brain homogenate, this is not responsible for the absence of PrPC, as it is clearly present in untreated brain homogenate of CJD unaffected individuals. It is unclear why recombinant PrPC has been found to associate with plasminogen in vitro, but no studies using brain homogenate have found evidence of this binding. The physiological role of PrP is still poorly understood, so it is unknown whether the in vitro observations are physiologically relevant. There have been reports that the binding of PrP to plasminogen is a result of the detergent conditions of the experiment, and does not have any physiological role [38]. Despite several suggestions of an in vivo role, a direct in vivo interaction between plasminogen and PrP has never been proven. Therefore, a difference may exist between recombinant human PrP and the macromolecular environment in which PrP exists in the brain. Binding to other proteins, for example, could prevent the binding of plasminogen to PrP in this situation, even though such an interaction is theoretically possible.
With the continuing emergence of cases of both BSE and vCJD around the world, it is important that molecules that may show a preference for binding to PrPSc are investigated for diagnostic utility. There have been several reports of antibodies that have been shown to bind to PrPSc, but not PrPC, including the anti-DNA antibody OCD4, and an antibody raised against the YYR motif [31, 39–42]. In addition, peptide-based assays, using peptides based on the prion protein itself, have previously been proposed [43, 44]. The advantage of using short synthetic peptides is that they are cheaper and easier to produce than monoclonal antibodies, yet demonstrate a high degree of sensitivity to PrPSc. The ultimate goal of prion diagnostics is to develop a blood test detecting PrPSc. Such a test requires a high degree of sensitivity, as the concentration of PrPSc in blood is estimated to be extremely low. The buffy coat of whole blood, containing leukocytes and platelets, is estimated to have the highest concentration of PrPSc at 1 pg/ml during the symptomatic phase, and 0.1 pg/ml during the pre-symptomatic phase [45]. In addition, there is a possibility that some of the PrPSc present in blood may be PK sensitive. Therefore, an ideal diagnostic reagent for PrPSc in the blood would not require the PK digestion step.
The peptides show a high level of sensitivity in detecting PrPSc, and have been shown to capture PK sensitive PrPSc, so they provide a promising basis for the development of such a test, especially in combination with an appropriate concentration method and highly sensitive detection. They also have a substantial advantage over full length plasminogen, an abundant plasma protein. Plasminogen undergoes many interactions with other blood proteins in vivo, as part of the fibrinolytic pathway, and these interactions may interfere with a blood based diagnostic test. However, our results indicate that the short kringle based peptides can capture PrPSc spiked in human plasma, and that interactions with other plasma proteins do not represent a significant source of inhibition. The peptides have no natural interacting partners in the blood, potentially enabling a more specific reaction with PrPSc. In addition, the peptides are easily modified, so biotinylation can be performed for simple incorporation into capture assays, or other diagnostic assays, such as ELISA based methods. With availability of sensitive prion-specific detection such as this, new avenues are opened for the earlier diagnosis, and possible treatment, of prion diseases.
Acknowledgements
This work was supported in part by United States Department of Defense Grant DAMD17-03-1-0283, Department of Agriculture Grant 2002-35201-2608 and National Institutes of Health Grant AG-14359.
The abbreviations used are
- BSE
bovine spongiform encephalopathy
- CJD
Creutzfeldt-Jakob disease
- GSS
Gerstmann-Sträussler-Scheinker disease
- PrPC
the normal cellular prion protein
- PrPSc
the abnormal, scrapie isoform of prion protein
- tPA
tissue plasminogen activator
- PK
proteinase K
- PBS
phosphate-buffered saline; SDS, sodium dodecyl sulfate
- SDS
sodium dodecyl sulfate.
