Summary
In addition to the inherent interest stemming from their ecological and human health impacts, microbes have many advantages as model organisms, including ease of growth and manipulation and relatively simple genomes. However, the imaging of bacteria via light microscopy has been limited by their small sizes. Recent advances in fluorescence microscopy that allow imaging of structures at extremely high resolutions are thus of particular interest to the modern microbiologist. In addition, advances in high-throughput microscopy and quantitative image analysis are enabling cellular imaging to finally take advantage of the full power of bacterial numbers and ease of manipulation. These technical developments are ushering in a new era of using fluorescence microscopy to understand bacterial systems in a detailed, comprehensive, and quantitative manner.
Introduction
The power and beauty of direct observation has long ensured microscopy a special place in the scientific toolbox. Amongst microscopy methods, light microscopy is the most versatile and accessible due to its relative ease of implementation and ability to image cells under native living conditions, without the need for harsh treatments such as fixatives or vacuum environments. By the turn of the 20th century, the quest for the proverbial “picture worth a thousand words” helped drive the development of optics to the point of nearly perfecting light microscopes capable of imaging at resolutions bounded only by the physics of diffraction. The next major breakthrough came in the ability to fluorescently label specific proteins of interest to determine their distributions and abundance. This labeling was first performed using dye-conjugated antibodies and immunofluorescence, and later with genetically-encoded fluorescent proteins. Different labeling strategies have different advantages and disadvantages. For example, immunofluorescence enables visualization of endogenous native proteins but only in fixed samples, whereas fluorescent proteins enable imaging of dynamics in live cells but invoke potential complications due to perturbations caused by the protein fusion. The development of new labeling methods is an active area (reviewed in detail in [1,2]) that is constantly improving our ability to simultaneously image multiple structures, reduce the disruptiveness of the label, report on specific protein conformations, and image additional macromolecules such as DNA, RNA, lipids, and carbohydrates.
In addition to advances in labeling methods, the past decade witnessed an explosion of fluorescence microscopy-based approaches to image protein dynamics and interactions. For example, protein mobility and maturation are now routinely assayed by Fluorescence Recovery After Photobleaching (FRAP) or with photoconvertible fluorescent proteins [3], and physical intramolecular or intermolecular associations can be monitored in both space and time through Förster Resonance Energy Transfer (FRET) [4] or Bimolecular Fluorescent Complementation (BiFC) [5]. Though advances in imaging protein dynamics and interactions are still being made, these methods are now in widespread use in both microbes and other cellular systems and have been previously reviewed elsewhere [3–5], such that they will not be discussed further here. Rather, I will focus on three rapidly developing areas that promise to be of particular use for studying free-living microbes, namely advances in improving the resolution, throughput, and quantitative analysis of fluorescence microscopy.
Super-resolution methods image protein localization and dynamics at near-molecular scales
In the past decade conventional fluorescence microscopy has been used to demonstrate that bacterial cells, like their eukaryotic counterparts, are highly organized at the subcellular level. Bacteria thus need to deposit specific components at specific places in the cell in a dynamic fashion [6,7]. For example, we now know that cytoskeletal filaments extend through the bacterial cell and the cell poles are home to large macromolecular assemblies that function in chemotaxis and motility (reviewed in [6,7]). But how exactly are these structures organized at the molecular scale? The primary limitation of optical imaging methods such as fluorescence microscopy is that diffraction prevents structures closer than approximately half the wavelength of light from being resolved from one another. This physical diffraction limit of ~200 nm was long thought to be an absolute barrier to using light microscopy to understand the organization of individual proteins that are on the scale of ~1–10 nm. The last generation of microscopy advances such as confocal, 2-photon, and Total Internal Reflection (TIRF) microscopy focused on reducing the impact of out-of-focus light in order to approach the diffraction limit in thick samples like mammalian tissue [8]. Though powerful, these methods provided little to no advantages for very thin samples such as microbes. However, a series of methods, collectively referred to as super-resolution imaging or “nanoscopy”, have recently been implemented to image proteins at the sub-diffraction-limited resolutions that would be particularly advantageous for studying small microbes. Of the super-resolution imaging methods reported to date, two general approaches appear to be emerging as harboring the most practical and immediate potential. One of these approaches, Stimulated Emission Depletion (STED), involves a new type of multi-laser microscope, while the other approach, single molecule localization microscopy, combines conventional optics with clever use of repeated activation of photo-switchable fluorescent molecules.
