Abstract
Characterization of how IFNs mediate their biological response led to identification of the JAK-STAT signaling cascade, where JAKs are receptor-associated kinases and STATs the transcription factors they activate. Today, 4 JAKs and 7 STATs are known to transduce pivotal signals for the over 50 members of the four helix bundle family of cytokines. This review will provide an overview and historical perspective of the JAK-STAT paradigm.
Keywords: Interferons (IFNs), cytokines, JAKs, STATs and signal transduction
1. Introduction
Interferons (IFNs), founding members of the cytokine family, were first described by Isaacs and Lindenmann more than 50 years ago [1]. Over the subsequent 25 years, these four-helix bundle cytokines were purified to reveal a surprising biochemical diversity [2]. Concomitant developments in cloning technologies provided both the nascent biotechnology industry with one of its first products and revealed that IFNs can be divided into two major families. Type I IFNs, which included fibroblast (a.k.a – IFN-β) and leukocyte (a.k.a. – IFN-α’s) IFNs, was the larger and more pleiotropic family, whereas type II IFN was represented by a single member, immune IFN (a.k.a. – IFN-γ).
The early availability of recombinant IFNs afforded an opportunity to investigate how cytokines mediate their potent biological responses. Initial cDNA expression studies identified a unique set of IFN stimulated genes (ISGs), as well as distinct type I and II receptors [2–4]. Characterization of the ability of IFN-α to drive ISG expression led to the identification of Signal transducers and activators of transcription (Stat)-1 and Stat2 [5–7]. Subsequent studies implicated Tyk2 (a Janus kinase; a.k.a., JAK) and tyrosine phosphorylation in STAT dependent signaling [8–10]. Over the next several years 7 STATs and 4 JAKs were identified, providing important insight into how the ~50 members of the four-helix bundle cytokine family transduce their potent biological responses (see Table 1). Parallel, but more difficult studies on STAT signal decay identified several families of negative regulators, most notably members of the SOCS (Suppressors of Cytokine Signaling) family (see [154, 155] this issue; reviewed in [11–14]).
Table 1. JAK-STAT signaling by the four-helix bundle cytokines†.
Genetic and biochemical studies have determined that four-helix bundle cytokines transduce their signals through specific JAKs and STATs. Assignments with the highest level of confidence are shown in bold. Those with less confidence are shown in plain lettering, and those with the least confidence are shown in brackets.
| Ligands | JAKs | STATs | |||||
|---|---|---|---|---|---|---|---|
| IFN family | |||||||
| IFN-I (Type I)† | Jak1, | Tyk2 | Stat1, | Stats2, | Stats3 | Stat4 | |
| (Stats5–6) | |||||||
| IFN-γ (Type II) | Jak1, | Jak2 | Stat1 | ||||
| IFN-l (IL-28a,b,-29) | Jak1, | Tyk2 | Stat1, | Stat2, | Stat3 | ||
| IL-10 | Jak1, | Tyk2 | Stat3, | Stat1 | |||
| IL-19 | Jak1, | Jak2 | Stat3, | Stat1 | |||
| IL-20 | Jak1, | Jak2 | Stat3, | Stat1 | |||
| IL-22 (IL-TIF) | Jak1, | Jak2 | Stat3, | Stat1, | (Stat5) | ||
| IL-24 (mda7) | Jak1, | Jak2 | Stat3, | Stat1 | |||
| IL-26 (AK155) | Jak1, | Tyk2 | Stat3, | Stat1 | |||
| gp130 family | |||||||
| IL-6 | Jak1, | (Jak2) | Stat3, | Stat1 | |||
| IL-11 | Jak1 | Stat3, | Stat1 | ||||
| LIF | Jak1, | (Jak2) | Stat3, | Stat1 | |||
| CNTF | Jak1, | (Jak2) | Stat3, | Stat1 | |||
| CLC/CLF§ | Jak1, | (Jak2) | Stat3, | Stat1 | |||
| NP | Jak1, | (Jak2) | Stat3 | ||||
| CT-1 | Jak1, | (Jak2) | Stat3 | ||||
| OSM | Jak1, | (Jak2) | Stat3, | Stat1 | |||
| IL-31 | Jak1, | (Jak2) | Stat3, | Stat5, | Stat1 | ||
| G-CSF | Jak1, | (Jak2) | Stat3 | ||||
| Leptin | Jak2 | Stat3 | |||||
| IL-12 (p35+p40) | Tyk2, | Jak2 | Stat4 | ||||
| IL-23 (p19+p40) | Tyk2, | Jak2 | Stat3, | Stat4 | Stat1 | ||
| IL-27# (p28+EBI3) | Jak2 | Stat1, | Stat3 | Stat4, | (Stat5) | ||
| IL-35 (p35+EBI3) | |||||||
| γC family | |||||||
| IL-2 | Jak1 | Jak3 | Stat5, | (Stat3) | |||
| IL-7 | Jak1 | Jak3 | Stat5, | (Stat3) | |||
| TSLP¶ | Stat5 | ||||||
| IL-9 | Jak1 | Jak3 | Stat5, | Stat3 | |||
| IL-15 | Jak1 | Jak3 | Stat5, | (Stat3) | |||
| IL-21 | Jak1 | Jak3 | Stat3, | Stat5, | |||
| (Stat1) | |||||||
| IL-4 | Jak1 | Jak3 | Stat6 | ||||
| IL-13¶ | Jak1 | Jak2 | Stat6, | (Stat3) | |||
| IL-3 family | |||||||
| IL-3 | Jak2 | Stat5 | |||||
| IL-5 | Jak2 | Stat5 | |||||
| GM-CSF | Jak2 | Stat5 | |||||
| Single Chain family | |||||||
| Epo | Jak2 | Stat5 | |||||
| GH | Jak2 | Stat5, | (Stat3) | ||||
| Prl | Jak2 | Stat5 | |||||
| Tpo | Jak2 | Stat5 | |||||
In humans this family consists of 12 IFN-αs, IFN-β.ω and Limitin.
a.k.a. NNT-1/BSF-3
IL-30 is the p28 subunit of IL-27
Bind to related, but γC independent receptors.
Several Interleukins are not members of this family (e.g., IL-1, IL-8, IL-14, IL-16, IL-17, IL-18, IL-25, IL-32, IL-33, IL-34).
2. Discovery of the JAK-STAT Signaling Paradigm
Shortly after the isolation of the first ISGs, an IFN-I specific enhancer, the ISRE (IFN Stimulated Response Element; AGTTTN3TTTCC), was identified [4, 6, 15]. Analysis of IFN-α stimulated nuclear extracts revealed three distinct ISRE binding complexes: IFN-I Stimulated Gene Factor 1 (ISGF-1; a.k.a. IRF-2); ISGF-2 (a.k.a. IRF-1); and ISGF-3, whose activation correlated directly with the expression of immediate early ISGs [6, 15, 16]. Purification of ISGF-3 led to identification of four component proteins of 113 kDa, 91 kDa, 84 kDa and 48 kDa [6, 7, 16, 17]. The 48 kDa protein (p48), previously shown to be the DNA binding component, was recognized as a member of the IRF (IFN-I response factor) family of transcription factors and subsequently named IRF-9 [18]. The 84 kDa (p84) and 91 kDa (p91) proteins were found to be alternative mRNA splice products of a single gene. They also exhibited significant homology with p113, but not other proteins (see Fig. 1). The term STAT was subsequently coined, with p84 becoming Stat1β, p91 Stat1α and p113 Stat2 [5, 7].
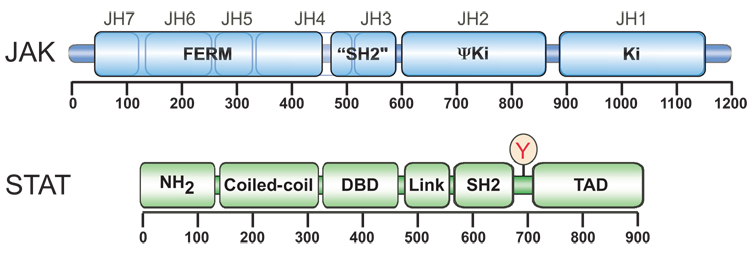
Figure 1. Structural organization of the JAK and STAT families.
