Abstract
The SR proteins are essential factors that control the splicing of precursor mRNA by regulating multiple steps in spliceosome development. The prototypical SR protein ASF/SF2 contains two N-terminal RRMs (RRM1 and RRM2) and a 50-residue C-terminal RS (arginine-serine rich) domain that can be phosphorylated at numerous serines by the protein kinase SRPK1. The RS domain is further divided into N-terminal (RS1) and C-terminal (RS2) segments whose modification guides the nuclear localization of ASF/SF2. While previous studies revealed that SRPK1 phosphorylates RS1, regio- and temporal-specific control within the largely redundant RS domain is not well understood. To address this issue, engineered footprinting and single turnover experiments were performed to determine where and how SRPK1 initiates phosphorylation within the RS domain. The data show that local sequence elements in the RS domain control the strong kinetic preference for RS1 phosphorylation. SRPK1 initiates phosphorylation in a small region of serines (initiation box) in the middle of the RS domain at the C-terminal end of RS1 and then proceeds in an N-terminal direction. This initiation process requires both a viable docking groove in the large lobe of SRPK1 and one RRM (RRM2) on the N-terminal flank of the RS domain. Thus, while local RS/SR content steers regional preferences in the RS domain, distal contacts with SRPK1 guide initiation and directional phosphorylation within these regions.
Keywords: protein kinase, regiospecificity, phosphorylation, splicing, SR protein
Most genes contain noncoding sequences (introns) that are removed in the mature mRNA in a process known as splicing. It is anticipated that more than 90% of human genes are alternatively spliced, a process in which exons or portions of exons may be included or excluded in the final product.1 Alternative splicing provides a robust mechanism for increasing protein diversity and for regulating various stages of organism development such as the case for sex determination in Drosophila.2 The processing of early mRNA transcripts (pre-mRNA) to mature forms for protein translation is conducted at the spliceosome, a macromolecular assembly composed of both RNA and numerous protein factors. While a series of snRNPs conducts the transesterification reaction and intron removal, selection of the correct splice sites and further maturation of the spliceosome are dependent on a family of splicing factors known as SR proteins.3 SR proteins are composed of one or two RNA recognition motifs (RRMs)¶ and a C-terminal domain rich in Arg-Ser dipeptide repeats (RS domain) from which they derive their name. The SR proteins recognize exonic splicing enhancer (ESEs) sequences through their RRMs and then facilitate the recruitment of other key components early in the development of the spliceosome. The RS domains are likely to play a prominent role at these stages as it has been shown that they enhance interactions with U1-70K (a component of the U1 snRNP) and the U2AF heterodimer at the 5′ and 3′ splice sites, respectively.4; 5
SR proteins isolated from cell extracts are highly phosphorylated in their RS domain. Early studies have demonstrated that while phosphorylation of SR proteins is required for initial developmental stages of the spliceosome, dephosphorylation is also critical for attainment of the fully active, mature spliceosome.6; 7 Two protein kinase families (SRPK and Clk) are known to phosphorylate the RS domains of SR proteins and affect their cellular localization and splicing function.8; 9; 10 Although it can enter the nucleus under certain conditions (cell cycle, osmotic stress), SRPK1, one of two SRPKs in mammals, is largely a cytoplasmic kinase that phosphorylates numerous serines in the RS domain.11; 12 This modification allows the SR proteins to interact with a carrier protein, SR-transportin, that escorts the splicing factors into the nucleus where they mostly reside in speckles.10 Speckles are highly dynamic, membrane-free structures containing SR proteins and many other factors involved in transcription, processing, and export of mRNAs.13 The cytoplasmic localization of SRPK1 is controlled by a large insert (ca 300 aa) that bifurcates the kinase domain. While the removal of this insert does not significantly impact catalysis, it leads to nuclear localization of the kinase and increased population of SR proteins in speckles.12 Recently, it was found that the insert serves as a docking region for co-chaperones that regulate nuclear entry of SRPK1.14 The Clk family of kinases can further phosphorylate the SR proteins and disperse these speckles.8; 15 While phosphorylation is essential for nuclear localization and speckle formation, the splicing activities of the SR proteins appear highly sensitive to the total phosphoryl content of the RS domains. For example, it has been shown that both hypo- and hyperphosphorylation of the prototypical SR protein ASF/SF2 inhibits splicing.16 Thus, precise control of phosphorylation is essential not only for the cytoplasmic and nuclear localization but also for the splicing function of SR proteins.
Although SRPK1 and Clk/Sty catalyze multi-site phosphorylation of ASF/SF2, they display distinctive specificities important for cellular function of this SR protein. While Clk/Sty can phosphorylate all serines in the RS domain of ASF/SF2 (20 serines), SRPK1 is more selective, phosphorylating those largely in the N-terminal portion of the domain based on MALDI-TOF footprinting experiments.11 Also, both enzymes appear to function cooperatively as the addition of Clk/Sty to ASF/SF2 pre-phosphorylated by SRPK1 leads to the further modification of all serines in the RS domain. Clk/Sty is likely to modify more residues in the C-terminal portion of the RS domain in the cell compared to SRPK1 as a deletion construct lacking these residues cannot be effectively released from nuclear speckles.15 These studies suggest that the RS domain of ASF/SF2 can be separated into two functional segments (Fig. 1), where SRPK1 largely modifies RS1 allowing nuclear entry and Clk/Sty then modifies RS2 leading to the dispersion of nuclear speckles and splicing recruitment. Single turnover analyses reveal that SRPK1 catalyzes a semi-processive reaction where the enzyme stays attached to ASF/SF2 for 5-8 rounds of phosphorylation before dissociating and re-binding to modify the remaining residues.17; 18
Figure 1. Structure of the SRPK1:ASF/SF2 Complex.
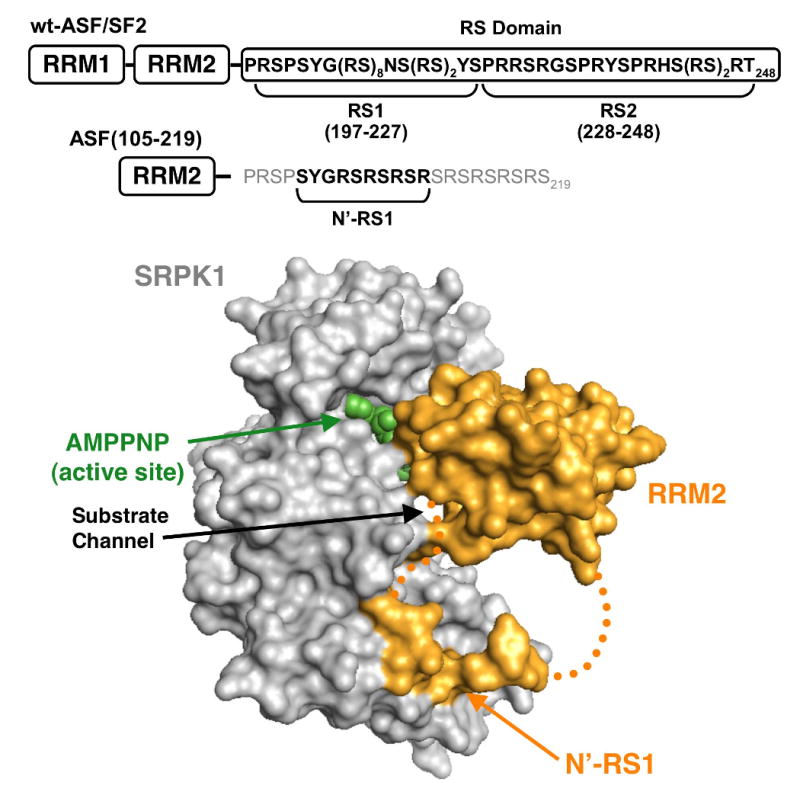
SRPK1 lacking the insert domain (gray) is shown bound with a truncated form of ASF/SF2 [ASF(105-219)] (gold). Density for RRM2 and a short piece of the RS domain (N′-RS1) are visible. Residues 211-219 and the PRSP sequence linking RRM2 and N′-RS1 are disordered (dotted gold line). Active site is designated by AMPPNP.
Footprinting studies reveal that the phosphorylation of ASF/SF2 by SRPK1 occurs in a highly ordered fashion where the kinase moves in a general C-to-N-terminal direction along the RS domain.18 A recent X-ray structure of SRPK1 with a truncated form of ASF/SF2 provides potential insights into how such a reaction could be facilitated.19 Although the substrate construct lacks RRM1 and RS2 sequences and a portion of RS1 is disordered in the structure, the RRM2 and an N-terminal piece of RS1 (N′-RS1: SYSGRSRSRSR) are visible (Fig. 1). RRM2 binds to SRPK1 at 3 distinct loci that encompass helices D and F and the glycine-rich loop whereas N′-RS1 occupies an electronegative docking groove in the large lobe of SRPK1 away from the active site (AMPPNP). The disordered C-terminal end of RS1 is likely to be positioned in the active site near ATP at the onset of phosphorylation. The positioning of N′-RS1 in a docking groove outside the active site is consistent with SRPK1 possessing a preference for initially modifying C-terminal prior to N-terminal residues in RS1. Cross-linking experiments further support this idea and reveal that the RS domain slides through the active site toward the N-terminal end of RS1, in line with the proteolytic footprinting data.18; 19 In this manuscript we addressed the molecular underpinnings of regiospecific and directional phosphorylation of the ASF/SF2 RS domain by SRPK1. We showed that the strong temporal preference for RS1 phosphorylation is guided solely by local RS/SR content. SRPK1 initiates phosphorylation in a narrow sequence (initiation box) in the center of the RS domain, moves in an N-terminal direction in RS1 before dissociating and modifying weaker serines in RS2. While temporal-specific phosphorylation is controlled at the local residue level, initiation within RS1 is controlled by distal protein-protein interactions. The results of these studies provide a template for understanding the general phosphorylation preferences of other members of the SR protein family.