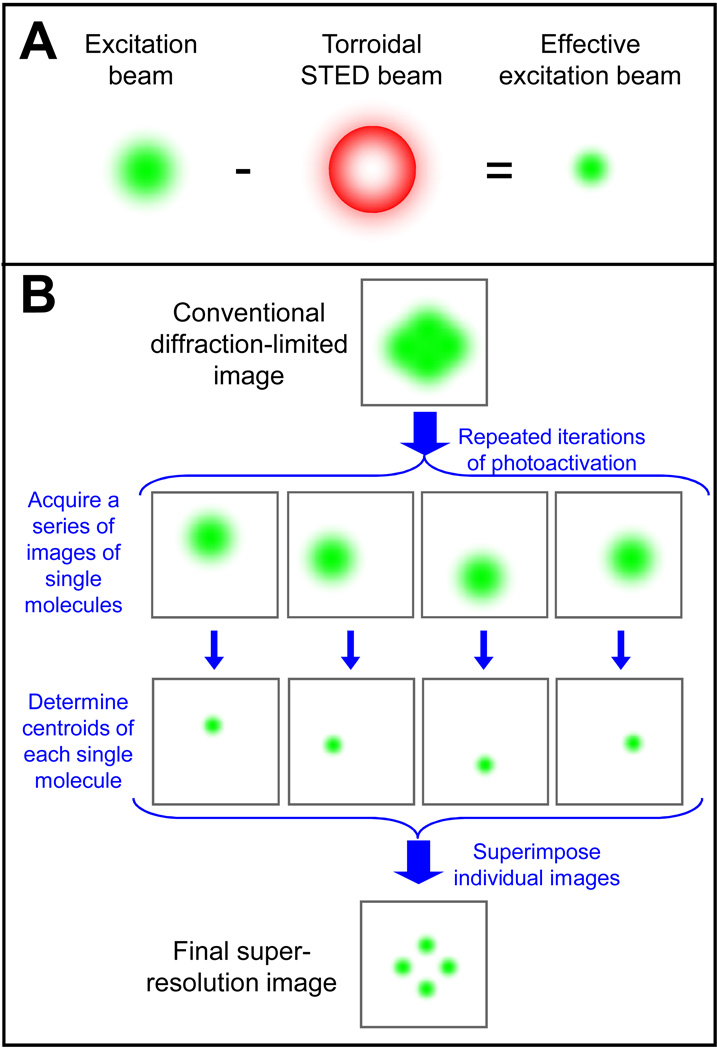
STED fluorescence microscopy combines two lasers to break the diffraction limit [9]. One laser, the excitation laser, is tuned to a wavelength that activates the fluorophore of interest. The second, the STED laser, is tuned to a longer wavelength that very rapidly de-activates the fluorophore. The idea is that each laser is diffraction-limited, but by carefully timing a doughnut-shaped STED beam of saturating intensity immediately after activation by the excitation beam, only the fluorescent molecules in the very center of the beams escape rapid de-activation and fluoresce in a detectable manner (Figure 1A) [10]. This small zone of effective fluorescence activation is then scanned over the entire sample to produce the final image. STED has been successfully applied to a number of animal cell types and has been pushed to lateral resolutions of below 20 nm [10]. Most excitingly for microbiologists, STED was recently used to image two different protein complexes in mitochondria, demonstrating that the TOM complex forms distinct nanoscale particles [11]. At only 200–500 nm in size, mitochondria are even smaller than most bacteria such that STED should be readily applicable to nearly all microbes of interest. Using four lasers instead of two, nanoscale colocalization of two different proteins was recently performed by STED [11], and a variant called IsoSTED enables isotropic super-resolution in all three dimensions [12]. STED has the advantages of being fast (a recent study was able to image synaptic vesicle dynamics at video-rate [13]), suitable for live-cell imaging, unburdened by complicated image processing, and compatible with any type of fluorescent molecule. The disadvantages of STED largely stem from the fact that it works by only imaging the fluorescence of a small fraction of the fluorophores it excites, leading to relatively high rates of bleaching and phototoxicity. The cost and complexity of the multi-laser system have limited STED’s widespread appeal, but its recent commercialization should soon reduce the barrier to its implementation.
Two different approaches to breaking the diffraction limit. A. STED microscopy uses two different lasers, a conventional excitation laser (left) and a torroidal doughnut-shaped STED laser (middle). The STED laser deactivates the fluorescence outside of a small central zone, thereby reducing the effective excitation area (right). B. Single molecule localization microscopy methods such as PALM and STORM use photoactivatable fluorophores to successively image the localization of a small number of molecules at a time at high precision by finding the molecule’s centroid. The many resulting images (4 are shown in the example illustrated here) are then superimposed to generate the final super-resolution image.