The JAKs share seven JAK homology (JH) domains, JH1-JH7. JH1 serves as the catalytic domain, whereas JH2 represents a pseudokinase domain. JH4 includes an SH2-like domain of unknown function and JH4-JH7 comprise a FERM domain that is responsible for association with cytokine receptors. The six STATs share 7 functionally conserved domains. They include the amino terminal domain (NH2), the coiled-coiled domain (Coiled-Coil), the DNA binding domain (DBD), the Linker domain (LK), the SH2 domain, the tyrosine activation domain, and the transcriptional activation domain (TAD).
Antibodies developed against Stat1 and Stat2 revealed that both proteins were localized to the cytoplasm in resting cells. Upon IFN-α stimulation, however, Stat1 (both isoforms) and Stat2 were rapidly phosphorylated on a single conserved tyrosine. This led to the formation of a stable Stat1:Stat2 heterodimer and nuclear translocation (see Fig. 1; [8]). Exploiting these antibodies, it was determined that IFN-γ stimulated the rapid activation of just Stat1 [19]. Moreover, the IFN-γ stimulated Stat1 homodimers were found to directly bind to a distinct palindromic sequence, the Gamma-IFN activated site (GAS; TTTCCNGGAAA; [20–23]). Consistent with their ability to activate Stat1, type I IFNs (IFN-Is) were also subsequently determined to direct the formation of Stat1 homodimers and drive expression of GAS target genes, albeit more transiently. Ensuing biochemical and structural studies determined that the stable interaction between the phosphotyrosine of one STAT with the SH2 domain of the corresponding STAT was responsible for dimerization [24–27].
During the same period, founding members of the unique JAK family (i.e., Tyk2, Jak1 and Jak2) were identified through tyrosine kinase homology screens (see Fig. 1; [28, 29]). Subsequent elegant genetic and biochemical studies linked Tyk2 with the IFN-I response, and then Jak1 and Jak2 with a number of other cytokines, providing important insight into how cytokine receptors stimulate tyrosine phosphorylation [10, 30–33]. The identification of four additional STATs and one JAK provided the tools to determine that all ~50 members of the four-helix bundle family of cytokines transduce their biological responses through this pathway (see Table 1 & Fig. 2; [34–43]). Intriguingly, except for the ~20 type I IFNs (and 3 recently identified type III IFN-Is; [44, 45]), all other members of this family transduce their signals through the simpler JAK-STAT pathway associated with GAS target genes (see Fig. 2; [46]).
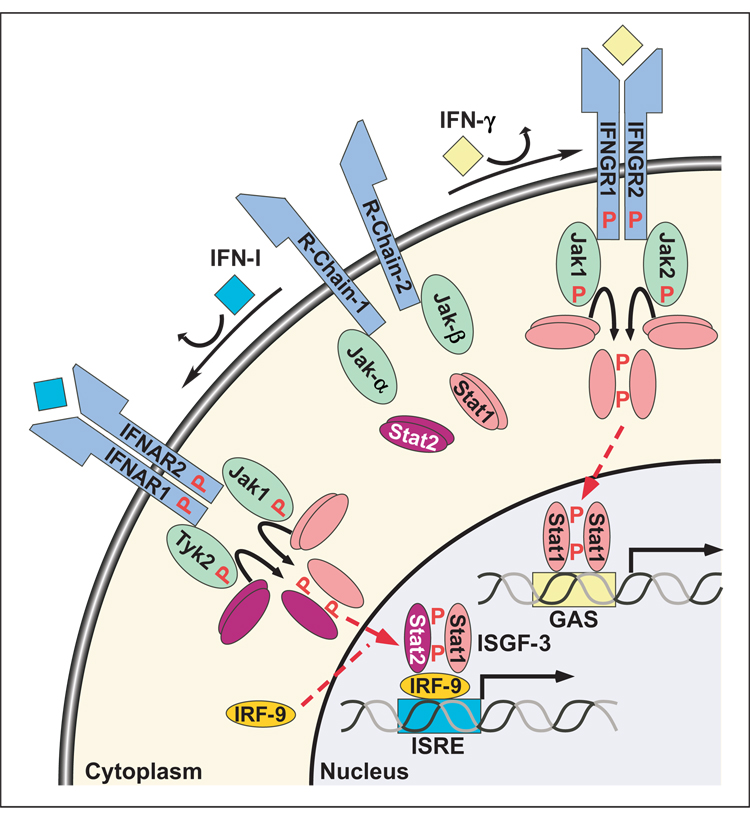
Figure 2. The IFN-I and IFN-γ signaling paradigm.
Upon binding to its dimeric receptor (IFNAR1 & IFNAR2), type I IFN promotes the apposition of two receptor associated JAKs (Jak1 & Tyk2), directing transphosphorylation and activation. The activated JAKs then phosphorylate receptor tyrosine(s), promoting a SH2 domain dependent recruitment of Stat1 and Stat2. At the receptor, Stat1 and Stat2 are activated by phosphorylation, they heterodimerizes, translocate into the nucleus, and associate with IRF-9 to from ISGF-3, which binds to ISREs to drive the expression of corresponding target genes. Only type I and type III IFNs signal through this pathway. IFN-γ directs the activation of a unique dimeric receptor (i.e., IFNGR1 & IFNGR2) by promoting the activation of two receptor associated JAKs (i.e., Jak1 and Jak2). These JAKs phosphorylate a single IFNGR1 tyrosine, which directs the SH2 domain dependent recruitment and activation of Stat1. Activated Stat1 homodimers translocate to the nucleus, bind to the members of the GAS family of enhancers and drive the expression of target genes. All four-helix bundle cytokines family, including type I and III IFNs transduce signals through this pathway.
3. The Janus Kinases (JAK) Family
The four JAK family members, Jak1, Jak2, Jak3 and Tyk2, range in size from 120 to 140 kDa, and except for Jak3 (leukocyte-JAK; [36]), are expressed in most tissues (reviewed in; [46, 47]). This kinase family features seven conserved JAK homology (JH) domains (see Fig. 1), notably including a tandem set of carboxy terminal kinase domains, where only JH1 has bona fide catalytic activity (Ki). JH2 is referred to as the pseudo kinase (ψKi) domain. Reminiscent of other kinases, activation is driven by phosphorylation of critical tyrosines in the inactivation loop, which releases its blockade of the catalytic site. Although no function has been assigned to the SH2-related domain (“SH2”; JH5 and half of JH4), the amino terminal domains (JH1-3 and half of JH4) constitute a FERM (four point one, ezrin, radixin, moesin) domain, which directs stable association with membrane proximal receptor motifs. As illustrated in Figure 2, Jak1 stably associates with IFNAR2 and IFNGR1; Jak2 with IFNGR2; and Tyk2 with IFNAR1.
Gene targeting studies have underscored the critical role JAKs play in the biological response to cytokines. Jak1 knockout mice die perinatally due to a failure to nurse (ascribed to a LIF defect). Their tissues are defective in response to cytokines from the IL-2, IL-6, IFN and IL-10 families ([48]; see also Table 1). Jak2 knockout mice exhibit an earlier lethality (i.e., E12.5), reflecting the critical role this kinase plays in definitive erythropoiesis [49, 50]. Ex vivo studies on Jak2[−/−] tissues have highlighted the important role this kinase plays in directing signals stimulated by the IFN-γ, as well as the single-chain, IL-2 and IL-3 receptor families (see Table 1). Notably, several myeloproliferative disorders, including the majority of cases of polycythemia vera, essential thrombocythemia and primary myelofibrosis have been attributed to a single Stat5-activating point mutation in the JH2 domain of Jak2, V617F, underscoring a potential therapeutic target (see [51, 156–158]).