Results
Phosphoryl Content of RS Domain Depends on Arg-Ser Content
Using footprinting and MALDI-TOF analyses in a previous study, we were able to identify 10 serines in three proteolytic peptide fragments that cover the N-terminal region of the RS domain of ASF/SF2 from Ser-199 to Ser-227.11 Since this reflects only 2/3 of the total phosphorylation sites based on mass spectrometric analyses of the full-length substrate, we investigated whether SRPK1 could phosphorylate residues beyond Ser-227 in RS2 (Fig.1). The full-length ASF/SF2 and a deletion construct lacking RS2 sequences [ASF(1-226)] were incubated with SRPK and ATP for 3 hr and analyzed by MALDI-TOF (Fig. 2A). The decrease from 15 to 12 sites upon RS2 deletion suggests that SRPK1 could possibly modify several serines beyond Ser-227. However, since this difference in phosphorylation could be the result of an unexpected change in accessibility of RS1 upon deletion, we measured the phosphorylation of shorter peptide fragments based on the RS domain to explore a potential relationship between serine and observed phosphoryl contents. We expressed and purified 6 constructs lacking both RRMs based on the C-terminal residues in ASF/SF2 (Fig. 2B). All constructs were made with N-terminal His tags except for ASF(197-248) and ASF(220-248) which contain C-terminal tags. We showed in prior studies that placement of the His tag does not impact total phosphoryl content.20 In general, we found a direct linear correlation between the observed phosphoryl and Arg-Ser contents of the peptides (Fig.2B). Interestingly a construct which contains only 3 serines from RS1 [ASF(220-248)] is phosphorylated at a total of 6 sites suggesting that SRPK1 may have the capacity to modify serines in RS2 (Fig 2B). Overall, the deletion analyses suggest that while SRPK1 largely phosphorylates N-terminal serines in the RS domain, serines outside the RS1 region may be targeted as well (vide infra).
Figure 2. Phosphorylation of the RS domain is dependent on Arg-Ser content.
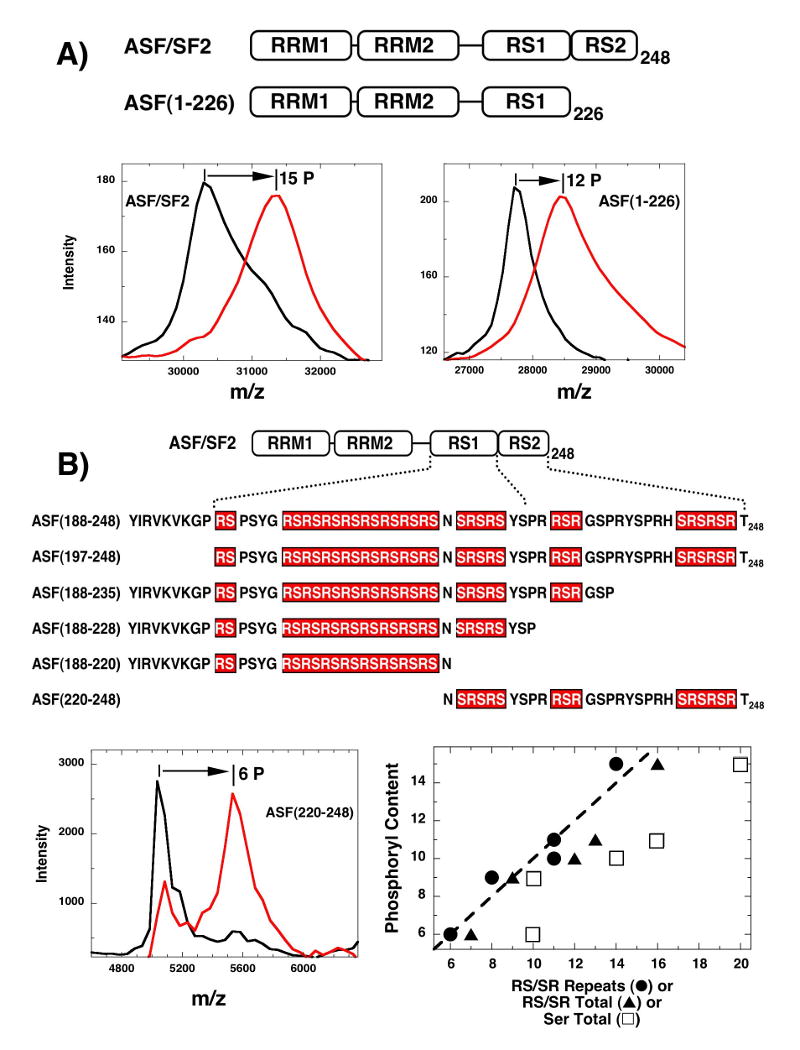
A) RS2 deletion in ASF/SF2. MALDI-TOF spectra of wild-type ASF/SF2 and an RS2-deleted substrate, ASF(1-226), are collected in the presence of SRPK1 and the absence (black curve) and presence of ATP (red line). B) Phosphoryl contents of RS domain constructs. MALDI-TOF spectra for all constructs are collected in the presence of SRPK1 and the absence and presence of ATP [e.g., ASF(220-248)]. Total phosphoryl contents for each construct are plotted as a function of both RS/SR repeats (●), total RS/SR content (▲) and total number of serines (□). RS/SR repeats are a series of contiguous Arg-Ser or Ser-Arg whereas total RS/SR content reflects all serines directly flanked on the N- or C-terminal side by an arginine. Dotted line represents the expected correlation (slope=1, y-intercept=0) between observed ordinate and abscissa.
SRPK1 Can Phosphorylate Serines in the RS2 Segment
To provide evidence that SRPK1 is capable of modifying serines in RS2 in context of the full-length substrate, we mapped potential phosphorylation sites in this region using an engineered LysC footprinting method.18 We replaced 5 lysines with arginine in RRM2 and one arginine at the RS1/RS2 border with lysine (R229K) in a C-terminal His-tagged ASF/SF2 so that upon phosphorylation by SRPK1 and LysC cleavage, two fragments (N- & C-terminal fragments) containing RS1 and RS2 segments could be readily identified by SDS-PAGE (Fig.3A). We showed in separate experiments that this mutant substrate [cl-ASF(229)] was phosphorylated at a similar rate and phosphoryl content as the wild-type ASF/SF2 (Supplementary Fig.1). Upon complete phosphorylation of cl-ASF(229), LysC cleavage generates three major species: an intermediate band (24 kDa), the N-terminal band (19 kDa), and a diffuse C-terminal band (5 kDa) (Fig.3B). The molecular weight of the intermediate band is consistent with a substrate form cleaved at the N-terminal lysines in RRM1 but not at Lys-229. To confirm the identity of these fragments, Ni-resin pull-downs were performed taking advantage of the C-terminal His tag (Fig.3A). As expected, while the N-terminal fragment lacking a His tag was not efficiently pulled down, the C-terminal and intermediate bands along with uncleaved substrate were strongly pulled down. The ratio of the N- and C-terminal fragments prior to pull down is 3.3 which is consistent with the phosphorylation of about 3-4 sites in RS2 and 11-12 sites in RS1. Small impurity bands in the cleaved and uncleaved reactions were detected but they represent a small fraction of the 32P counts (<5% relative to uncleaved substrate & C-terminal fragment) and, therefore, were not considered in the analysis. The general fragmentation pattern is similar to previous reports for related cleavage substrates of ASF/SF2.18; 20 Overall, the data show that the RS domain can be separated into RS1- and RS2-containing LysC fragments and that approximately 3-4 serines in RS2 can be phosphorylated by SRPK1.
Figure 3. Serines in RS2 are phosphorylated by SRPK1.