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95(23):13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gambetti P, et al. Sporadic and familial CJD: classification and characterisation. Br Med Bull. 2003;66:213–239. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- 3.Will RG, et al. A new variant of Creutzfeldt-Jakob disease in the UK. Lancet. 1996;347(9006):921–925. doi: 10.1016/s0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- 4.Collen D, Lijnen HR. Basic and clinical aspects of fibrinolysis and thrombolysis. Blood. 1991;78(12):3114–3124. [PubMed] [Google Scholar]
- 5.Hayden SM, Seeds NW. Modulated expression of plasminogen activator system components in cultured cells from dissociated mouse dorsal root ganglia. J Neurosci. 1996;16(7):2307–2317. doi: 10.1523/JNEUROSCI.16-07-02307.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shushakova N, et al. The urokinase/urokinase receptor system mediates the IgG immune complex-induced inflammation in lung. J Immunol. 2005;175(6):4060–4068. doi: 10.4049/jimmunol.175.6.4060. [DOI] [PubMed] [Google Scholar]
- 7.Muller CM, Griesinger CB. Tissue plasminogen activator mediates reverse occlusion plasticity in visual cortex. Nat Neurosci. 1998;1(1):47–53. doi: 10.1038/248. [DOI] [PubMed] [Google Scholar]
- 8.Sappino AP, et al. Plasminogen activators in tissue remodeling and invasion: mRNA localization in mouse ovaries and implanting embryos. J Cell Biol. 1989;109(5):2471–2479. doi: 10.1083/jcb.109.5.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen ZL, Strickland S. Neuronal death in the hippocampus is promoted by plasmin-catalyzed degradation of laminin. Cell. 1997;91(7):917–925. doi: 10.1016/s0092-8674(00)80483-3. [DOI] [PubMed] [Google Scholar]
- 10.Ledesma MD, et al. Brain plasmin enhances APP alpha-cleavage and Abeta degradation and is reduced in Alzheimer's disease brains. EMBO Rep. 2000;1(6):530–535. doi: 10.1093/embo-reports/kvd107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiman B, Collen D. Molecular mechanism of physiological fibrinolysis. Nature. 1978;272(5653):549–550. doi: 10.1038/272549a0. [DOI] [PubMed] [Google Scholar]
- 12.Fischer MB, et al. Binding of disease-associated prion protein to plasminogen. Nature. 2000;408(6811):479–483. doi: 10.1038/35044100. [DOI] [PubMed] [Google Scholar]
- 13.Maissen M, et al. Plasminogen binds to disease-associated prion protein of multiple species. Lancet. 2001;357(9273):2026–2028. doi: 10.1016/S0140-6736(00)05110-2. [DOI] [PubMed] [Google Scholar]
- 14.Castellino FJ, McCance SG. The kringle domains of human plasminogen. Ciba Found Symp. 1997;212:46–60. doi: 10.1002/9780470515457.ch4. discussion 60-5. [DOI] [PubMed] [Google Scholar]
- 15.Plow EF, et al. The cell biology of the plasminogen system. Faseb J. 1995;9(10):939–945. doi: 10.1096/fasebj.9.10.7615163. [DOI] [PubMed] [Google Scholar]
- 16.Praus M, et al. Stimulation of plasminogen activation by recombinant cellular prion protein is conserved in the NH2-terminal fragment PrP23-110. Thromb Haemost. 2003;89(5):812–819. [PubMed] [Google Scholar]
- 17.Ryou C, Prusiner SB, Legname G. Cooperative binding of dominant-negative prion protein to kringle domains. J Mol Biol. 2003;329(2):323–333. doi: 10.1016/s0022-2836(03)00342-5. [DOI] [PubMed] [Google Scholar]
- 18.Ellis V, et al. Plasminogen activation is stimulated by prion protein and regulated in a copper-dependent manner. Biochemistry. 2002;41(22):6891–6896. doi: 10.1021/bi025676g. [DOI] [PubMed] [Google Scholar]
- 19.Deininger MH, et al. Cortical neurons of Creutzfeldt-Jakob disease patients express the urokinase-type plasminogen activator receptor. Neurosci Lett. 2002;324(1):80–82. doi: 10.1016/s0304-3940(02)00168-4. [DOI] [PubMed] [Google Scholar]
- 20.Bonomo RP, et al. Copper(II) binding modes in the prion octapeptide PHGGGWGQ: a spectroscopic and voltammetric study. Chemistry. 2000;6(22):4195–4202. doi: 10.1002/1521-3765(20001117)6:22<4195::aid-chem4195>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 21.Epple G, et al. Prion protein stimulates tissue-type plasminogen activator-mediated plasmin generation via a lysine-binding site on kringle 2. J Thromb Haemost. 2004;2(6):962–968. doi: 10.1111/j.1538-7836.2004.00675.x. [DOI] [PubMed] [Google Scholar]
- 22.Epple G, et al. Both lysine-clusters of the NH2-terminal prion-protein fragment PrP23-110 are essential for t-PA mediated plasminogen activation. Thromb Haemost. 2004;91(3):465–472. doi: 10.1160/TH03-06-0382. [DOI] [PubMed] [Google Scholar]
- 23.Zerr I, et al. Plasminogen activities and concentrations in patients with sporadic Creutzfeldt-Jakob disease. Neurosci Lett. 2004;371(2–3):163–166. doi: 10.1016/j.neulet.2004.08.063. [DOI] [PubMed] [Google Scholar]