Jak3 expression is limited to lymphoid tissues. Biochemical and genetic studies have genetically and physically linked Jak3 to the common gamma chain (γC), which is associated with members of the lymphoid predominant IL-2 family of cytokine receptors (e.g., receptors for IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21; see Table 1). Consistent with this, Jak3 and γC knockout mice both exhibit Severe Combined Immunodeficiency (SCID)-like defects, highlighting Jak3 as an appealing therapeutic target [51–54].
As outlined above, Tyk2 was initially associated with IFN-I response. However, subsequent biochemical and genetic studies implicated this kinase in the response to IL-12 and IL-23, as well as several members of the IL-6 and IL-10 receptor families. Intriguingly, loss of Tyk2 has been associated with distinct phenotypes in humans and mice. Whereas Tyk2 knockout mice feature modest cytokine defects and a proclivity towards type 2 (i.e., allergic) T-cell responses [55–57], Tyk2 deficient humans exhibit a severe allergic phenotype that has been attributed to an impaired antimicrobial response [58]. In mice, Tyk2 may play a more important role in integrating the response to multiple cytokines [59].
4. The STAT Family of Transcription Factors
The seven mammals STAT (Stats1–6, 5a & 5b) range in size from 750 and 900 amino acids (see Fig. 1). Both their chromosomal distribution and homologues in model eukaryotes, suggest this family arose from a single primordial gene, as the need for cell-to-cell communication increased [60, 61]. Stat3 and Stat5 are most closely related to those homologues found in model eukaryotes, like Dictyostelium, C elegans and Drosophila (see article number 4; [60]). Notably, the single Drosophila STAT transduces signals through a “classic” JAK-STAT pathway, whereas homologues in Dictyostelium and C. elegans appear to signal through alternative pathway(s).
STATs can be divided into 7 structurally and/or functionally conserved domains (see Fig. 1; [24, 25, 62, 63]). (1) The Amino Terminal Domain (NH2; ~125 amino acids) is well conserved and promotes the formation of homotypic dimers among unphosphorylated STATs [63–66]. This not only assures that STATs remain in an “off” conformation in the resting state, but also facilitates delivery and subsequent activation of STAT pairs at the receptor. (2) The Coiled-Coil Domain (amino acids ~135 to ~315) consists of a potentially dynamic four-helix bundle that protrudes laterally (~80 Å) from the core. This domain associates with regulatory proteins and has also been implicated in controlling the process of nuclear import and export (see article number 3; [46, 67]. (3) The DNA Binding Domain (DBD; amino acids ~320 to ~480) is also well conserved and mediates a robust binding to GAS palindromes. All activated STAT homodimers, except Stat2, directly bind GAS elements. The DBD has also been implicated in the regulation of nuclear import and export (see also article number 3; [24, 25, 67, 68]. (4) The Linker Domain (amino acids ~480 to ~575) structurally translates active dimerization to the DNA binding motif. Studies also suggest that it regulates a process of continual basal (i.e., in resting cells) nuclear export [69]. (5) The SH2 Domain (amino acids ~575 to ~680) is the most highly conserved motif. It mediates specific recruitment to receptor chains, as well as the formation of active STAT dimers [26, 27, 46]. It has been argued that this domain may represent the primordial SH2 domain [25]. (6) The Tyrosine Activation Motif consists of a conserved tyrosine along with 5–7 specific carboxy terminal amino acids, usually near residue 700. Like the corresponding SH2 domain, this motif resides on the exposed surface of the inactive homodimer, facilitating its JAK dependent phosphorylation during receptor recruitment [63, 64]. Upon phosphorylation, this motif is recognized and bound by the corresponding SH2 domain of the partner STAT, directing the critical structural changes required for an active conformation [62–64]. (7) The Transcriptional Activation Domain (TAD) resides at the carboxy terminus and is highly variable in length and sequence between STAT family members. However, for each specific STAT, except Stat2, this sequence is conserved in humans and mice [46, 70]. Many TADs include conserved serine phosphorylation sites that facilitate the recruitment of coactivators (e.g., CBP, p300 and the MCM complex; see below; [71, 72]). STAT TADs also recruit pol II, HATs (e.g., PCAF, GCN5 and NcoA-1 [73–75]), chromatin modifying complexes (e.g., BAF and SWI2-SNF2; [76–79]) and HDACs [80, 81]. The STAT TAD also appears to regulate protein stability. Specifically, Stat4, Stat5 and Stat6 can be targeted for ubiquitin-dependent destruction, whereas Stat1, Stat2 and Stat3 are more stable [82, 83]. Finally, a number of native carboxy terminally truncated STAT isoforms have been shown to direct unique programs of gene expression through their association with other transcription factors (e.g., Stat1β with Stat2 in ISGF-3 and Stat3β with c-jun; [5, 84, 85]).
4.1 Stat1
Consistent with biochemical studies linking Stat1 activation with the biological response to IFN-α (i.e., ISGF-3; [7]) and IFN-γ (i.e., a Stat1 homodimer; see Fig. 2; [9]), Stat1 knockout mice exhibit profound defects in their biological response to type I and type II IFNs [86, 87]. However, defects in the biological response to other Stat1 activating ligands (e.g., IL-6 and EGF) are considerably more modest. In humans, defects in IFN-γ—Stat1 axis have been intimately linked with increased susceptibility to mycobacterial infection [88, 89]. In addition to their role in directing an effective innate response to intracellular bacteria, Stat1 target genes have been associated with suppression of cellular proliferation [46]. This contrasts the pro-proliferative and anti-inflammatory activities linked to Stat3 (see below; see also article numbers 4, 11 & 12), and raises the possibility that Stat1 and Stat3 serve to functionally antagonize each other (e.g., [90]). Finally, Stat1β, which arises as an alternate splice isoform, is missing the 39 amino acid carboxy terminal TAD [7]. Although Stat1β appears to be fully functional in the response to IFN-Is, it is defective in the response to IFN-γ, where it may actually antagonize Stat1α (i.e., full length) activity [91].
4.2 Stat2
Most divergent in sequence and function, Stat2 neither appears to homodimerize nor bind DNA directly. Rather, in an exclusive response to type I (and type III; [44, 45]) IFNs, Stat2 forms an active heterodimer with either Stat1α or Stat1β. This Stat1-Stat2 dimer associates with a unique DNA binding protein, IRF-9, to drive a Stat2-TAD dependent expression of target genes [5, 92, 93]. This appears to include the expression of microRNAs [94]. Curiously, the Stat2 TAD is not conserved between humans and mice [70]. Consistent with these biochemical studies, Stat2 knockout mice exhibit profound defects in their biological response to type I IFNs and likely type III IFNs [95]. More detailed analysis of IFN-I response in Stat2[−/−] tissues has revealed a loss in IFN-I autocrine activity, abnormal DC maturation and a loss in Socs-1 expression [95–97]. This latter response appears to assure that the response to IFN-Is is considerably more transient than the response to IFN-γ [97, 154].
4.3 Stat3
Initially identified as an IL-6 dependent transcription factor [98], two alternate and functionally distinct splice isoforms have been rigorously characterized, Stat3α (full length) and Stat3β (missing the carboxy terminal TAD; [84, 99]). Reflecting Stat3’s ancient lineage [61], biochemical and genetic studies have underscored the important role this transcription factor plays in transducing signals for the IL-6 family (IL-6, IL-11, IL-31, LIF, CNTF, CLC/CLF, NP, CT1, OSM), the IL-10 family (IL-10, IL-19, IL-20, IL-22, IL-24, IL-26), as well as G-CSF, Leptin, IL-21, IL-27 and potentially IFN-Is (see Table 1; see also article numbers 4 & 12; [47, 60, 100]). Consistent with a broad range in activity, Stat3 knockout mice exhibit an early embryonic lethal phenotype (at E6.5–7.5 [101]). Tissue specific Stat3 knockouts have been associated with an increased inflammatory response, altered energy homeostasis, developmental defects and a decreased oncogenic potential [102–111]. The inflammatory phenotype associated with Stat3 deficiency likely reflects its role in directing the response to anti-inflammatory cytokines (e.g., IL-10 family & IL-27), as well as Th17 and regulatory T-cells activity (see article number 10; [100, 112–114]). In contrast, Stat3 hyper-activation has been associated with immune suppression and transformation [115, 116]. Thus, Stat3’s role in transformation is likely to be complex [110, 117, 156].