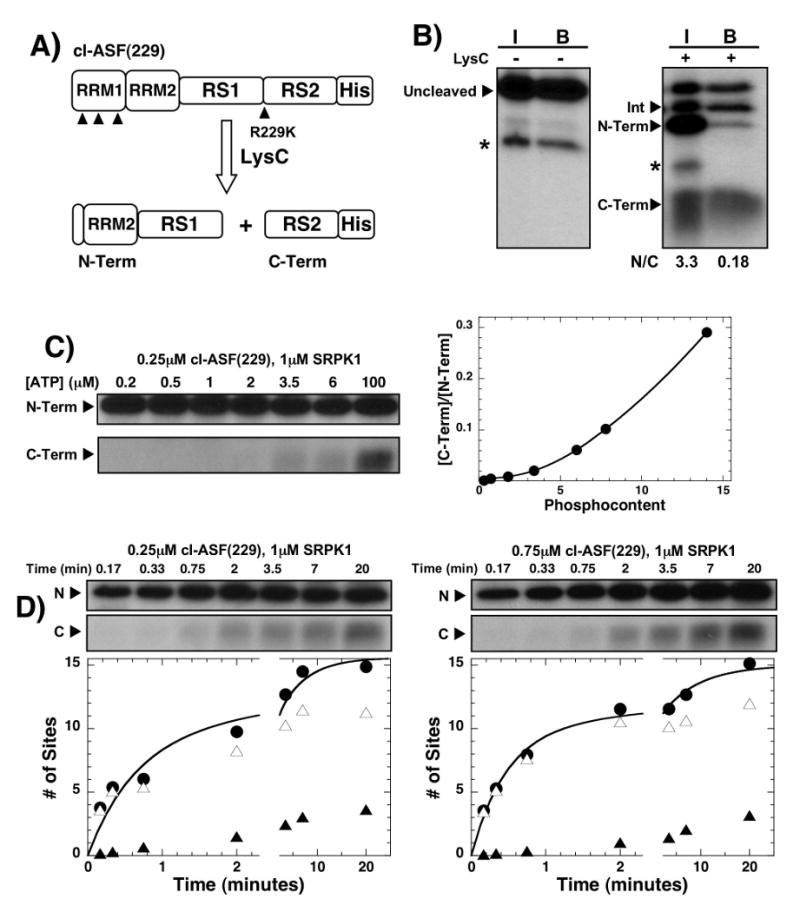
A) Cleavage construct. C-terminal His-tagged cleavage construct contains a single Arg-to-Lys mutation at position 229 in the RS domain and five Lys-to-Arg mutations in RRM2. All lysines in cl-ASF(229) are designated with solid arrows. LysC cleavage produces two major N- and C-terminal fragments. B) Pull-down assays. SRPK1, 32P-ATP and cl-ASF(229) react for 20 minutes before LysC treatment. The reaction is then bound to a Ni-resin and washed. The samples before (I) and after (B) resin binding are displayed in the SDS-PAGE autoradiogram. N/C is the ratio of 32P counts in the N- and C-terminal bands. Asterisks mark minor impurity bands. C) ATP limitation experiment. A complex of SRPK1 and cl-ASF(229) is mixed with varying amounts of 32P-ATP for 20 min before LysC treatment. The N- and C-terminal fragments are displayed in the autoradiogram and the C/N ratio (ratio of C- and N-terminal bands) is plotted as a function of total phosphoryl content (determined prior to LysC). D) Pulse-chase experiments. Complexes of SRPK1 and cl-ASF(229) in ratios of 4:1 and 1.3:1 are reacted with 100 μM 32P-ATP (pulse) for varying times before 4 mM cold ATP (chase) is added to stop the reaction. The samples are then treated with LysC to generate N- and C-terminal fragments. Phosphorylation of cl-ASF(229) prior to LysC cleavage is plotted as a function of time (●) for both enzyme-substrate stoichiometries. The number of sites phosphorylated in the N- (△) and C-terminal (▲) fragments at each stoichiometry is calculated from the C/N ratio and total phosphoryl contents at each time point. The time-dependent data are fit to a double exponential function with rate constants of 1.6 and 0.18 min-1 and amplitudes of 9 and 6 sites for 250 nM cl-ASF(229) and rate constants of 2.1 and 0.13 min-1 and amplitudes of 10 and 5 sites for 750 nM cl-ASF(229), respectively.
Phosphorylation of RS1 Occurs Prior to RS2
Having shown that SRPK1 is capable of phosphorylating some residues in RS2 we next wished to ask at what point these serines become modified. To address this question we monitored the phosphorylation of RS2 as a function of reaction progress. We showed in previous studies that the phosphoryl content of the RS domain can be precisely controlled by varying the total nucleotide concentration in ATP limitation experiments.18 Using this method, we phosphorylated cl-ASF(229) to completion (20 min) using various ATP concentrations in single turnover experiments, treated the samples with LysC and separated the fragments on SDS-PAGE. In control experiments we showed that complete phosphorylation of ASF/SF2 is attained in 15 minutes at limiting ATP concentrations (Supplementary Fig.2). We found that the C-terminal fragment was phosphorylated only at higher ATP concentrations (Fig. 3C), consistent with later phosphorylation of the RS2 region. In these studies, the total phosphoryl content was determined in control reactions where LysC was not added. By converting ATP concentration to total phosphoryl content we found that no significant RS2 modification occurs until after 8 serines in RS1 are modified (Fig.3C). At the highest ATP concentration (100 μM), all sites were phosphorylated and a C/N ratio of 0.28 was achieved consistent with the modification of about 12 serines in RS1 and 3 serines in RS2. These data suggest that SRPK1 phosphorylates RS2 serines late in the reaction cycle.
Although SRPK1 phosphorylates RS1 prior to RS2 in ATP limitation experiments, the preference for RS1 at low ATP concentration could be the result of phosphorylation-dependent changes in the Km for the nucleotide. To address this possibility, we performed pulse-chase experiments in which the reaction is initiated with a constant, high amount of 32P-ATP (100 μM) and then stopped with 4 mM cold ATP (chase) prior to LysC treatment. This experiment was performed at 4:1 and 1.3:1 ratios of SRPK1 to cl-ASF(229) to ensure that the observed RS2 phosphorylation is not the result of reaction priming at more than one site in higher order complexes. As shown in the gel panels in Fig.3D, the appearance of the C-terminal fragment is not dependent on initial enzyme-substrate stoichiometry implying that SRPK1 does not bind simultaneously at two positions in the RS domain. In the absence of proteolytic degradation, cl-ASF(229) is phosphorylated in a biphasic manner where the fast and slow phase rates differ by about 20-fold. As shown in the autoradiogram in Fig. 3D, RS2 phosphorylation occurs late in the progress curve. Using the C/N ratios and total phosphoryl contents, we found that in the initial phase of the reaction (< 2 min), most of the 32P is attached to the N-terminal RS1 with very little in RS2. Overall, both thermodynamic and kinetic data reveal that while RS1 phosphorylation is strongly favored, SRPK1 can also modify serines in RS2.
Cleavage Mutants Establish An Initiation Box for SRPK1 Phosphorylation
Although prior mapping studies showed that SRPK1 prefers to phosphorylate in a general C-to-N-terminal direction in RS1,18 we asked whether the enzyme uses a preferred start residue or region within RS1. To accomplish this we sequentially moved the cleavage site in the RS domain from position 229 outside RS1 to more N-terminal residues within RS1 (Fig. 4A). All RS domain mutations (R214K, R218K, R222K, or R224K) were made in the C-terminal His-tagged cleavage vector containing the five Lys-to-Arg replacements as described in Fig.3. We then performed ATP limitation and LysC cleavage experiments on these mutants to establish their phosphorylation preferences. The N- and C-terminal fragments were identified based on their molecular weights and through pull-down assays (Supplementary Fig.3). Pull-down analyses for clASF(214) were published previously.18 Unlike the other cleavage constructs, the C-terminal band for cl-ASF(224) appeared as a doublet that could be pulled down by the Ni-resin. For all cleavage mutants, while a broad range of ATP concentrations was employed in ATP limitation/LysC experiments, our goals were to compare the N/C ratios at low phosphoryl contents (≤ 1 phosphate) so that an initial phosphorylation preference could be established. While cl-ASF(229) strongly prefers N- vs C-terminal initiation by greater than 100-fold in ATP limitation studies (Fig. 3), cl-ASF(224) shows a lower preference of about 8-fold at low ATP (Fig. 4B,C). In contrast, cl-ASF(214) and cl-ASF(218) show strong preferences for initial C-terminal phosphorylation. Interestingly, cl-ASF(222) demonstrates little preference for N- or C-terminal phosphorylation initiation. For this mutant, initial N-terminal phosphorylation was favored by only a modest 2-fold at low ATP (Fig. 4B,C). In all cases the N/C ratios converge on expected values at high ATP concentrations based on the position of the cleavage sites in the RS domain. The final, experimental N/C ratios for cl-ASF(229), cl-ASF(224), cl-ASF(222), cl-ASF(218) and cl-ASF(214) are 3.4, 2.4, 1.7, 0.9 and 0.7, respectively, and compare favorably with the calculated ratios of 3, 1.8, 1.4, 1, 0.6 based on RS/SR content in the RS domain. In general, we found that preferred initial N-terminal phosphorylation was replaced by preferred initial C-terminal phosphorylation as the cleavage site was moved from position 229 to 214. Also, since we find cleavage sites where a mixture of N- and C-terminal phosphorylation is observed at the lowest phosphoryl contents, there does not appear to be a specific initiation serine. Overall, the data imply that SRPK1 may not initiate at a single serine in the RS domain but rather initiates in a defined region between Ser-221 and Ser-225 (referred to as initiation box; Fig. 4D).
Figure 4. Effects of RS domain cleavage site position on phosphorylation initiation.
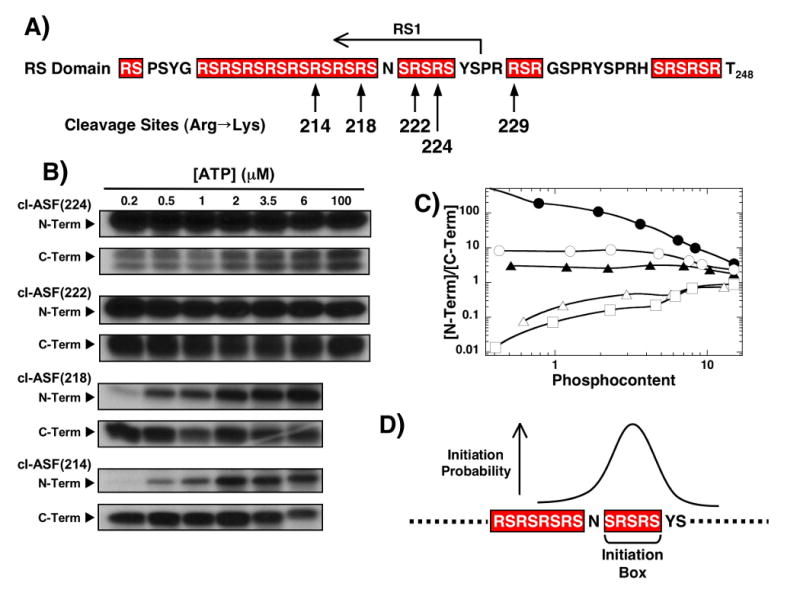
A) Cleavage sites. Alternate cleavage sites in the RS domain are made within the ASF/SF2 cleavage vector containing five Lys-to-Arg mutants in RRM2. B) ATP limitations studies. Complexes of SRPK1 (1 μM) and the cleavage mutants (250 nM) are reacted with varying amounts of 32P-ATP before the N- and C-terminal fragments are resolved by SDS-PAGE. C) N/C plot. Ratio of 32P in the N- and C-terminal fragments are plotted against the total phosphoryl content for cl-ASF(229) (●), cl-ASF(224) (○), cl-ASF(222) (▲), cl-ASF(218) (△) and cl-ASF(214) (□). Data for cl-ASF(229) are taken from Fig.3C. D) Initiation box. N- and C-terminal phosphorylation preferences at low ATP are used to establish the probable start region for SRPK1.