Single molecule localization microscopy uses a fundamentally different approach to break the diffraction limit and is referred to as Photoactivation Localization Microscopy (PALM) [14], Fluorescence PALM (FPALM) [15], or Stochastic Optical Reconstruction Microscopy (STORM) [16]. The idea is that if one knows that a given diffraction-limited area contains only a single fluorescent molecule, then the position of that molecule can be determined as the center of the resulting diffracted sphere. The number of photons collected from the molecule establishes the accuracy with which the molecule’s position can be determined and can approach single nanometers [17]. Photoactivatable fluorescent molecules that can be switched on and off are used to stochastically activate only a few molecules at a time, thereby ensuring that only one fluorescent molecule is excited in each diffraction-limited area. By iteratively re-imaging the same sample, each time determining the positions of a few molecules, and then superimposing the positions determined in each image, a final nanoscale image can be reconstructed (Figure 1B). Though conceptually and technically similar, PALM has generally employed genetically encoded fusions to photoactivatable fluorescent proteins such as EosFP, PA-GFP, and Dronpa, while STORM has used antibodies coupled to photo-switchable organic dye pairs such as Cy3-Cy5 [2]. By combining photoactivatable molecules that can be discriminated from one another, two-color PALM and STORM have recently been reported [18–20].
PALM/STORM can achieve lateral resolutions in the tens of nanometers and has been used to image bacteria-scale structures such as lysosomes and mitochondria, as well as both microtubule and actin filament cytoskeletal arrays [14]. Several methods using optical astigmatism, multi-plane imaging, or interferometry have also extended the nanoscale resolution of PALM/STORM to three dimensions [21–23]. Since single molecule localization microscopy depends on the superimposition of many iterated images, it is generally performed on fixed samples to assure that the molecules do not move between images. The need to collect, process, and integrate many raw images to generate a final super-resolution image also takes a long time; the initial implementation of PALM required 2–12 hours per final image, though newer implementations can reduce this time to the order of several minutes [14]. By foregoing the effort to reconstruct the population’s localization organization and instead focusing on the dynamics of individual molecules, several studies have used photoactivatable single molecule localization approaches to image protein movements in living cells [24]. These single particle tracking methods have been applied to the bacterial cytoskeleton, namely the Caulobacter crescentus MreB actin homolog [25] and the Escherichia coli FtsZ tubulin homolog [26], and promise to shed new light on the mechanisms of protein trafficking in all cell types. The relative simplicity of the PALM/STORM optical set-up has led to its rapid adoption. In addition, this approach has the exciting yet largely untapped potential to probe multi-subunit complex organization and stoichiometry via its imaging of single molecules.
High-throughput microscopy enables imaging-based screens and systems-level analysis
Genomic methods such as microarray analysis and deep sequencing have demonstrated the power of surveying the effects of a large number of conditions on large numbers of genes. The counterpart to super-resolution imaging’s goal of increasing spatial resolution is thus to increase the throughput of fluorescence microscopy to determine the localization of every cellular protein under many different conditions. Automated high-throughput imaging systems for imaging large animal cells are now commercially developed and routinely used in high-content screens [27]. These assays simultaneously examine the effects of small molecules or RNAi-mediated gene knockdowns on cell morphology and the localization and levels of both proteins and DNA. For example, such screens have been applied to a number of pathogen-host interactions to identify mutants that are defective in microbial pathogenesis [28,29]. However, conventional high-throughput microscopy is performed with air immersion objectives whose spatial resolution is far below the diffraction limit, and is therefore insufficient for imaging small microbial cells. While small cells would clearly benefit from the super-resolution imaging methods discussed above, even high-throughput diffraction-limited resolution would be a significant advance.
The first generation of solutions to the problem of high-throughput diffraction-limited imaging came through brute force, laboriously repeating conventional imaging thousands of times over. In this manner, genomic studies determined the protein localization of C-terminal GFP fusions to most of the proteins of Saccharomyces cerevisiae [30], Schizosaccharomyces pombe [31], and E. coli [32]. Similarly, genomic deletion libraries of S. cerevisiae and E. coli were imaged to identify genes involved in cell shape determination [33,34]. These pioneering efforts yielded a series of exciting discoveries. To focus on E. coli, for example, the genomic localization screen mentioned above revealed that the Tol/Pal pathway is localized to the division plane. Subsequent analysis demonstrated that this pathway represents a previously unappreciated division regulator [35]. Thus, despite decades of intensive study of E. coli cell division by other methods, it was the high-throughput observation of Tol/Pal localization that identified the long-elusive coordinator of inner and outer membrane constriction. Likewise, a screen of E. coli deletion mutants recently identified RodZ as a novel conserved member of the bacterial cell shape determination pathway [33].