4.4 Stat4
Identified in a search for Stat1 homologues [36, 37], Stat4 was mapped adjacent to Stat1 on murine chromosome 2 [118]. Subsequently, Stat4 was found to transduce signals for IL-12 (consisting of p40 + p35 subunits) and more recently IL-23 (consisting of p19 + p35 subunits; [119, 120]). Specifically, Stat4 directs the IL-12 dependent polarization of naïve CD4+ T-cells towards IFN-γ secreting Th1 cells, as well as the activation of IFN-γ secreting NK cells [121–123]. Stat4 also plays an important role in the IL-23 dependent polarization of naïve CD4+ T-cells into Th17 cells (see article number 10; [100, 113]). Finally, studies have highlighted the ability of other cytokines to synergize with IL-12 in stimulating potent Stat4 activation through their capacity to promote Stat4 serine phosphorylation [124].
4.5 Stat5
Initially identified as prolactin and IL-3 stimulated transcription factor [40–42], Stat5a and Stat5b arise from a set of tandemly duplicated genes adjacent to Stat3 on murine chromosome 17 [61, 118]. Like Stat3, these two STATs exhibit the highest degree of homology with invertebrate STATs and are functionally pleiotropic (see article number 4; [46, 47, 61]). Biochemical and genetic studies have revealed that Stat5a and Stat5b direct the biological response for the IL-3 family (IL-3, IL-5 and GM-CSF), single chain family (e.g., GH, Prl, Tpo and Epo) and γC family (i.e., the IL-2, IL-7, IL-9, IL-15 and IL-21) of cytokine receptors (see Table 1; [46, 47]. At 96% amino acid identity, these two STATs appear to be functionally redundant, excepting the response to Prl, which favors Stat5a, and GH, which favors Stat5b (see article number 6; [40, 125–127]). Deletion of the entire Stat5a+Stat5b locus has underscored the critical role Stat5 plays in driving both erythropoiesis and lymphopoiesis [128].
4.6 Stat6
Initially identified as IL-4Stat, Stat6 was subsequently shown to transduce signals for IL-13, which shares a receptor chain with IL-4 [38, 39, 46, 47, 121]. Reflecting its evolutionary juxta-position with Stat2 on murine chromosome 12 [118], Stat6 is divergent in sequence and features a large (~150 residue), functionally unique TAD [118, 129]. Intriguingly, Stat6 homodimers bind to a GAS element that features an additional central nucleotide, providing an opportunity to activate a distinct subset of GAS-driven genes. Stat6 knockout mice have underscored the critical role this STAT plays in directing IL-4/IL-13 dependent: Th2-cell polarization; B-cell function (e.g., proliferation, maturation, MHC-II and IgE expression); and mast cell activity [121, 130–132].
5. Regulating STAT Activity
A characteristic feature of JAK-STAT signaling is its rapid onset and decay. Consistent with this, STATs associate with several classes of regulators, including those that promote covalent modifications in addition to canonical tyrosine phosphorylation. The best-characterized negative regulators include phosphatases, nuclear import/export machinery and members of the SOCS (Suppressors of Cytokine Signaling) family. However, other negative regulators like PIAS and Nmi have been reported [133, 134].
5.1 Covalent STAT modifications
STATs appear to undergo covalent modifications in addition to canonical tyrosine phosphorylation, most notably serine phosphorylation (see below). However, there are also several reports of acetylation and O-glycosylation [135–138]. Intriguingly, there is biochemical and genetic evidence for the ubiquitin-directed decay of Stat4, Stat5 and Stat6 [82, 83]. However, evidence supporting a role for R-methylation (Stat1) and SUMOylation (Stat1) in controlling STAT activity remains more controversial [139–141].
With the potential exception of Stat2, all STATs are phosphorylated on at least one serine residue in their TAD, a modification that promotes transcriptional activity (see above; [71, 72, 124, 142, 143]. However, it is not clear where this modification occurs. Conserved phosphorylation sites include, a PMS*P motif (S727 in STATs 1 & 3; S721 in Stat4), a PS*P motif (S725 in Stat5a; S730 in Stat5b) and a SS*PD motif (S756 in Stat6; [82]). Stat1 and Stat5 feature at least one additional serine phosphorylation site in their TAD, S708 and S779, respectively. Biochemical studies have implicated a number of kinases in STAT serine phosphorylation, including MAP kinases (e.g., p38: STATs 1, 3 & 4; ERK: Stat3 & Stat5; JNK: Stat3), PKCδ (Stat1 & Stat3), mTOR (Stat3), NLK (Stat3), CaMKII and IKKε (Stat1; [71, 144, 145]). However, genetic evidence supporting a role for theses kinases in regulating STAT activity is considerably more limited.
5.2 Phosphatases
Since kinases play an important role in cytokine response, it is not surprising that phosphatases, including SHP-1, SHP-2 and CD45 have been implicated in returning cytokine receptors and JAKs back to their basal unphosphorylated state ([60, 146]; see also article number 12). Likewise, SHP-2, PTP1B, TC-PTP and PTP-BL have been implicated in restoring STATs to basal unphosphorylated state [146–151]. However, only two of these phosphatases, SHP-2 and TC-PTP, appear to exhibit robust activity in the nucleus, where STAT desphosphorylation has been linked to STAT nuclear export [67, 152, 153].
5.3 Nuclear Import-Export
The dramatic nuclear accumulation of activated STATs belie the complex process regulating STAT localization (see article number 3; [67, 69, 152]). The predominately cytosolic localization of inactive STATs reflects a steady state, where continuous basal nuclear import is balanced by continuous basal export. Upon stimulation, this balance is shifted towards STAT nuclear accumulation, and then in the opposite direction during the process of signal decay. Of note, multiple NES (nuclear export sequence) and NLS (nuclear localization sequence) elements have been implicated in this dynamic process [67, 69, 152].
The SOCS Family
A subset of the SOCS (Suppressors of Cytokine Signaling) proteins constitute a classic negative “feedback loop” [14, 154]. In the basal state SOCS proteins are expressed at low levels. However, upon stimulation the expression of these STAT target genes is rapidly induced, whereupon they function to antagonize further STAT activation. Gene targeting studies have underscored important roles for Socs-1, Socs-2 and Socs-3 in antagonizing the IFN-γ–Stat1, IL-12–Stat4, IL-4–Stat6, GH–Stat5 and IL-6–Stat3 axes.