A Flanking RRM Is Important for Phosphorylation Initiation in the RS Domain
Having established that SRPK1 selects a small region in the RS domain for phosphorylation initiation of RS1 (Fig.4), we next addressed the factors that control this initiation mechanism. To investigate a possible contribution of the RRMs, we made a series of deletion and domain swap mutations (Fig.5A). We deleted RRM1 in the cleavage construct cl-ASF(214) [cl-ΔRRM1(214)] to address whether two RRMs are necessary for C-terminal phosphorylation initiation. We also swapped the positions of the RS domain and RRM2 in this new construct [cl-RS-RRM2(214)] to address the role of RRM2 in the absence of RRM1 (Fig.5A). The latter mutant is expected to remove contacts between RRM2 and SRPK1 defined by the X-ray structure (Fig.1). To address the specificity of these interactions, we made 4 alanine mutations in the RRM2 of cl-ΔRRM1(214) at W134, Q135, E184, R154 to generate the construct cl-RRM2-4M(214). These residues comprise three docking surfaces for RRM2 to SRPK1 found in the X-ray structure.19 To address phosphorylation regiospecificity in these new constructs we also shifted the cleavage site in the RS domain in cl-RS-RRM2(214) to position 224 [cl-RS-RRM2(224)] near the RS1/RS2 boundary (Fig.5A). All of these constructs are phosphorylated to the same approximate level as wild-type ASF/SF2 (13-15 sites) and possess high catalytic activity (Supplementary Fig.4). To identify the new N- and C-terminal fragments resulting from LysC cleavage, Ni-resin pull downs were performed. While the LysC fragments for cl-ΔRRM1(214) and cl-RRM2-4M(214) have similar relative positions as that for cl-ASF(214),18 the N- and C-terminal fragments for cl-RS-RRM2(214) migrate in reverse order owing to their different molecular weights. As shown in Fig.5B, the N-terminal fragment migrated beneath the C-terminal fragment which was not pulled down as it lacks the His tag. Also, for all constructs that lacked RRM1, no intermediate fragment was observed since these constructs now only contain one cleavable lysine.
Figure 5. Deletion and swap mutants establish the role of RRMs for phosphorylation initiation.
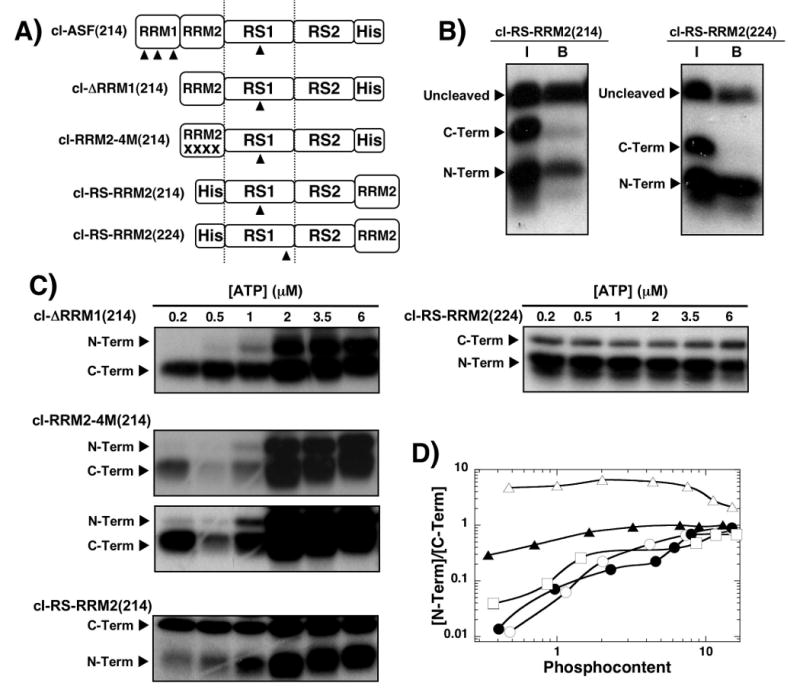
A) Deletion and swap mutants. Numbers in parentheses define the Arg-to-lys cleavage site in the RS domain. The same numbering scheme for the RS domain in wild-type ASF/SF2 is used for the swap and deletion mutants. B) Ni-resin pull-down assays. SRPK1 (1 μM) is used to phosphorylate 250 nM cl-RS-RRM2(214) and cl-RS-RRM2(224) before cleavage by LysC. Samples before binding to the N-resin (I) and after binding and washing (B) are displayed in the autoradiogram. C) ATP limitation experiments. Complexes of SRPK1 (1 μM) and mutants (250 nM) are phosphorylated for 20 min using varying 32P-ATP, cleaved with LysC and separated on SDS-PAGE. Two time exposures (20 & 70 min) for cl-RRM2-4M(214) gel were performed for better visualization of low and high ATP concentrations. D) N/C plot. The ratios of 32P in the N- and C-terminal fragments for cl-ΔRRM1(214) (●), cl-ASF(214) (○), cl-RRM2-4M(214) (□), cl-RS-RRM2(214) (▲), and cl-RS-RRM2(224) (△) are plotted against the total phosphoryl content
After identifying the LysC fragment patterns in these new constructs, ATP limitation experiments were performed to establish phosphorylation initiation preferences. While cl-ΔRRM1(214) showed a strong preference for C-terminal initiation, cl-RS-RRM2(214) showed a significantly reduced preference (Fig.5C). The ATP-dependent N/C ratios for cl-ΔRRM1(214) are similar to those for cl-ASF(214) suggesting that removal of RRM1 does not impact the C-to-N-terminal preference of wild-type SRPK1 when RRM2 is still present on the N-terminal side of the RS domain (Fig.5D). In contrast, whereas cl-ΔRRM1(214) and cl-ASF(214) prefer initial C- vs. N- phosphorylation by about 80-fold, cl-RS-RRM2(214) prefers C-terminal initiation by only 3-fold. Thus, moving RRM2 from an N-terminal to a C-terminal position relative to the RS domain reduces the initial C-terminal preference by about 25-fold. To determine whether cl-RS-RRM2(214) exhibits the same preference for RS1 compared to RS2 phosphorylation as the wild-type substrate, ATP limitation experiments were performed on a swap construct where the cleavage site was moved closer to the RS1/RS2 boundary [cl-RS-RRM2(224)] (Fig.5A). The N/C ratio of 5 at low phosphoryl content for this cleavage vector is similar to that for cl-ASF(224) (N/C=8; Fig.4C) and is consistent with a strong preference for RS1 phosphorylation. Also, the N/C ratio of 2 at high ATP is the same as that for cl-ASF(224) (Fig.4C), suggesting that SRPK1 still phosphorylates serines in RS2 even though RRM2 was moved relative to the RS domain. Finally, the observation that SRPK1 still initiates phosphorylation of cl-RRM2-4M(214) in the C-terminal fragment (30-fold preference at low ATP) indicates that the interaction of RRM2 with the enzyme is flexible and not entirely reliant on the discrete contacts found in the X-ray structure. Overall, the data imply that an RRM directly flanking the N-terminus of the RS domain while not controlling RS1/RS2 regiospecificity is important for controlling phosphorylation initiation in the RS domain.
Interdependent Docking Surfaces Control Initiation
Since the above deletion and swap mutants suggest that an N-terminal RRM flanking the RS domain assists in phosphorylation initiation, we next investigated the role of interactions of the N-terminal portion of the RS1 segment with its docking groove in SRPK1. We performed ATP limitation studies on cl-ΔRRM1(214) and cl-RS-RRM2(214) using a docking-defective mutant of SRPK1. This mutant [SRPK1(6M)] contains six charge-to-alanine mutations (Asp-564,-548, Glu-557,-558,-571, & Lys-615) in the docking groove in the large lobe of SRPK1. We showed previously that SRPK1(6M) phosphorylates 15 sites in the RS domain of ASF/SF2 using a fully distributive mechanism and shows weaker initiation preference20 similar to that for wild-type SRPK1 and cl-RS-RRM2(214) where C-terminal phosphorylation is favored by only 3-fold (Fig.5D). Using SRPK1(6M), we found that cl-ΔRRM1(214) is phosphorylated in a fully random manner with an N/C ratio of 1 at low ATP concentrations (Fig.5A,B) suggesting that the removal of both the docking groove residues and RRM1 leads to a further reduction in initiation preference. Interestingly, SRPK1(6M) shows a preference for N-terminal phosphorylation initiation with cl-RS-RRM2(214) (N/C=2). Thus, while wild-type SRPK1 prefers C-terminal phosphorylation initiation of cl-ΔRRM1(214) by 80-fold, the docking defective mutant enzyme is fully random for this substrate. To determine whether mutations in the docking groove affect the regiospecificity of the enzyme, we performed LysC cleavage of cl-ASF(229) at low (0.2 μM) and high (100 μM) ATP and analyzed the LysC fragments (Fig.6A). While SRPK1(6M) phosphorylates exclusively in the N-terminal fragment (RS1) at low phosphoryl content, the mutant displays the same overall regiospecificity as wild-type SRPK1 at high phosphoryl content (N/C=3.8). Based on total phosphoryl content and the N/C ratio at high ATP, SRPK1(6M) modifies about 3 serines in RS2, similar to that for the wild-type enzyme (Fig.3B). Taken together, the data imply that no single contact surface between SRPK1 and ASF/SF2 can adequately emulate the strong preference for C-terminal phosphorylation initiation. Rather, contacts between SRPK1 and the N-terminal portion of the RS1 and its flanking RRMs function in an interdependent manner to control phosphorylation initiation (Fig. 6C).