The routine imaging of thousands of strains without expending unrealistic time and effort presents the need for more efficient methods for high-throughput diffraction-limited imaging. Several sophisticated high-throughput imaging solutions using either microfluidics or printing technologies have been proposed [36,37]. However, the complexity and difficulty of implementing these approaches have limited their widespread use. We recently developed a simple “pedestal slide” system to image 48 separate samples at diffraction-limited resolution on one slide, each pedestal slide containing a 6 × 8 array of agarose pedestals [38]. The 48 strains can be stamped onto the pedestal slide and covered with a single large coverslip, since the pedestal spacing prevents cross-contamination of neighboring strains. The cells are immobilized between the coverslip and the agarose pedestal. By making the pedestals with growth media, the cells are allowed to robustly grow and divide on the slide, enabling high-throughput live-cell time-lapse imaging. The pedestal slides can be imaged on virtually any type of microscope and can increase the throughput of the traditional process of imaging at maximal diffraction-limited resolution nearly 50-fold, without any sacrifice in image quality [38]. Moreover, since the optics in this system are identical to those of a conventional slide preparation, the pedestal slide system could be readily combined with any advanced imaging methods such as STED or PALM/STORM. Regardless of which method for high-throughput diffraction-limited microscopy becomes widely adopted, the power of combining genomic and imaging approaches ensures that its use will soon be pervasive.
Quantitative analysis enhances the information content and utility of fluorescence microscopy
Fluorescence microscopy images contain a wealth of readily-apparent qualitative information. But these images generally contain much more information that can be extracted via quantitative image analysis. Fluorescence intensity linearly correlates with fluorophore abundance. By calibrating fluorescence intensity to different types of standards, fluorescence microscopy can thus provide an assay for macromolecule concentrations in living cells. These kinds of single-cell measurements have been used in a wide variety of experimental systems including assays of stochastic gene expression or protein-protein interactions [39,40].
While total cellular fluorescence measurement is a powerful advance, even more insight can be gleaned from measuring fluorescence intensity subcellularly and tracking its dynamics over time. One nice illustration of the power of this approach comes from yeast, where careful quantitation of the localization dynamics of a series of proteins related to actin patches showed that different proteins associate with endocytic events at different points in the process, thereby establishing an ordered pathway for actin-mediated endocytosis [41]. Spatially-resolved quantitation can also shed light on the mechanisms of protein targeting. For example, an analysis of the patterns of IcsA localization in Shigella flexneri demonstrated that this polarly-localized bacterial virulence factor is initially secreted at the cell poles and that a polar protein gradient is maintained by the uniformly-localized IcsP protease [42].
One challenge in image quantitation is that digital images are pixilated such that all of the points within a given pixel are binned to produce a single intensity value. This problem becomes significant when analyzing small cells like microbes that are generally on the scale of just tens of pixels. Interpolation methods can subdivide pixilated images into arbitrarily fine grids to increase the effective image resolution. Two-dimensional interpolated contouring was used to identify heterogeneities in E. coli elongation [43]. A recently developed image analysis software suite termed Projected System of Internal Coordinates from Interpolated Contours (PSICIC) both automates the use of interpolated contouring to determine cell boundaries with sub-pixel precision, and generates an internal coordinate system that allows fluorescence data from variably-shaped cells to be readily compared and analyzed [44]. Experiments comparing PSICIC-analyzed fluorescence micrographs to data collected by electron microscopy (EM) indicated that PSICIC provides a nearly 15-fold improvement in the precision of cell border determination as compared to conventional pixel-limited analysis methods [44]. This result suggests that information that was once thought to be exclusive to EM could be accessed through careful analysis of light microscopy.