6. A Bright Future
Characterization of the ability of IFNs to direct an antiviral response led to the identification of the JAK-STAT signaling cascade, and provided insight into how the more than 50 members of the four-helix bundle cytokine family transduce their biological response (see Table 1). Future studies are likely to exploit conditional gene targeting, as well as improving pharmaceutical agents to explore how these pathways regulate immune homeostasis in vivo. This is not only likely to include the discovery of additional regulators, ligands and receptors, but also provide new insight into “crosstalk” with other signaling cascades.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 2.Pestka S. The human interferon alpha species and receptors. Biopolymers. 2000;55:254–287. doi: 10.1002/1097-0282(2000)55:4<254::AID-BIP1001>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 3.Levy D, Larner A, Chaudhuri A, Babiss LE, Darnell JE., Jr. Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc Natl Acad Sci U S A. 1986;83:8929–8933. doi: 10.1073/pnas.83.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reich N, Evans B, Levy D, Fahey D, Knight E, Jr., Darnell JE., Jr. Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc Natl Acad Sci U S A. 1987;84:6394–6398. doi: 10.1073/pnas.84.18.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu X-Y, Schindler C, Improta T, Aebersold R, Darnell JE. The proteins of ISGF-3, the IFN-α induced transcription activator, define a new family of signal transducers. Proceedings of the National Academy of Sciences, USA. 1992;89:7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kessler DS, Levy DE, Darnell JE., Jr. Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc Natl Acad Sci U S A. 1988;85:8521–8525. doi: 10.1073/pnas.85.22.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr. Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci U S A. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 9.Shuai K, Schindler C, Prezioso V, Darnell JE. Activation of transcription by IFN-γ: Tyrosine phosphorylation of a 91-kDa DNA binding protein. Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 10.Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 11.Levy DE, Darnell JE., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 12.Decker T, Muller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- 13.O'Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 2002;109 Suppl:S121–S131. doi: 10.1016/s0092-8674(02)00701-8. [DOI] [PubMed] [Google Scholar]
- 14.Alexander WS, Hilton DJ. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu Rev Immunol. 2004;22:503–529. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- 15.Levy D, Reich N, Kessler D, Pine R, Darnell JE., Jr. Transcriptional regulation of interferon-stimulated genes: a DNA response element and induced proteins that recognize it. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 2):799–802. doi: 10.1101/sqb.1988.053.01.090. [DOI] [PubMed] [Google Scholar]
- 16.Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE., Jr. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc Natl Acad Sci U S A. 1990;87:8555–8559. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy DE, Kessler DS, Pine R, Darnell JE., Jr. Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3:1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 18.Veals SA, Schindler C, Leonard D, Fu XY, Aebersold R, Darnell JE, Jr., Levy DE. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol Cell Biol. 1992;12:3315–3324. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shuai K, Schindler C, Prezioso VR, Darnell JE., Jr. Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 20.Lew DJ, Decker T, Strehlow I, Darnell JE. Overlapping elements in the guanylate-binding protein gene promoter mediate transcriptional induction by alpha and gamma interferons. Mol Cell Biol. 1991;11:182–191. doi: 10.1128/mcb.11.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eilers A, Seegert D, Schindler C, Baccarini M, Decker T. The response of gamma interferon activation factor is under developmental control in cells of the macrophage lineage. Mol Cell Biol. 1993;13:3245–3254. doi: 10.1128/mcb.13.6.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanno Y, Kozak CA, Schindler C, Driggers PH, Ennist DL, Gleason SL, Darnell JE, Jr., Ozato K. The genomic structure of the murine ICSBP gene reveals the presence of the gamma interferon-responsive element, to which an ISGF3 alpha subunit (or similar) molecule binds. Mol Cell Biol. 1993;13:3951–3963. doi: 10.1128/mcb.13.7.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearse RN, Feinman R, Shuai K, Darnell JE, Jr., Ravetch JV. Interferon gamma-induced transcription of the high-affinity Fc receptor for IgG requires assembly of a complex that includes the 91-kDa subunit of transcription factor ISGF3. Proc Natl Acad Sci U S A. 1993;90:4314–4318. doi: 10.1073/pnas.90.9.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr., Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 26.Gupta P, Blazar BR, Gupta K, Verfaillie CM. Human CD34(+) bone marrow cells regulate stromal production of interleukin-6 and granulocyte colony-stimulating factor and increase the colony-stimulating activity of stroma. Blood. 1998;91:3724–3733. [PubMed] [Google Scholar]
- 27.Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE., Jr. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 28.Firmbach-Kraft I, Byers M, Shows T, Dalla-Favera R, Krolewski JJ. tyk2, prototype of a novel class of non-receptor tyrosine kinase genes. Oncogene. 1990;5:1329–1336. [PubMed] [Google Scholar]
- 29.Wilks AF, Harpur AG, Kurban RR, Ralph SJ, Zurcher G, Ziemiecki A. Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Mol Cell Biol. 1991;11:2057–2065. doi: 10.1128/mcb.11.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silvennoinen O, Ihle JN, Schlessinger J, Levy DE. Interferon-induced nuclear signalling by Jak protein tyrosine kinases. Nature. 1993;366:583–585. doi: 10.1038/366583a0. [DOI] [PubMed] [Google Scholar]
- 31.Watling D, Guschin D, Muller M, Silvennoinen O, Witthuhn BA, Quelle FW, Rogers NC, Schindler C, Stark GR, Ihle JN, et al. Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-gamma signal transduction pathway. Nature. 1993;366:166–170. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- 32.Argetsinger LS, Campbell GS, Yang X, Witthuhn BA, Silvennoinen O, Ihle JN, Carter-Su C. Identification of JAK2 as a growth hormone receptor-associated tyrosine kinase. Cell. 1993;74:237–244. doi: 10.1016/0092-8674(93)90415-m. [DOI] [PubMed] [Google Scholar]
- 33.Müller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, Harpur AG, Barbieri G, Witthuhn BA, Schindler C, Pellegrini S, Wilks AF, Ihle IN, Stark GR, Kerr IM. The protein tyrosine kinase JAK1 complements defects in interferon- alpha/beta and -gamma signal transduction. Nature. 1993;366:129–135. doi: 10.1038/366129a0. [DOI] [PubMed] [Google Scholar]
- 34.Bonni A, Frank DA, Schindler C, Greenberg ME. Characterization of a pathway for ciliary neurotrophic factor signaling to the nucleus. Science. 1993;262:1575–1579. doi: 10.1126/science.7504325. [DOI] [PubMed] [Google Scholar]
- 35.Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto v, Quelle FW, Thierfelder v, Kreider BL, Gilbert DJ, Jenkins NA, Copeland NG, Silvennoinen O, Ihle JN. Stat4, a novel gamma interferon activation site-binding protein expressed in early myeloid differentiation. Mol Cell Biol. 1994;14:4342–4349. doi: 10.1128/mcb.14.7.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong Z, Wen Z, Darnell JE., Jr. Stat3 and Stat4: members of the family of signal transducers and activators of transcription. Proc Natl Acad Sci U S A. 1994;91:4806–4810. doi: 10.1073/pnas.91.11.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schindler C, Kashleva H, Pernis A, Pine v, Rothman P. STF-IL4: A novel IL-4 induced signal transducing factor. EMBO Journal. 1994;13:1350–1356. doi: 10.1002/j.1460-2075.1994.tb06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou J, Schindler U, Henzel WJ, Ho TC, Brasseur M, McKnight SL. An interleukin-4-induced transcription factor: IL-4 Stat. Science. 1994;265:1701–1706. doi: 10.1126/science.8085155. [DOI] [PubMed] [Google Scholar]