Figure 6. SRPK1 docking mutations impact phosphorylation initiation.
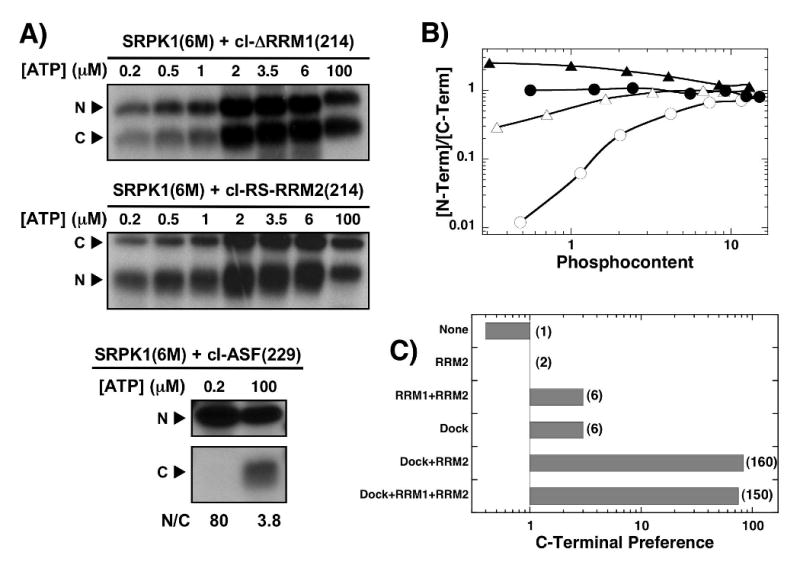
A) ATP limitation experiments. Complexes of SRPK1(6M) (3 μM) and 250 nM cl-ΔRRM1(214), cl-RS-RRM2(214) and cl-ASF(229) are reacted with varying 32P-ATP for 20 min, cleaved with LysC and N- and C-terminal fragments are separated on SDS-PAGE. B) N/C plot. The ratios of 32P in the N- and C-terminal fragments for SRPK1(6M) with cl-ΔRRM1(214) (●) and cl-RS-RRM2(214) (▲) and for wild-type SRPK1 with cl-ΔRRM1(214) (○) and cl-RS-RRM2(214) (△) are plotted against the total phosphoryl content. The data for the wild-type enzyme are taken from Fig. 5D. C) Effects of contacts on C-terminal preference. N/C ratios at low ATP are plotted in the bar graph as a function of added contacts. Numbers in parentheses represent the fold increase in C-terminal initiation preference relative to SRPK1(6M) and cl-RS-RRM2(214).
Sequence Requirements for RS2 Phosphorylation
Since the kinetic preference of SRPK1 for RS1 over RS2 is neither associated solely with the RRMs nor a property of the docking site on the enzyme (Fig.5C & 6A), we next investigated local factors within the RS domain that might govern this regiospecificity. We introduced a lengthy RS/SR repeat region into RS2 [(RS)8] and found that this new mutant [cl-RS8-RS8 (224)] was phosphorylated to a higher extent (20 vs. 15 sites) than the wild-type construct, cl-ASF(224) (Fig. 7A,B). Removing eight consecutive RS/SR repeats in RS1 from this construct to generate cl-SA8-RS8(224) lowers the phosphoryl content by about 9 sites consistent with a direct correlation between Arg-Ser and phosphoryl contents. However, we found that the high phosphoryl content of cl-RS8-RS8 (224) is dependent on a consecutive array of RS/SR repeats and not on total serine content as a mutant which contains the same number of serines but fewer arginines [cl-RS8-RA4 (224)] is phosphorylated to about the same level as cl-ASF(224) (Fig.7A). To determine how changes to RS2 affect regiospecificity, we phosphorylated the substrates at low (0.2 μM) and high ATP (100 μM) and separated the N- and C-terminal fragments upon LysC treatment (Fig.7C). At high ATP concentrations, both the N- and C-terminal fragments of cl-RS8-RS8 (224) are phosphorylated to the same extent (N/C=1; Fig.7C) and based on the total phosphoryl content, suggests that about 10 serines in the C-terminal fragment are modified in this construct (Fig.7A). This high level of phosphorylation in the mutant RS2 is not dependent on phosphorylation in RS1 as cl-SA8-RS8(224) is also phosphorylated at 10 serines in its C-terminal fragment. As expected, LysC treatment of cl-SA8-RS8(224) produces a very low N/C ratio consistent with mostly C-terminal phosphorylation (Fig.7C). Interestingly, disruption of the consecutive RS/SR repeats in the RS2 region in cl-RS8-RA4(224) leads to a very high N/C ratio at both low and high ATP consistent with the phosphorylation of only two sites in RS2 (Fig.7A,C). SRPK1 does not appear to show any preference at initiating phosphorylation in either RS1 or RS2 in cl-RS8-RS8(224) as the N/C ratio is near unity at both low and high ATP concentrations. Single turnover kinetic analyses were performed to determine whether modifications to RS2 impacted the rate of RS domain phosphorylation (Fig.7D). Adding RS/SR repeats to RS2 did not negatively impact the phosphorylation kinetics although the reaction end points varied in line with the total number of phosphorylatable serines. Overall, the data imply that the kinetic preference for RS1 phosphorylation is dependent on a long, uninterrupted RS/SR repeat.
Figure 7. Effects of altering RS/SR content on RS2 phosphorylation.
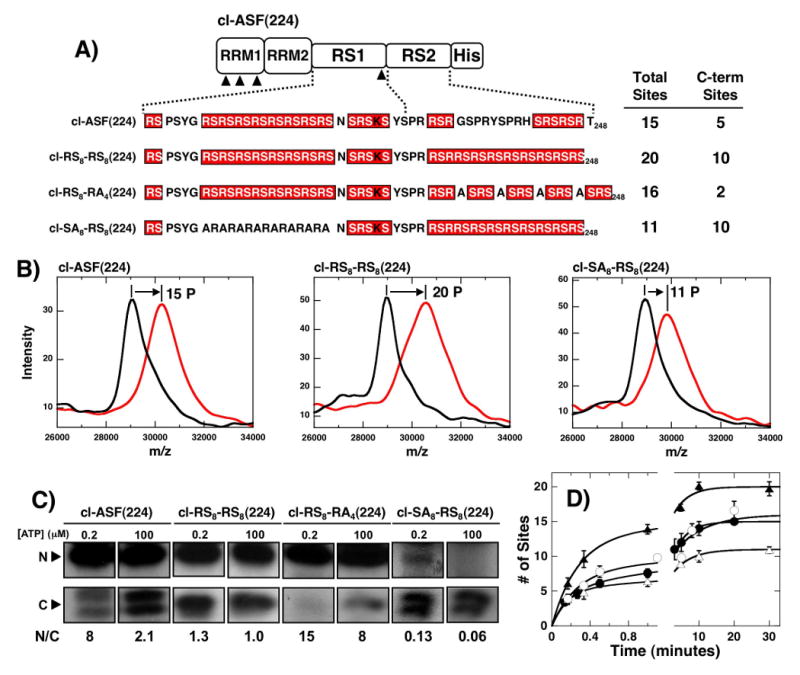
A) Mutations in RS2. Mutations in RS2 segment are made in context of the parent construct cl-ASF(224). MALDI-TOF analyses of the total phosphoryl contents of each mutant are displayed along with estimates of the phosphoryl contents of the C-terminal fragments based on LysC treatment (see panel C). B) MALDI-TOF spectra. Mass spectrometric data were recorded for cl-ASF(224), cl-RS8-RS8(224) and cl-SA8-RS8(224) in the presence of SRPK1 and in the absence (black) and presence (red) of ATP. C) ATP limitation experiments. Complexes of SRPK1 (1 μM) and mutants (250 nM) are phosphorylated for 20 min using 0.2 and 100 μM 32P-ATP, cleaved with LysC and separated on SDS-PAGE. N/C ratios at each ATP are shown. D) Single turnover kinetic analyses. Complexes of 1μM SRPK1 and 250 nM cl-ASF(224) (●), cl-RS8-RS8(224) (▲), cl-RS8-RA4(224) (○) and cl-SA8-RS8(224) (△) are reacted with 100 μM 32P-ATP, quenched with SDS loading buffer and the total phosphoryl contents are plotted as a function of time. Amplitude and rate constants for the initial phases are 5.5 ± 0.48 and 5.4 ± 1.0 min-1 for cl-ASF(224), 12 ± 1.2 and 3.7 ± 0.7 min-1 for cl-RS8-RS8(224), 8.2 ± 1.9 and 3.8 ± 1.9 min-1 for cl-RS8-RA4(224) and 5.3 ± 0.31 and 6.3 ± 0.93 min-1 for cl-SA8-RS8(224), respectively. All progress curves are performed in triplicate and the error bars for each time point are displayed.