Perhaps the most powerful aspect of quantitative image analysis is that it produces data that can be integrated with other computational approaches. Quantitative data from imaging experiments has fueled a wide range of theoretical modeling studies, including analyses of E. coli chemotaxis signaling [45], cell wall architecture [46], and division constriction [47]. Combined with the types of high-throughput imaging discussed above, image quantitation also enables the use of powerful data analysis methods like hierarchical clustering and principal components analysis. For example, clustering methods have been used to identify distinct cell shape determination pathways in animal cells treated with RNAi libraries [48], and PCA has been used to statistically compare irregularly-shaped bacteria cells [49,50]. In the future, the ability to integrate quantitative cell shape and protein localization datasets with other genomic datasets should significantly advance our ability to understand protein function in a systematic fashion.
Conclusions
Fluorescence microscopy can provide information about protein levels, distributions, dynamics, and interactions and in both living and fixed cell samples, and has thus emerged as one of the most powerful tools of the modern microbiologist. New advances that improve image resolution, imaging throughput, and quantitative image analysis are pushing fluorescence microscopy to unprecedented depth, breadth, and utility. These advances potentially can be combined to generate a comprehensive analysis pipeline. Excitingly, developments such as super-resolution’s ability to break the diffraction limit suggest that the capabilities of fluorescence microscopy can be pushed to levels that were considered impossible only a few years ago. In the future it should be interesting to see how these methods evolve and how the microbiology community uses them to generate new insights into fundamental biological questions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 2. Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol. 2008;9:929–943. doi: 10.1038/nrm2531. * Excellent current review on super-resolution methods and photoactivatable fluorescent probes.
- 3.Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 4.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 5.Ciruela F. Fluorescence-based methods in the study of protein-protein interactions in living cells. Curr Opin Biotechnol. 2008;19:338–343. doi: 10.1016/j.copbio.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Gitai Z. The new bacterial cell biology: moving parts and subcellular architecture. Cell. 2005;120:577–586. doi: 10.1016/j.cell.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 7.Collier J, Shapiro L. Spatial complexity and control of a bacterial cell cycle. Curr Opin Biotechnol. 2007 doi: 10.1016/j.copbio.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halbhuber KJ, Konig K. Modern laser scanning microscopy in biology, biotechnology and medicine. Ann Anat. 2003;185:1–20. doi: 10.1016/S0940-9602(03)80002-X. [DOI] [PubMed] [Google Scholar]
- 9.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 10.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 11. Donnert G, Keller J, Wurm CA, Rizzoli SO, Westphal V, Schonle A, Jahn R, Jakobs S, Eggeling C, Hell SW. Two-color far-field fluorescence nanoscopy. Biophys J. 2007;92:L67–L69. doi: 10.1529/biophysj.107.104497. ** Beautiful demonstration of both two-color STED and applying STED to mitochondria.
- 12.Schmidt R, Wurm CA, Jakobs S, Engelhardt J, Egner A, Hell SW. Spherical nanosized focal spot unravels the interior of cells. Nat Methods. 2008;5:539–544. doi: 10.1038/nmeth.1214. [DOI] [PubMed] [Google Scholar]
- 13. Westphal V, Rizzoli SO, Lauterbach MA, Kamin D, Jahn R, Hell SW. Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science. 2008;320:246–249. doi: 10.1126/science.1154228. ** Demonstrates the power of rapid STED imaging to follow protein dynamics in living cells.
- 14.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 15. Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. ** First published description of single molecule localization microscopy (PALM).
- 16. Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. ** First publication of STORM.
- 17.Bobroff N. Position measurement with a resolution and noise-limited instrument. Rev. Sci. Instrum. 1986;57:1152–1157. [Google Scholar]
- 18. Shroff H, Galbraith CG, Galbraith JA, White H, Gillette J, Olenych S, Davidson MW, Betzig E. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci U S A. 2007;104:20308–20313. doi: 10.1073/pnas.0710517105. * First publication of two-color PALM.
- 19.Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. * First publication of two-color STORM.
- 21. Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. * First publication of three-dimensional super-resolution STORM imaging.
- 22. Juette MF, Gould TJ, Lessard MD, Mlodzianoski MJ, Nagpure BS, Bennett BT, Hess ST, Bewersdorf J. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5:527–529. doi: 10.1038/nmeth.1211. * First publication of three-dimensional super-resolution PALM imaging.
- 23.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman CM, Kanchanawong P, Davidson MW, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci U S A. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 25. Biteen JS, Thompson MA, Tselentis NK, Bowman GR, Shapiro L, Moerner WE. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat Methods. 2008;5:947–949. doi: 10.1038/NMETH.1258. ** Demonstrated that even commonly-used fluorescent proteins like EYFP can be photoactivated. Using this method, the dynamics of Caulobacter MreB were analyzed.