- 40.Azam M, Erdjument-Bromage H, Kreider BL, Xia M, Quelle F, Basu R, Saris C, Tempst P, Ihle JN, Schindler C. Interleukin-3 signals through multiple isoforms of Stat5. EMBO J. 1995;14:1402–1411. doi: 10.1002/j.1460-2075.1995.tb07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mui AL, Wakao H, O'Farrell AM, Harada N, Miyajima A. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologs. EMBO J. 1995;14:1166–1175. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1994;13:2182–2191. doi: 10.1002/j.1460-2075.1994.tb06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawamura M, McVicar DW, Johnston JA, Blake TB, Chen YQ, Lal BK, Lloyd AR, Kelvin DJ, Staples JE, Ortaldo JR, et al. Molecular cloning of L-JAK, a Janus family protein-tyrosine kinase expressed in natural killer cells and activated leukocytes. Proc Natl Acad Sci U S A. 1994;91:6374–6378. doi: 10.1073/pnas.91.14.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 45.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 46.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 47.Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 48.Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM, Jr., Schreiber RD. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 49.Neubauer H, Cumano A, Mueller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 50.Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G, Ihle JN. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 51.Tefferi A. JAK and MPL mutations in myeloid malignancies. Leuk Lymphoma. 2008;49:388–397. doi: 10.1080/10428190801895360. [DOI] [PubMed] [Google Scholar]
- 52.Nosaka T, vanDeursen JM, Tripp RA, Thierfelder WE, Witthuhn BA, McMickle AP, Doherty PC, Grosveld GC, Ihle JN. Defective lymphoid development in mice lacking Jak3. Science. 1995;270:800–802. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]
- 53.Park SY, Saijo K, Takahashi T, Osawa M, Arase H, Hirayama N, Miyake K, Nakauchi H, Shirasawa T, Saito T. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3:771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 54.Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- 55.Seto Y, Nakajima H, Suto A, Shimoda K, Saito Y, Nakayama KI, Iwamoto I. Enhanced Th2 cell-mediated allergic inflammation in Tyk2-deficient mice. J Immunol. 2003;170:1077–1083. doi: 10.4049/jimmunol.170.2.1077. [DOI] [PubMed] [Google Scholar]
- 56.Karaghiosoff M, Neubauer H, Lassnig, C C, Kovarik P, Schindler H, Pircher H, McCoy B, Bogdan C, Decker T, Brem G, Pfeffer K, Muller M. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity. 2000;13:549–560. doi: 10.1016/s1074-7613(00)00054-6. [DOI] [PubMed] [Google Scholar]
- 57.Shimoda K, Kato K, Aoki K, Matsuda T, Miyamoto A, Shibamori M, Yamashita M, Numata A, Takase K, Kobayashi S, Shibata S, Asano Y, Gondo H, Sekiguchi K, Nakayama K, Nakayama T, Okamura T, Okamura S, Niho Y. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity. 2000;13:561–571. doi: 10.1016/s1074-7613(00)00055-8. [DOI] [PubMed] [Google Scholar]
- 58.Minegishi Y, Saito M, Morio T, Watanabe K, Agematsu K, Tsuchiya S, Takada H, Hara T, Kawamura N, Ariga T, Kaneko H, Kondo N, Tsuge I, Yachie A, Sakiyama Y, Iwata T, Bessho F, Ohishi T, Joh K, Imai K, Kogawa K, Shinohara M, Fujieda M, Wakiguchi H, Pasic S, Abinun M, Ochs HD, Renner ED, Jansson A, Belohradsky BH, Metin A, Shimizu N, Mizutani S, Miyawaki T, Nonoyama S, Karasuyama H. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25:745–755. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 59.Karaghiosoff M, Steinborn R, Kovarik P, Kriegshauser G, Baccarini M, Donabauer B, Reichart U, Kolbe T, Bogdan C, Leanderson T, Levy D, Decker T, Muller M. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat Immunol. 2003;4:471–477. doi: 10.1038/ni910. [DOI] [PubMed] [Google Scholar]
- 60.Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002;285:1–24. doi: 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 61.Miyoshi K, Cui Y, Riedlinger G, Lehoczky J, Zon L, Oka T, Dewar K, Hennighausen L. Structure of the mouse stat 3/5 locus: evolution from drosophila to zebrafish to mouse. Genomics. 2001;71:150–155. doi: 10.1006/geno.2000.6433. [DOI] [PubMed] [Google Scholar]
- 62.Neculai D, Neculai AM, Verrier S, Straub K, Klumpp K, Pfitzner E, Becker S. Structure of the unphosphorylated STAT5a dimer. J Biol Chem. 2005;280:40782–40787. doi: 10.1074/jbc.M507682200. [DOI] [PubMed] [Google Scholar]
- 63.Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, McMurray JS, Demeler B, Darnell JE, Jr., Chen X. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005;17:761–771. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 64.Mertens C, Zhong M, Krishnaraj R, Zou W, Chen X, Darnell JE., Jr. Dephosphorylation of phosphotyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain. Genes Dev. 2006;20:3372–3381. doi: 10.1101/gad.1485406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhong M, Henriksen MA, Takeuchi K, Schaefer O, Liu B, ten Hoeve J, Ren Z, Mao X, Chen X, Shuai K, Darnell JE., Jr. Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc Natl Acad Sci U S A. 2005;102:3966–3971. doi: 10.1073/pnas.0501063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. J Biol Chem. 2003;278:34133–34140. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- 67.McBride KM, Reich NC. The ins and outs of STAT1 nuclear transport. Sci STKE. 2003;2003:RE13. doi: 10.1126/stke.2003.195.re13. [DOI] [PubMed] [Google Scholar]
- 68.Meyer T, Marg A, Lemke P, Wiesner B, Vinkemeier U. DNA binding controls inactivation and nuclear accumulation of the transcription factor Stat1. Genes Dev. 2003;17:1992–2005. doi: 10.1101/gad.268003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhattacharya S, Schindler C. Regulation of Stat3 nuclear export. J Clin Invest. 2003;111:553–559. doi: 10.1172/JCI15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park C, Lecomte MJ, Schindler C. Murine Stat2 is uncharacteristically divergent. Nucleic Acids Res. 1999;27:4191–4199. doi: 10.1093/nar/27.21.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Decker T, Müller M, Kovarik P. Regulation of Stats by posttranslational modification. In: Seghal PB, Hirano T, Levy DE, editors. Signal transducers and activators of transcription (Stats): Activation and biology. Kluwer Academic; 2003. [Google Scholar]
- 72.Ramsauer K, Farlik M, Zupkovitz G, Seiser C, Kroger A, Hauser H, Decker T. Distinct modes of action applied by transcription factors STAT1 and IRF1 to initiate transcription of the IFN-{gamma}-inducible gbp2 gene. Proc Natl Acad Sci U S A. 2007;104:2849–2854. doi: 10.1073/pnas.0610944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Litterst CM, Pfitzner E. An LXXLL motif in the transactivation domain of STAT6 mediates recruitment of NCoA-1/SRC-1. J Biol Chem. 2002;277:36052–36060. doi: 10.1074/jbc.M203556200. [DOI] [PubMed] [Google Scholar]
- 74.Giraud S, Bienvenu F, Avril S, Gascan H, Heery DM, Coqueret O. Functional interaction of STAT3 transcription factor with the coactivator NcoA/SRC1a. J Biol Chem. 2002;277:8004–8011. doi: 10.1074/jbc.M111486200. [DOI] [PubMed] [Google Scholar]
- 75.Paulson M, Press C, Smith E, Tanese N, Levy DE. IFN-Stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat Cell Biol. 2002;4:140–147. doi: 10.1038/ncb747. [DOI] [PubMed] [Google Scholar]
- 76.Letimier FA, Passini N, Gasparian S, Bianchi E, Rogge L. Chromatin remodeling by the SWI/SNF-like BAF complex and STAT4 activation synergistically induce IL-12Rbeta2 expression during human Th1 cell differentiation. Embo J. 2007;26:1292–1302. doi: 10.1038/sj.emboj.7601586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pattenden SG, Klose R, Karaskov E, Bremner R. Interferon-gamma-induced chromatin remodeling at the CIITA locus is BRG1 dependent. Embo J. 2002;21:1978–1986. doi: 10.1093/emboj/21.8.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu H, Kang H, Liu R, Chen X, Zhao K. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol Cell Biol. 2002;22:6471–6479. doi: 10.1128/MCB.22.18.6471-6479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang M, Qian F, Hu Y, Ang C, Li Z, Wen Z. Chromatin-remodelling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nat Cell Biol. 2002;4:774–781. doi: 10.1038/ncb855. [DOI] [PubMed] [Google Scholar]