Phosphorylation Mechanism Outside RS1
While C-terminal residues in the RS domain of wild-type ASF/SF2 are slowly phosphorylated by SRPK1, the introduction of a long RS/SR repeat in this region leads to rapid and high level phosphorylation of RS2 (Fig.8). We wished to now address whether changes in regiospecificity could lead to changes in processive phosphorylation. To accomplish this, we performed start-trap experiments using substrate forms with altered RS/SR contents and a kinase-inactive form of SRPK [kdSRPK1] that can trap free intermediates. kdSRPK1 is engineered to bind ASF/SF2 with high affinity but is unable to support catalysis owing to a single mutation in the active site.11 A complex of SRPK1 and ASF/SF2 is preformed before reaction initiation with ATP (start) and kdSRPK1 (trap) (Fig.8A). If ASF/SF2 is processively phosphorylated, kdSRPK1 will not trap any free substrate intermediates and will not stop reaction progress. On the other hand, if ASF/SF2 is phosphorylated in a distributive manner, kdSRPK1 will trap early intermediates and effectively stop the reaction. In this study we performed start-trap experiments on three ASF/SF2 constructs similar to those in Fig.7 but containing N-terminal His tags and lacking LysC cleavage sites. For wild-type ASF/SF2, SRPK1 rapidly phosphorylates about 11 serines in the first minute in the absence of kdSRPK1 (control) and phosphorylates about 8 serines in this time frame when kdSRPK1 is added with ATP (start-trap) (Fig.8B). Addition of kdSRPK1 before ATP (trap-start) leads to significant reaction inhibition indicating that the inactive enzyme is an effective trap. The decrease in the amplitude of the initial phase in the start-trap data implies that SRPK1 adds about 8 phosphates onto ASF/SF2 before dissociating from the RS domain, in line with prior reports.17; 18; 20 Whether SRPK1 can re-bind and perform further processive phosphorylation after this initial reaction is unknown since the trap-start rate is similar to that for the slow phase and, thus, cannot be used to establish mechanism. However, since the second phase is more than 100-fold slower than the initial phase, we presume that it is phosphorylated in a distributive manner. For the substrate containing an additional (RS)8 repeat in RS2 (RS8-RS8), a higher level of processivity is achieved suggesting that added arginines have a beneficial effect on processive phosphorylation (Fig.8C). Interestingly, removal of the (RS)8 segment from RS1 (SA8-RS8) does not impact processivity compared to wild-type ASF/SF2 (Fig.8D). The data imply that SRPK1 can support the processive phosphorylation of RS/SR repeats that are outside RS1 and that the stretch need not be directly C-terminal to RRM2.
Figure 8. Start-trap experiments using ASF/SF2 with modified RS domains.
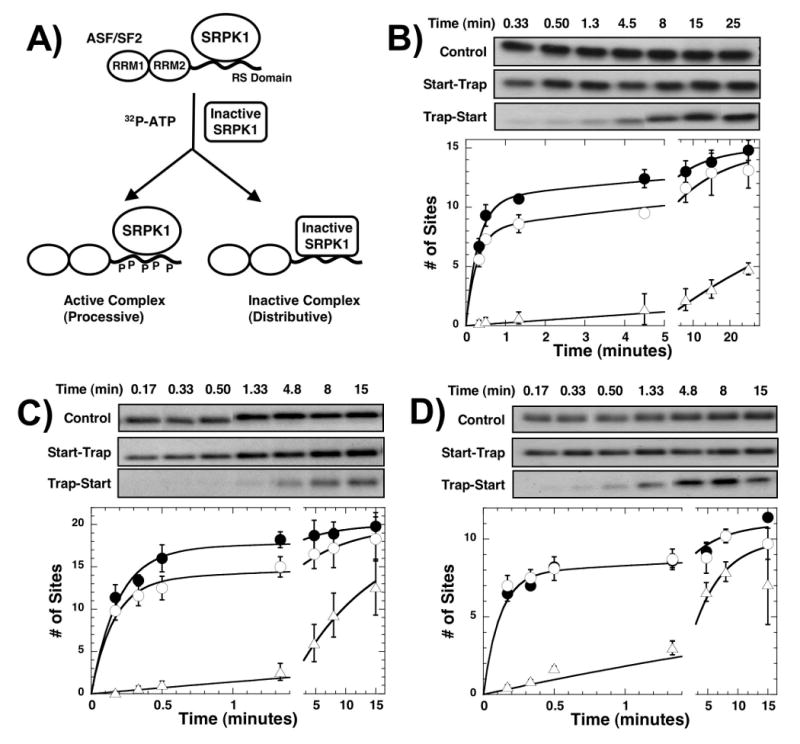
A) Start-trap protocol. (B-D) Start-trap experiments. SRPK1 (1μM) is preincubated with 200 nM of wt-ASF/SF2 (B), RS8-RS8 (C), and SA8-RS8 (D) and then the reaction is started with 100 μM 32P-ATP in the absence (control, ●) and presence of 30 μM kdSRPK1 (start-trap, ○). In trap-start experiments, the complex is first mixed with kdSRPK1 before 32P-ATP is added (△). The experiments are performed in triplicate and error bars for each time point are displayed. One representative kinetic experiment for each substrate is shown in the gel panels. The amplitudes of the fast phases are 11 ± 0.60 and 8.0 ± 0.36 sites in the absence and presence of kdSRPK1 for wild-type ASF/SF2, 17 ± 1.0 and 14 ± 0.80 sites in the absence and presence of kdSRPK1 for RS8-RS8, and 7.8 ± 0.56 and 7.8 ± 0.48 sites in the absence and presence of kdSRPK1 for SA8-RS8.
Discussion
SRPK1 displays unique enzymatic properties within the protein kinase family as it catalyzes multi-site, directional phosphorylation.18; 20 Previous studies showed that the enzyme rapidly adds between 5-8 phosphates onto ASF/SF2 in a processive manner, dissociates in a distributive step and then re-binds and modifies the remaining serines in a distributive (‘hit and release’) phase. This mechanism is promoted through initially high affinity interactions (Kd ≈ 50-100 nM) that likely maintain a high forward rate of catalysis compared to substrate dissociation in the early stages of the reaction.18; 20 In the present study we addressed how these interactions promote both regio- and temporal-specific phosphorylation. While peptide studies demonstrate that SRPK1 will phosphorylate serine (but not threonine) when flanked by arginine,21 the enzyme is unusually specific for the RS domain and will not readily modify serines outside this domain in ASF/SF2.18 Prior footprinting studies suggest that SRPK1 is further restricted to phosphorylating mostly N-terminal serines in the RS domain of ASF/SF2 raising the question of how certain serines next to arginines are selected and others avoided. Although incomplete, the X-ray structure of the kinase-splicing factor complex offers both local and distal residues that could support regiospecific control. The active site and substrate channel contain many electronegative side chains that could recognize local arginines near the phosphorylatable serines in the RS domain (Fig.1). In addition, the docking groove constitutes a distal feature that can recognize arginines further N-terminal from the phosphorylatable serines in the active site. In combination with RRM2 contacts that could further modulate initiation preferences, a broad range of both local and non-local structural elements may play a role in the observed residue preferences for this kinase and its SR protein targets.
SRPK1 Is A Residue-Specific Protein Kinase With Regio-Temporal Preferences
Our earlier MALDI-TOF analyses of ASF/SF2 suggested that SRPK1 not only is specific for the RS domain but also may limit itself to only serines in the RS1 segment.11 Since these findings are based on the detection of phosphoserines in N-terminal peptide fragments in the mass spectrometer, the absence of observable phosphoserines in the C-terminal region does not exclude a broader specificity for SRPK1. By monitoring the incorporation of 32P into N- and C-terminal fragments of the RS domain upon LysC proteolysis, we found that SRPK1 is not only capable of phosphorylating peptides based on the C-terminal region of the RS domain (Fig.2B), but also can modify RS2 in the full-length substrate in a time-dependent manner (Fig.3). While SRPK1 rapidly phosphorylates 10-12 serines in RS1 in about 2 minutes, RS2 can be phosphorylated at another 3-4 sites within 15-20 minutes. The kinetic preference for RS1 over RS2 (∼20-fold) is linked directly to the overall length of the RS/SR repeat. Inserting long Arg-Ser repeats into the C-terminal end of the RS domain leads to rapid, high level phosphorylation of RS2 that no longer displays preference for RS1 (Fig.7). Interestingly, phosphorylation of RS2 is dependent on the continuity of the RS/SR repeat as four alanine mutations at every other arginine disable most modifications in RS2. Such findings suggest that SRPK1 may limit itself to the RS domain and not modify RS sequences in the RRMs as the serines in the RS domain are displayed in strong continuous repeats. SRPK1 does not require an N-terminal flanking RRM for rapid, complete phosphorylation of the RS domain as the swap substrate cl-RS-RRM2(224) is readily modified despite possessing a C-terminal RRM. Overall, these studies show that SRPK1 is strictly residue-specific but kinetically favors certain regions owing to high RS/SR content.
SRPK1 Starts Phosphorylation At the Edge of RS1 in the Initiation Box
Although SRPK1 moves in a general C-to-N-terminal direction along the RS domain of ASF/SF2,18 the X-ray model does not define the initiation region (Fig.1). Using an itinerant cleavage site we addressed this issue and showed that SRPK1 does not initiate phosphorylation at a specific serine but rather selects a small group of serines at the C-terminal edge of RS1, a small region referred to as the initiation box (Fig. 4D). Such plasticity in the start serine helps to explain some earlier observations that small clusters of serines in RS1 may be converted to alanine without impacting phosphorylation efficiency. Interestingly, mutation of all three serines in the initiation box to alanine did not halt or negatively affect the kinetics of phosphorylation at other serines in the RS domain.18 The data imply that while SRPK1 has a preference for initiation here, it readily adapts to changes in the initiation box and modifies other residues in RS1. Thus, while SRPK1 targets the initiation box, this region provides a “soft” edge for the kinase. While we cannot dismiss a broadening of initiation preference upon Arg-to-Lys mutation, these findings suggest that SRPK1 has flexibility in binding the RS domain and is not likely to form a unique enzyme-substrate complex. Instead, the initial, unphosphorylated SRPK1-ASF/SF2 complex may exist in three major forms that differ by the position of the first serine in the active site (Ser-221, -223, or -225). Clearly, this narrow set of serines in the initiation box can be expanded depending on the nature of the RS domain. By adding more RS/SR repeats in the C-terminal portion of the RS domain, SRPK1 has equal access to both RS1 and RS2 segments and no longer shows sole preference for the initiation box. Furthermore, these modified RS/SR repeats in RS2 serines can be processively phosphorylated to the same extent as RS1 (Fig.8) implying that there is no requirement for a directly co-linear RRM and RS/SR repeat. Overall, such findings suggest that other SR proteins may be phosphorylated using a mechanism where phosphorylation initiation at the residue- and region-specific levels may vary depending on the position and length of the RS/SR repeats.