- 26.Niu L, Yu J. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys J. 2008;95:2009–2016. doi: 10.1529/biophysj.108.128751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Howell BJ. High-content screening: emerging hardware and software technologies. Methods Enzymol. 2006;414:468–483. doi: 10.1016/S0076-6879(06)14025-2. [DOI] [PubMed] [Google Scholar]
- 28.Agaisse H. Investigating the involvement of host factors involved in intracellular pathogen infection by RNAi in Drosophila cells. Methods Mol Biol. 2008;415:395–402. doi: 10.1007/978-1-59745-570-1_23. [DOI] [PubMed] [Google Scholar]
- 29.Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science. 2005;309:1248–1251. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- 30.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 31.Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 32.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 33.Shiomi D, Sakai M, Niki H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. Embo J. 2008;27:3081–3091. doi: 10.1038/emboj.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohya Y, Sese J, Yukawa M, Sano F, Nakatani Y, Saito TL, Saka A, Fukuda T, Ishihara S, Oka S, et al. High-dimensional and large-scale phenotyping of yeast mutants. Proc Natl Acad Sci U S A. 2005;102:19015–19020. doi: 10.1073/pnas.0509436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. ** Using the data from the E. coli localized protein library, Tol/Pal were identified as localized to the division plane and involved in coordinating inner and outer membrane constriction.
- 36.Neumann B, Held M, Liebel U, Erfle H, Rogers P, Pepperkok R, Ellenberg J. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- 37.Chung K, Crane MM, Lu H. Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat Methods. 2008;5:637–643. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- 38. Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z. Quantitative Genome-scale Analysis of Protein Localization in an Asymmetric Bacterium. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0901781106. In Press. ** Development of pedestal slides and a high-throughput high-resolution imaging pipeline enabled the rapid analysis of the localization of most Caulobacter proteins as both N- and C-terminal mCherry fusions.
- 39.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 40.Sourjik V, Vaknin A, Shimizu TS, Berg HC. In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol. 2007;423:365–391. doi: 10.1016/S0076-6879(07)23017-4. [DOI] [PubMed] [Google Scholar]
- 41.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 42.Robbins JR, Monack D, McCallum SJ, Vegas A, Pham E, Goldberg MB, Theriot JA. The making of a gradient: IcsA (VirG) polarity in Shigella flexneri. Mol Microbiol. 2001;41:861–872. doi: 10.1046/j.1365-2958.2001.02552.x. [DOI] [PubMed] [Google Scholar]
- 43. Reshes G, Vanounou S, Fishov I, Feingold M. Cell shape dynamics in Escherichia coli. Biophys J. 2008;94:251–264. doi: 10.1529/biophysj.107.104398. * Early use of sub-pixel interpolation image analysis to extract detailed information about cell shape and growth dynamics in bacteria.
- 44. Guberman JM, Fay A, Dworkin J, Wingreen NS, Gitai Z. PSICIC: noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLoS Comput Biol. 2008;4:e1000233. doi: 10.1371/journal.pcbi.1000233. * Established a complete package for microbial image analysis by implementing automated interpolation methods for high-resolution cell border identification and internal coordinate system construction for intercellular comparisons.
- 45.Keymer JE, Endres RG, Skoge M, Meir Y, Wingreen NS. Chemosensing in Escherichia coli: two regimes of two-state receptors. Proc Natl Acad Sci U S A. 2006;103:1786–1791. doi: 10.1073/pnas.0507438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang KC, Wen BN, Gitai Z, Wingreen NS. Organization of the cell wall of Gram-negative bacteria. Proc Natl Acad Sci U S A. 2008;105:19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lan G, Daniels BR, Dobrowsky TM, Wirtz D, Sun SX. Condensation of FtsZ filaments can drive bacterial cell division. Proc Natl Acad Sci U S A. 2009;106:121–126. doi: 10.1073/pnas.0807963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bakal C, Aach J, Church G, Perrimon N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 2007;316:1753–1756. doi: 10.1126/science.1140324. ** Elegant use of clustering quantitative image data for systems-level analysis of cell shape determination pathways.
- 49.Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z. Two independent spiral structures control cell shape in Caulobacter. Proc Natl Acad Sci U S A. 2005;102:18608–18613. doi: 10.1073/pnas.0507708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pincus Z, Theriot JA. Comparison of quantitative methods for cell-shape analysis. J Microsc. 2007;227:140–156. doi: 10.1111/j.1365-2818.2007.01799.x. [DOI] [PubMed] [Google Scholar]