- 80.Nusinzon I, Horvath CM. Unexpected roles for deacetylation in interferon- and cytokine-induced transcription. J Interferon Cytokine Res. 2005;25:745–748. doi: 10.1089/jir.2005.25.745. [DOI] [PubMed] [Google Scholar]
- 81.Rascle A, Johnston JA, Amati B. Deacetylase activity is required for recruitment of the basal transcription machinery and transactivation by STAT5. Mol Cell Biol. 2003;23:4162–4173. doi: 10.1128/MCB.23.12.4162-4173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang D, Moriggl R, Stravopodis D, Carpino N, Marine JC, Teglund S, Feng J, Ihle JN. A small amphipathic alpha-helical region is required for transcriptional activities and proteasome-dependent turnover of the tyrosine-phosphorylated Stat5. Embo J. 2000;19:392–399. doi: 10.1093/emboj/19.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tanaka T, Soriano MA, Grusby MJ. SLIM is a nuclear ubiquitin E3 ligase that negatively regulates STAT signaling. Immunity. 2005;22:729–736. doi: 10.1016/j.immuni.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 84.Maritano D, Sugrue ML, Tininini S, Dewilde S, Strobl B, Fu X, Murray-Tait V, Chiarle R, Poli V. The STAT3 isoforms alpha and beta have unique and specific functions. Nat Immunol. 2004;5:401–409. doi: 10.1038/ni1052. [DOI] [PubMed] [Google Scholar]
- 85.Ivanov VN, Bhoumik A, Krasilnikov M, Raz R, Owen-Schaub LB, Levy D, Horvath CM, Ronai Z. Cooperation between STAT3 and c-jun suppresses Fas transcription. Mol Cell. 2001;7:517–528. doi: 10.1016/s1097-2765(01)00199-x. [DOI] [PubMed] [Google Scholar]
- 86.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 87.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 88.Chapgier A, Boisson-Dupuis S, Jouanguy E, Vogt G, Feinberg J, Prochnicka-Chalufour A, Casrouge A, Yang K, Soudais C, Fieschi C, Santos OF, Bustamante J, Picard C, de Beaucoudrey L, Emile JF, Arkwright PD, Schreiber RD, Rolinck-Werninghaus C, Rosen-Wolff A, Magdorf K, Roesler J, Casanova JL. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet. 2006;2:e131. doi: 10.1371/journal.pgen.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jouanguy E, Lamhamedi-Cherradi S, Lammas D, Dorman SE, Fondaneche MC, Dupuis S, Doffinger R, Altare F, Girdlestone J, Emile JF, Ducoulombier H, Edgar D, Clarke J, Oxelius VA, Brai M, Novelli V, Heyne K, Fischer A, Holland SM, Kumararatne DS, Schreiber RD, Casanova JL. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat Genet. 1999;21:370–378. doi: 10.1038/7701. [DOI] [PubMed] [Google Scholar]
- 90.Costa-Pereira AP, Tininini S, Strobl B, Alonzi T, Schlaak JF, Is'harc H, Gesualdo I, Newman SJ, Kerr IM, Poli V. Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc Natl Acad Sci U S A. 2002;99:8043–8047. doi: 10.1073/pnas.122236099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Muller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell JE, Jr., Stark GR, Kerr IM. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J. 1993;12:4221–4228. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Improta T, Schindler C, Horvath CM, Kerr IM, Stark GR, Darnell JE., Jr. Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc Natl Acad Sci U S A. 1994;91:4776–4780. doi: 10.1073/pnas.91.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qureshi SA, Leung S, Kerr IM, Stark GR, Darnell JE., Jr. Function of Stat2 protein in transcriptional activation by alpha interferon. Mol Cell Biol. 1996;16:288–293. doi: 10.1128/mcb.16.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 96.Hahm B, Trifilo MJ, Zuniga EI, Oldstone MB. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 2005;22:247–257. doi: 10.1016/j.immuni.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 97.Zhao W, Cha EN, Lee C, Park CY, Schindler C. Stat2-Dependent Regulation of MHC Class II Expression. J Immunol. 2007;179:463–471. doi: 10.4049/jimmunol.179.1.463. [DOI] [PubMed] [Google Scholar]
- 98.Akira S, Nishio Y, Inoue M, Wang X-J, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 99.Zhang X, Wrzeszczynska MH, Horvath CM, Darnell JE., Jr. Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol Cell Biol. 1999;19:7138–7146. doi: 10.1128/mcb.19.10.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jankovic D, Trinchieri G. IL-10 or not IL-10: that is the question. Nat Immunol. 2007;8:1281–1283. doi: 10.1038/ni1207-1281. [DOI] [PubMed] [Google Scholar]
- 101.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yoshimatsu T, Kawaguchi D, Oishi K, Takeda K, Akira S, Masuyama N, Gotoh Y. Non-cell-autonomous action of STAT3 in maintenance of neural precursor cells in the mouse neocortex. Development. 2006;133:2553–2563. doi: 10.1242/dev.02419. [DOI] [PubMed] [Google Scholar]
- 103.Hokuto I, Ikegami M, Yoshida M, Takeda K, Akira S, Perl AK, Hull WM, Wert SE, Whitsett JA. Stat-3 is required for pulmonary homeostasis during hyperoxia. J Clin Invest. 2004;113:28–37. doi: 10.1172/JCI200419491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gao Q, Wolfgang MJ, Neschen S, Morino K, Horvath TL, Shulman GI, Fu XY. Disruption of neural signal transducer and activator of transcription 3 causes obesity, diabetes, infertility, and thermal dysregulation. Proc Natl Acad Sci U S A. 2004;101:4661–4666. doi: 10.1073/pnas.0303992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alonzi T, Maritano D, Gorgoni B, Rizzuto G, Libert C, Poli V. Essential role of STAT3 in the control of the acute-phase response as revealed by inducible gene inactivation [correction of activation] in the liver. Mol Cell Biol. 2001;21:1621–1632. doi: 10.1128/MCB.21.5.1621-1632.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Levy DE, Lee CK. What does Stat3 do? J Clin Invest. 2002;109:1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee CK, Raz R, Gimeno R, Gertner R, Wistinghausen B, Takeshita K, DePinho RA, Levy DE. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002;17:63–72. doi: 10.1016/s1074-7613(02)00336-9. [DOI] [PubMed] [Google Scholar]
- 108.Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K, Akira S, Clarke AR, Watson CJ. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes and Development. 1999;13:2604–2616. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Levy DE, Loomis CA. STAT3 signaling and the hyper-IgE syndrome. N Engl J Med. 2007;357:1655–1658. doi: 10.1056/NEJMe078197. [DOI] [PubMed] [Google Scholar]
- 110.de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ, Levy DE, Depinho RA, Bonni A. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008 doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Inghirami G, Chiarle R, Simmons WJ, Piva R, Schlessinger K, Levy DE. New and old functions of STAT3: a pivotal target for individualized treatment of cancer. Cell Cycle. 2005;4:1131–1133. doi: 10.4161/cc.4.9.1985. [DOI] [PubMed] [Google Scholar]
- 112.Chen Z, Laurence A, O'Shea JJ. Signal transduction pathways and transcriptional regulation in the control of Th17 differentiation. Semin Immunol. 2007;19:400–408. doi: 10.1016/j.smim.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mathur AN, Chang HC, Zisoulis DG, Stritesky GL, Yu Q, O'Malley JT, Kapur R, Levy DE, Kansas GS, Kaplan MH. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol. 2007;178:4901–4907. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- 114.Owaki T, Asakawa M, Morishima N, Mizoguchi I, Fukai F, Takeda K, Mizuguchi J, Yoshimoto T. STAT3 Is Indispensable to IL-27-Mediated Cell Proliferation but Not to IL-27-Induced Th1 Differentiation and Suppression of Proinflammatory Cytokine Production. J Immunol. 2008;180:2903–2911. doi: 10.4049/jimmunol.180.5.2903. [DOI] [PubMed] [Google Scholar]
- 115.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 116.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG,C, Albanese Darnell JE. Stat3 as an Oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 117.Naugler WE, Karin M. The wolf in sheep's clothing: the role of interleukin-6 in immunity, inflammation and cancer. Trends Mol Med. 2008;14:109–119. doi: 10.1016/j.molmed.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 118.Copeland NG, Gilbert DJ, Schindler C, Zhong Z, Wen Z, Darnell JE, Jr., Mui AL, Miyajima A, Quelle FW, Ihle JN, et al. Distribution of the mammalian Stat gene family in mouse chromosomes. Genomics. 1995;29:225–228. doi: 10.1006/geno.1995.1235. [DOI] [PubMed] [Google Scholar]