Multiple Protein-Protein Interactions Drive Orderly Phosphorylation
SRPK1 has a strong preference for binding to the initiation box and then phosphorylating in an N-terminal direction. To establish the molecular factors underpinning this directional pathway, we removed several key interactions within the SRPK1-ASF/SF2 complex. To block contacts with the N-terminal residues in RS1 (e.g., N′-RS1) we made disruptive mutations in the docking groove of SRPK1. This strategy was chosen rather than making complimentary mutations in ASF/SF2 since the RS domain displays a highly flexible binding mode and is likely to reposition itself upon mutation. To block interactions between SRPK1 and the RRMs we deleted RRM1 and moved RRM2 relative to the RS domain to remove any specific contacts. We elected to remove/relocate the domains rather than to mutate the individual contacts for the RRMs with SRPK1 as these contacts may be catalytically important. As an example, the interactions of RRM2 with the enzyme encompass a total of 4 residues dispersed in three locations in helices αD and F and the glycine-rich loop.19 We showed that alanine mutations in two consecutive residues (Trp-His) in the glycine-rich loop designed to disrupt one of the binding surfaces between RRM2 and SRPK1 results in a mutant kinase that is about 100-fold slower than wild-type SRPK1 at phosphorylating a short peptide substrate (data not shown). This result is consistent with previously published results on glycine-rich loop mutants of PKA and v-Fps that display very low phosphoryl transfer rates and binding affinities for ATP.22; 23 Such findings suggest that probing the role of docking residues that involve catalytically important residues are likely to be problematic owing to secondary effects on the phosphoryl transfer and/or nucleotide binding steps.
By considering RS domain interactions in the docking groove and those made with RRM2 we were able to analyze the contributions of these distal contacts and determine how each group participates in phosphorylation initiation (Fig. 6D). Although SRPK1 strongly prefers to start at the initiation box, mutation of the docking groove and removal of both RRMs completely eliminates this preference and imparts a slight preference for N-terminal initiation. Adding back either the docking groove or both RRMs increases the preference for C-terminal initiation by about 6-fold suggesting that either group of contacts participates equally to the initiation process. Although the role of RRM1 in the enzyme-substrate complex is not known, this domain has a small impact on phosphorylation initiation (2-fold) suggesting that that it could make some direct interactions with SRPK1 or possibly stabilize/strengthen the RRM2 contacts with the enzyme. Nonetheless, the enhancement in C-terminal preference in RS1 through either the docking groove or RRMs is small suggesting that neither contact group is sufficient to explain the high preference for initiation box recognition in the wild-type enzyme-substrate complex. Instead, it is necessary to have both a functional docking groove and a neighboring RRM to illicit strong C-terminal initiation preference. In fact, these interactions between docking elements appears highly cooperative as the addition of the docking groove and the RRM contacts increases C-terminal preference by about 160-fold. A very interesting outcome of this analysis is that whatever the means by which RRM2 imposes a directional bias towards the C-terminal region of RS1, the molecular mechanism appears highly flexible. The removal of all four contact residues in RRM2 does not significantly impact C-terminal initiation. In this study we used a construct lacking RRM1 but we have shown in a prior study that these mutations also have no effect in a longer substrate containing RRM1.20 Without diffraction data it is not possible to surmise how the mutated RRM2 restricts phosphorylation initiation but it does suggest that SRPK1 may recognize more general topological features of the RRM rather than discrete residues.
Conclusion
The studies presented herein allow us to propose how SRPK1 phosphorylates the RS domain of ASF/SF2 and may then recognize and modify other SR protein family members. SRPK1 begins its phosphorylation cycle at the initiation box located at the C-terminal end of RS1 (Fig.9). The initiation event is confined by long-range contacts between the docking groove in SRPK1 and N-terminal residues in RS1 (N′-RS1) and between several loci on SRPK1 that stabilize RRM2. Removal of these contacts leads to severe misalignment of the RS domain and a shift from highly directional to random RS1 phosphorylation. Surprisingly, these guidance factors do not control regiospecific phosphorylation as constructs lacking RRMs and the docking groove residues still have a strong temporal preference for RS1 over the weaker RS2. Thus, the active site and substrate channel in SRPK1 (Fig.1) possess sufficient contacts to define regiospecific preferences, a phenomenon driven largely the RS/SR content of the substrate. Given the high level of processivity (Fig.8), RS1 likely stays attached to the enzyme in the extension phase and ‘slides’ through the active site for about half of the sites with new contacts being made with a secondary structural element (β4) from RRM2 that unfolds and occupies the docking groove at later phosphorylation steps (Fig.9). Given the observed level of processivity in the start-trap experiments (Fig.8B), not all the serines in RS1 are phosphorylated in a processive manner so that SRPK1 must re-bind to RS1 at later stages. Although RS2 in wild-type ASF/SF2 lacks a strong RS/SR stretch, it can be phosphorylated in a slower reaction that involves dissociative steps that reposition the kinase near the C-terminus of the RS domain (Fig.9). We suspect that these dissociative steps are driven by higher ‘off’ rates for the substrate as a function of phosphorylation. Furthermore, distributive phosphorylation is likely to play an essential dynamic role for completing the slower phosphorylation steps in RS2 and for some serines not originally phosphorylated in RS1 owing to alternative priming in the initiation box.
Figure 9. Proposed mechanism for regiospecific phosphorylation control.
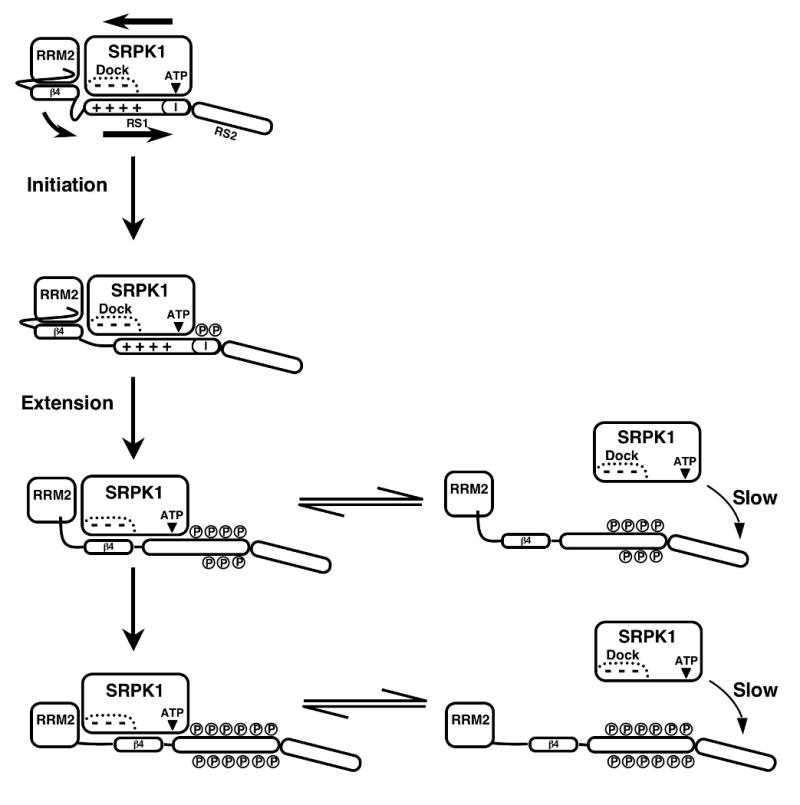
SRPK1 forms a high affinity complex with ASF/SF2 and starts the phosphorylates cycle in the “initiation box” at the C-terminal end of RS1 (I). Approximately seven serines are modified in the extension phase before the stability of the enzyme-substrate complex diminishes and SRPK1 can dissociate and re-bind to modify the remaining serines in RS1 or begin to modify serines in RS2.
Biological data suggest that RS1 phosphorylation by SRPK1 is sufficient for movement of ASF/SF2 from the cytoplasm to nuclear speckles while RS2 phosphorylation by the nuclear Clk/Sty is responsible for the dispersion of these speckles.15; 24 Recent studies suggest that SRPK1 enters the nucleus under certain stress conditions impacting splicing possibly through hyperphosphorylation of SR proteins.14 While a biological role for SRPK1-directed phosphorylation of RS2 under these or other conditions has not been established, the biochemical data provides valuable information on the regiospecific preferences in ASF/SF2 and can be used to explain how the enzyme may adapt to other SR proteins with diverse RS domains. Although the RS domain of ASF/SF2 is relatively short (50 aa), others are much longer with one exceeding 300 residues (SRp75). Furthermore, while ASF/SF2 has a long RS/SR repeat that is contiguous with the RRM (Fig.1), other SR proteins disperse these repeats and place some at the C-terminus of the RS domain away from the N-terminal RRMs (e.g., Srp38). The mechanism in Figure 9 allows for flexibility in positioning these different RS domains as we have shown that the weaker RS2 sequence can be rapidly phosphorylated if larger RS/SR repeats are present (Fig.8). We propose that long, intervening pieces of the RS domain, not contiguous with the RRM, can be ‘looped out’ such that C-terminal repeats may be phosphorylated without forgoing enzyme contacts with the RRM. Thus, we anticipate that SRPK1 may possess a strong temporal preference for long RS/SR repeats in these SR proteins, irrespective of position within the RS domain, and then may use the N-terminal RRM and docking groove for initiation and directional phosphorylation.