- 24.Xanthopoulos K, et al. Tissue plasminogen activator in brain tissues infected with transmissible spongiform encephalopathies. Neurobiol Dis. 2005;20(2):519–527. doi: 10.1016/j.nbd.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 25.Hill AF, et al. The same prion strain causes vCJD and BSE. Nature. 1997;389(6650):448–450. doi: 10.1038/38925. 526. [DOI] [PubMed] [Google Scholar]
- 26.Bruce ME, et al. Transmissions to mice indicate that 'new variant' CJD is caused by the BSE agent. Nature. 1997;389(6650):498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 27.Zou W, et al. Characterization of prion proteins. Methods Mol Biol. 2003;217:305–314. doi: 10.1385/1-59259-330-5:305. [DOI] [PubMed] [Google Scholar]
- 28.Kascsak RJ, et al. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol. 1987;61(12):3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li R, et al. Identification of an epitope in the C terminus of normal prion protein whose expression is modulated by binding events in the N terminus. J Mol Biol. 2000;301(3):567–573. doi: 10.1006/jmbi.2000.3986. [DOI] [PubMed] [Google Scholar]
- 30.Safar J, et al. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med. 1998;4(10):1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 31.Zou WQ, et al. Antibody to DNA detects scrapie but not normal prion protein. Proc Natl Acad Sci U S A. 2004;101(5):1380–1385. doi: 10.1073/pnas.0307825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolton DC, McKinley MP, Prusiner SB. Identification of a protein that purifies with the scrapie prion. Science. 1982;218(4579):1309–1311. doi: 10.1126/science.6815801. [DOI] [PubMed] [Google Scholar]
- 33.Parchi P, et al. Genetic influence on the structural variations of the abnormal prion protein. Proc Natl Acad Sci U S A. 2000;97(18):10168–10172. doi: 10.1073/pnas.97.18.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Safar J, Cohen FE, Prusiner SB. Quantitative traits of prion strains are enciphered in the conformation of the prion protein. Arch Virol Suppl. 2000;(16):227–235. doi: 10.1007/978-3-7091-6308-5_22. [DOI] [PubMed] [Google Scholar]
- 35.Tagliavini F, et al. Amyloid protein of Gerstmann-Straussler-Scheinker disease (Indiana kindred) is an 11 kd fragment of prion protein with an N-terminal glycine at codon 58. Embo J. 1991;10(3):513–519. doi: 10.1002/j.1460-2075.1991.tb07977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tagliavini F, et al. Amyloid fibrils in Gerstmann-Straussler-Scheinker disease (Indiana and Swedish kindreds) express only PrP peptides encoded by the mutant allele. Cell. 1994;79(4):695–703. doi: 10.1016/0092-8674(94)90554-1. [DOI] [PubMed] [Google Scholar]
- 37.Parchi P, et al. Different patterns of truncated prion protein fragments correlate with distinct phenotypes in P102L Gerstmann-Straussler-Scheinker disease. Proc Natl Acad Sci U S A. 1998;95(14):8322–8327. doi: 10.1073/pnas.95.14.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaked Y, Engelstein R, Gabizon R. The binding of prion proteins to serum components is affected by detergent extraction conditions. J Neurochem. 2002;82(1):1–5. doi: 10.1046/j.1471-4159.2002.00995.x. [DOI] [PubMed] [Google Scholar]
- 39.Korth C, et al. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature. 1997;390(6655):74–77. doi: 10.1038/36337. [DOI] [PubMed] [Google Scholar]
- 40.Korth C, Streit P, Oesch B. Monoclonal antibodies specific for the native, disease-associated isoform of the prion protein. Methods Enzymol. 1999;309:106–122. doi: 10.1016/s0076-6879(99)09010-2. [DOI] [PubMed] [Google Scholar]
- 41.Curin Serbec V, et al. Monoclonal antibody against a peptide of human prion protein discriminates between Creutzfeldt-Jacob's disease-affected and normal brain tissue. J Biol Chem. 2004;279(5):3694–3698. doi: 10.1074/jbc.M310868200. [DOI] [PubMed] [Google Scholar]
- 42.Paramithiotis E, et al. A prion protein epitope selective for the pathologically misfolded conformation. Nat Med. 2003;9(7):893–899. doi: 10.1038/nm883. [DOI] [PubMed] [Google Scholar]
- 43.Lau AL, et al. Characterization of prion protein (PrP)-derived peptides that discriminate full-length PrPSc from PrPC. Proc Natl Acad Sci U S A. 2007;104(28):11551–11556. doi: 10.1073/pnas.0704260104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan T, et al. Detection of misfolded prion protein in blood with conformationally sensitive peptides. Transfusion. 2007;47(8):1418–1425. doi: 10.1111/j.1537-2995.2007.01284.x. [DOI] [PubMed] [Google Scholar]
- 45.Brown P, et al. The distribution of infectivity in blood components and plasma derivatives in experimental models of transmissible spongiform encephalopathy. Transfusion. 1998;38(9):810–816. doi: 10.1046/j.1537-2995.1998.38998408999.x. [DOI] [PubMed] [Google Scholar]