- 119.Hunter CA. New IL-12-family members: IL-23 and IL-27, cytokines with divergent functions. Nat Rev Immunol. 2005;5:521–531. doi: 10.1038/nri1648. [DOI] [PubMed] [Google Scholar]
- 120.Zhong Z, Wen Z, Darnell JE., Jr. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 121.Wurster AL, Tanaka T, Grusby MJ. The biology of Stat4 and Stat6. Oncogene. 2000;19:2577–2584. doi: 10.1038/sj.onc.1203485. [DOI] [PubMed] [Google Scholar]
- 122.Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DA, Doherty PC, Grosveld GC, Ihle JN. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 123.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 124.Morinobu A, Gadina M, Strober W, Visconti R, Fornace V, Montagna C, Feldman GM, Nishikomori R, O'Shea JJ. STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc Natl Acad Sci U S A. 2002;99:12281–12286. doi: 10.1073/pnas.182618999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Teglund S, McKay C, Schuetz v, van Deursen JM, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle JN. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 126.Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW. Requirement of Stat5b for sexual dimorphism of body growth rates and liver gene expression. Proceedings of the National Academy of Sciences, USA. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, Hennighausen L, O'Shea JJ. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hebenstreit D, Wirnsberger G, Horejs-Hoeck J, Duschl A. Signaling mechanisms, interaction partners, and target genes of STAT6. Cytokine Growth Factor Rev. 2006;17:173–188. doi: 10.1016/j.cytogfr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 130.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 131.Shimoda K, van Deursen J, Sangster MY, Sarawar v, Carson RT, Tripp RA, Chu C, Quelle FW, Nosaka T, Vignali DA, Doherty PC, Grosveld G, Paul WE, Ihle JN. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 132.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 133.Shuai K. Modulation of STAT signaling by STAT-interacting proteins. Oncogene. 2000;19:2638–2644. doi: 10.1038/sj.onc.1203522. [DOI] [PubMed] [Google Scholar]
- 134.Zhu M, John S, Berg M, Leonard WJ. Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFNgamma-mediated signaling. Cell. 1999;96:121–130. doi: 10.1016/s0092-8674(00)80965-4. [DOI] [PubMed] [Google Scholar]
- 135.Kramer OH, Baus D, Knauer SK, Stein S, Jager E, Stauber RH, Grez M, Pfitzner E, Heinzel T. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev. 2006;20:473–485. doi: 10.1101/gad.364306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nadiminty N, Lou W, Lee SO, Lin X, Trump DL, Gao AC. Stat3 activation of NF-{kappa}B p100 processing involves CBP/p300-mediated acetylation. Proc Natl Acad Sci U S A. 2006;103:7264–7269. doi: 10.1073/pnas.0509808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 138.Gewinner C, Hart G, Zachara N, Cole R, Beisenherz-Huss C, Groner B. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J Biol Chem. 2004;279:3563–3572. doi: 10.1074/jbc.M306449200. [DOI] [PubMed] [Google Scholar]
- 139.Song L, Bhattacharya S, Yunus AA, Lima CD, Schindler C. Stat1 and SUMO modification. Blood. 2006;108:3237–3244. doi: 10.1182/blood-2006-04-020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Komyod W, Bauer UM, Heinrich PC, Haan S, Behrmann I. Are STATS arginine-methylated? J Biol Chem. 2005;280:21700–21705. doi: 10.1074/jbc.C400606200. [DOI] [PubMed] [Google Scholar]
- 141.Meissner T, Krause E, Lodige I, Vinkemeier U. Arginine methylation of STAT1: a reassessment. Cell. 2004;119:587–589. doi: 10.1016/j.cell.2004.11.024. discussion 589–590. [DOI] [PubMed] [Google Scholar]
- 142.Varinou L, Ramsauer K, Karaghiosoff M, Kolbe T, Pfeffer K, Muller M, Decker T. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-gamma-dependent innate immunity. Immunity. 2003;19:793–802. doi: 10.1016/s1074-7613(03)00322-4. [DOI] [PubMed] [Google Scholar]
- 143.Shen Y, Schlessinger K, Zhu X, Meffre E, Quimby F, Levy DE, Darnell JE., Jr. Essential role of STAT3 in postnatal survival and growth revealed by mice lacking STAT3 serine 727 phosphorylation. Mol Cell Biol. 2004;24:407–419. doi: 10.1128/MCB.24.1.407-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kojima H, Sasaki T, Ishitani T, Iemura S, Zhao H, Kaneko S, Kunimoto H, Natsume T, Matsumoto K, Nakajima K. STAT3 regulates Nemo-like kinase by mediating its interaction with IL-6-stimulated TGFbeta-activated kinase 1 for STAT3 Ser-727 phosphorylation. Proc Natl Acad Sci U S A. 2005;102:4524–4529. doi: 10.1073/pnas.0500679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tenoever BR, Ng SL, Chua MA, McWhirter SM, Garcia-Sastre A, Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 146.Mustelin T, Vang T, Bottini N. Protein tyrosine phosphatases and the immune response. Nat Rev Immunol. 2005;5:43–57. doi: 10.1038/nri1530. [DOI] [PubMed] [Google Scholar]
- 147.Nakahira M, Tanaka T, Robson BE, Mizgerd JP, Grusby MJ. Regulation of Signal Transducer and Activator of Transcription Signaling by the Tyrosine Phosphatase PTP-BL. Immunity. 2007;26:163–176. doi: 10.1016/j.immuni.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 148.Irie-Sasaki J, Sasaki T, Matsumoto W, Opavsky A, Cheng M, Welstead G, Griffiths E, Krawczyk C, Richardson CD, Aitken K, Iscove N, Koretzky G, Johnson P, Liu P, Rothstein DM, Penninger JM. CD45 is a JAK phosphatase and negatively regulates cytokine receptor signalling. Nature. 2001;409:349–354. doi: 10.1038/35053086. [DOI] [PubMed] [Google Scholar]
- 149.Haque SJ, Harbor P, Tabrizi M, Yi T, Williams BR. Protein-tyrosine phosphatase Shp-1 is a negative regulator of IL-4-and IL-13-dependent signal transduction. J Biol Chem. 1998;273:33893–33896. doi: 10.1074/jbc.273.51.33893. [DOI] [PubMed] [Google Scholar]
- 150.Aoki N, Matsuda T. A cytosolic protein tyrosine phosphatase PTP1B specifically dephosphorylates and deactivates prolactin-activated STAT5a and STAT5b. J Biol Chem. 2000 doi: 10.1074/jbc.M005615200. [DOI] [PubMed] [Google Scholar]
- 151.Klingmueller U, Lorenz U, Cantley V, Neel BG, Lodish HF. Specefic recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of Jak2 and termination of proliferative signals. Cell. 1995;80:729–738. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- 152.Vinkemeier U. Getting the message across, STAT! Design principles of a molecular signaling circuit. J Cell Biol. 2004;167:197–201. doi: 10.1083/jcb.200407163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.ten Hoeve J, de Jesus Ibarra-Sanchez M, Fu Y, Zhu W, Tremblay M, David M, Shuai K. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol. 2002;22:5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Croker BA, Kiu H, Nicholson SE. SOCS regulation of the JAK/STAT signalling pathway. Seminars in Cell and Developmental Biology. 2008;19 doi: 10.1016/j.semcdb.2008.07.010. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Müller P, Boutros M, Zeidler MP. Identification of JAK/STAT pathway regulators - insights from RNAi screens. Seminars in Cell and Developmental Biology. 2008;19 doi: 10.1016/j.semcdb.2008.06.001. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Groner B, Lucks P, Borghouts C. The function of Stat3 in tumor cells and their microenvironment. Seminars in Cell and Developmental Biology. 2008;19 doi: 10.1016/j.semcdb.2008.06.005. this issue. [DOI] [PubMed] [Google Scholar]
- 157.Wilks AF. The JAK kinases: Not just another kinase drug discovery target. Seminars in Cell and Developmental Biology. 2008;19 doi: 10.1016/j.semcdb.2008.07.020. this issue. [DOI] [PubMed] [Google Scholar]
- 158.Vainchenker W, Dusa A, Constantinescu SN. JAKs in Pathology: Role of Janus Kinases in Hematopoietic Malignancies and Immunodeficiencies. Seminars in Cell and Developmental Biology. 2008;19 doi: 10.1016/j.semcdb.2008.07.002. this issue. [DOI] [PubMed] [Google Scholar]