Material & Methods
Materials
Lysobacter enzymogenes endoproteinase Lys-C (LysC), adenosine triphosphate (ATP), 3-(N-morpholino)propanesulphonic acid (Mops), 2-[N-morpholino]ethanesulphonic acid (MES), Tris (hydroxymethyl) aminomethane (Tris), MgCl2, NaCl, EDTA, glycerol, acetic acid, Phenix imaging film, TFA, BSA, acetonitrile, and liquid scintillant were obtained from Fisher Scientific. [γ-32P] ATP was obtained from NEN Products, a division of Perkin-Elmer Life Sciences. α-cyano-4-hydroxy cinnamic acid matrix solution was obtained from Aldrich Chemicals and recrystallized once from ethanol.
Expression and purification of recombinant proteins
All mutations in ASF/SF2 were generated by single or sequential polymerase chain reactions using the QuikChange™ mutagenesis kit and relevant primers (Stratagene, La Jolla, CA). N-terminal deletion constructs were generated by PCR amplification and sub-cloning into pET28a (C-terminal His tag). Domain swap mutant constructs were generated by sequential sub-cloning steps into pET15b (N-terminal His tag). All mutations in SRPK1 were made from a wild-type vector containing an N-terminal His tag (pET15b). The docking groove mutant construct SRPK1(6M) was generated by 6 separate point mutations and was previously described.11 Cleavage vectors with alternative cleavage sites were constructed in procedures similar to cl-ASF(214) vector, which was referred to as the ASF(5R1K) vector in a previous paper.18 The plasmids for wild-type and mutant forms of ASF/SF2 and SRPK1 were transformed into the BL21 (DE3) E. coli strain and the cells were then grown at 37°C in LB broth supplemented with 100 μg/ml ampicillin or 50 μg/ml kanamycin depending on the type of plasmid vector. Protein expression was induced with 0.4 mM IPTG at room temperature for 5 hours for ASF constructs and 12 hours for SRPK constructs. SRPK1 was purified by Ni-resin affinity chromatography using a published procedure.17 All ASF/SF2 constructs were refolded and purified using a previously published protocol.18
Phosphorylation Reactions & LysC Proteolysis
The phosphorylation of wild-type and mutant forms of ASF/SF2 by SRPK1 was carried out in the presence of 50 mM Mops (pH 7.4), 10 mM free Mg2+, 5 mg/mL BSA, and [γ-32P]ATP (600-1000 cpm pmol-1) at 23 °C according to previously published procedures.18 Reactions were typically initiated with the addition of 32P-ATP (100 μM) in a total reaction volume of 10 μL and then were quenched with 10 μL SDS-PAGE loading buffer. Each quenched reaction was loaded onto a 12% SDS-PAGE gel and the dried gels were exposed with Kodak imaging film (Biomax MR). The protein bands corresponding to phosphorylated ASF/SF2 were excised and counted on the 32P channel in liquid scintillant. Control experiments, specific activity determination and time-dependent product concentrations were determined as previously described.17 For equilibrium ATP-dependent phosphorylation mapping (ATP limitation experiments), sample proteolysis with LysC (100 ng) was carried out for 1 h (0.2, 0.5, 1 and 2 μM ATP), 2 h (3.5 μM ATP) or 4 h (6 and 100 μM ATP) at 37 °C. For kinetic time-dependent phosphorylation mapping (pulse-chase experiments), reactions were initiated with [32P]ATP (100 μM) and then were cold-chased at varying times with 4 mM ATP. Proteolysis with LysC (100 ng) was then carried out for 4 h at 37 °C. The N- and C-terminal fragments were excised and counted on the 32P channel in liquid scintillant.
Mass Spectrometric Analyses
MALDI-TOF analyses were carried out using a PerSeptive Biosystems Voyager DE PRO spectrometer. In general, ASF/SF2 (1 μM) was incubated with SRPK1 (300 nM) and 0.3 mM ATP in the presence of 50mM Tris-HCL (pH 7.4) and 10 mM free Mg2+ for 3 hrs in a total volume of 200 μL at room temperature. Reactions then were quenched with 5% acetic acid, desalted with Zip-tip C18 and eluted with 80% acetonitrile, 2% acetic acid for MALDI-TOF analysis. Unphosphorylated sample controls were prepared in the same manner, without ATP. The matrix solution consisted of 5 mg/ml α-cyano-4-hydroxy cinnamic acid in 1:1:1 acetonitrile, ethanol, and 0.52% TFA. Final pH of the matrix solution was 2.0.
Data Analysis
The phosphoryl contents of the N- and C-terminal fragments upon LysC cleavage were calculated using the ratios of 32P in the bands and the total phosphoryl contents prior to cleavage using MALDI-TOF measurements. Progress curve data for ASF/SF2 phosphorylation were plotted as ratios of incorporated phosphate and the total substrate concentration (# of sites) as a function of time and were fit to a double-exponential function.
Supplementary Material
Acknowledgments
This work was supported by NIH grants to J.A.A. (GM67969) and X-D.F. (GM52872). C-T.M. and J.C.H. were supported by an NIH training grant (GM07752).
Footnotes
Abbreviations: ASF/SF2, human alternative splicing factor; LysC, lysyl endoproteinase; MALDI-TOF, Matrix assisted laser desorption ionization-time of flight mass spectroscopy; RS domain, C-terminal domain rich in arginine-serine repeats (residues 198-248); RS1, N-terminal portion of RS domain (residues 198-227); RS2, C-terminal portion of RS domain (residues 228-248); RRM, RNA recognition motif; SR protein: splicing factor containing arginine-serine repeats; SRPK, SR-specific protein kinase.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 3.Stojdl DF, Bell JC. SR protein kinases: the splice of life. Biochem Cell Biol. 1999;77:293–8. [PubMed] [Google Scholar]
- 4.Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–24. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 5.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–70. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 6.Mermoud JE, Cohen P, Lamond AI. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 1992;20:5263–9. doi: 10.1093/nar/20.20.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mermoud JE, Cohen PT, Lamond AI. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. Embo J. 1994;13:5679–88. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan PI, Stojdl DF, Marius RM, Scheit KH, Bell JC. The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp Cell Res. 1998;241:300–8. doi: 10.1006/excr.1998.4083. [DOI] [PubMed] [Google Scholar]
- 9.Yun CY, Fu XD. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J Cell Biol. 2000;150:707–18. doi: 10.1083/jcb.150.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc Natl Acad Sci U S A. 2001;98:10154–9. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velazquez-Dones A, Hagopian JC, Ma CT, Zhong XY, Zhou H, Ghosh G, Fu XD, Adams JA. Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J Biol Chem. 2005;280:41761–8. doi: 10.1074/jbc.M504156200. [DOI] [PubMed] [Google Scholar]
- 12.Ding JH, Zhong XY, Hagopian JC, Cruz MM, Ghosh G, Feramisco J, Adams JA, Fu XD. Regulated cellular partitioning of SR protein-specific kinases in mammalian cells. Mol Biol Cell. 2006;17:876–85. doi: 10.1091/mbc.E05-10-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:664–75. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009;23:482–95. doi: 10.1101/gad.1752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G. Interplay between SRPK and Clk/Sty Kinases in Phosphorylation of the Splicing Factor ASF/SF2 Is Regulated by a Docking Motif in ASF/SF2. Mol Cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 16.Prasad J, Manley JL. Regulation and substrate specificity of the SR protein kinase Clk/Sty. Mol Cell Biol. 2003;23:4139–49. doi: 10.1128/MCB.23.12.4139-4149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aubol BE, Chakrabarti S, Ngo J, Shaffer J, Nolen B, Fu XD, Ghosh G, Adams JA. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proc Natl Acad Sci U S A. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma CT, Velazquez-Dones A, Hagopian JC, Ghosh G, Fu XD, Adams JA. Ordered multi-site phosphorylation of the splicing factor ASF/SF2 by SRPK1. J Mol Biol. 2008;376:55–68. doi: 10.1016/j.jmb.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 19.Ngo JC, Giang K, Chakrabarti S, Ma CT, Huynh N, Hagopian JC, Dorrestein PC, Fu XD, Adams JA, Ghosh G. A Sliding Docking Interaction Is Essential for Sequential and Processive Phosphorylation of an SR Protein by SRPK1. Mol Cell. 2008;29:563–76. doi: 10.1016/j.molcel.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagopian JC, Ma CT, Meade BR, Albuquerque CP, Ngo JC, Ghosh G, Jennings PA, Fu XD, Adams JA. Adaptable Molecular Interactions Guide Phosphorylation of the SR Protein ASF/SF2 by SRPK1. J Mol Biol. 2008;382:894–909. doi: 10.1016/j.jmb.2008.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang HY, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu XD. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998;140:737–50. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant BD, Hemmer W, Tsigelny I, Adams JA, Taylor SS. Kinetic analyses of mutations in the glycine-rich loop of cAMP- dependent protein kinase. Biochemistry. 1998;37:7708–15. doi: 10.1021/bi972987w. [DOI] [PubMed] [Google Scholar]
- 23.Hirai TJ, Tsigelny I, Adams JA. Catalytic assessment of the glycine-rich loop of the v-Fps oncoprotein using site-directed mutagenesis [In Process Citation] Biochemistry. 2000;39:13276–84. doi: 10.1021/bi001216g. [DOI] [PubMed] [Google Scholar]
- 24.Koizumi J, Okamoto Y, Onogi H, Mayeda A, Krainer AR, Hagiwara M. The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs) J Biol Chem. 1999;274:11125–31. doi: 10.1074/jbc.274.16.11125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


