Abstract
In this study we targeted Olig2, a basic helix-loop-helix transcription factor that plays an important role in motoneuron and oligodendrocyte development, in human embryonic stem cell (hESC) line BG01 by homologous recombination. One allele of Olig2 locus was replaced by a GFP cassette with a targeting efficiency of 5.7%. Targeted clone R-Olig2 (like the other clones) retained pluripotency, a typical hESC morphology and a normal parental karyotype 46, XY. Most importantly, GFP expression recapitulated endogenous Olig2 expression when R-Olig2 was induced by sonic hedgehog and retinoic acid, and GFP+ cells could be purified by fluorescence-activated cell sorting (FACS). Consistent with previous reports on rodents, early GFP-expressing cells appeared biased to a neuronal fate whereas late GFP-expressing cells appeared biased to an oligodendrocytic fate. This was corroborated by myoblast coculture, transplantation into the rat spinal cords and whole genome expression profiling. The present work reports an hESC reporter line generated by homologous recombination targeting a neural lineage specific gene, which can be differentiated and sorted to obtain pure neural progenitor populations.
Keywords: Gene targeting, neurogenesis, gliogenesis, bHLH transcription factor, Olig2, ESC
INTRODUCTION
Early developmental events have been difficult to study in humans and much of our understanding of neuron and glial cell differentiation has come from studies of mice and rats. Human embryonic stem cells (hESCs) [1], capable of self-renewal and differentiating into cells of all three germ layers, are ideal to model human development and diseases, especially when combined with gene targeting technology [2]. Although homologous recombination has been achieved in hESCs for several genes including Oct4, Hprt1, Mixl1, the human Rosa locus, Fezf2 and several unspecified loci [3–10], the lack of hESC knockin reporters in the neural lineage prompted us to generate a neuroglial reporter line using gene targeting.
Olig2 is a basic helix loop helix (bHLH) transcription factor and an important fate regulator in neurogenesis and gliogenesis in the central nervous system (CNS) [11–17]. Double knockout of Olig2 and another Olig family member Olig1, results in ablation of oligodendrocytes and motoneurons in mice [12, 15]. Reporter genes knocked in at Olig loci in mice provided valuable lineage tracing data in vivo [12, 15, 18, 19] and in vitro [20, 21].
In the present study, we have generated a cell line designated R-Olig2 in which an enhanced GFP (EGFP) cassette was inserted to the Olig2 locus of hESC line BG01 via homologous recombination. R-Olig2 remained pluripotent, had a normal karyotype, and allowed for direct visualization of Olig2 expression by fluorescence microscopy and purification of Olig2+ precursors by fluorescence-activated cell sorting (FACS). This work provides a powerful tool to study motoneuron and oligodendrocyte lineage development in human, which will also facilitate research on cell based therapy for the treatment of several neurological conditions, including spinal cord injury, multiple sclerosis and amyotrophic lateral sclerosis.
MATERIALS AND METHODS
Construction of Olig2-EGFP knockin vector
A human BAC clone containing the Olig2 gene was purchased from Invitrogen (Clone No. RP11-585D4) and verified by PCR amplification of the Olig2 gene. The targeting vector was constructed in DH5α using red recombination as described [22]. The translational start codon of Olig2 was designated as the +1 position and was used throughout to describe the Olig2 gene. To generate the targeting construct, pStartK was used as the template to amplify the fragment outside attL1 and attL2. The primers contained two overhangs that were homologous to the flanking sequence of Olig2, so that when this PCR product was transformed into the ET competent Olig2 BAC, full-length Olig2 gene and ~2kb of its upstream and ~5.3kb of its downstream sequences were pulled out into pStartK as selected by kanamycin. The resultant plasmid was named pStartKhOlig2 and its sequence was verified by limited sequencing. Then, a fragment containing a total of 100 bp homology arm of Olig2 exon 2 (−19 to +30 and +976 to +1025, respectively), two AscI sites and cat, the gene encoding chloramphenicol was amplified and co-transfected with pStartK-hOlig2 into ET competent DH5α, seeking to replace the Olig2 exon 2 by cat, and the resultant plasmid was selected by chloramphenicol and ampicillin. The positive plasmid was verified by partial sequencing and named pStartKhOlig2cam. pStartK-hOlig2cam was then digested by AscI and an EGFP-neomycin fragment was purified and ligated into pStartK-hOlig2cam, resulting in pStartKhOlig2eGFP, in which cat was swapped with a sequence encoding EGFP and neomycin (expression of neomycin was driven by RNA Pol II promoter). In order to add a negative selection site to this vector, pStartK-hOlig2eGFP was incubated with a multisite gateway plasmid which contained attR1 and attR2 sites and Tk2, a thymidine kinase gene. After incubation with clonase (Invitrogen), the hOlig2eGFP fragment was exchanged via LR recombination and was ligated with the Tk2 gene. The final construct was selected with ampicillin and named pWSTK3_hOlig2eGFP. When delivered into hESCs, only homologous recombinants would have Tk2 gene excised and survive under negative selection with 2′-Deoxy-2′-fluoro-β-D-arabinofuranosyl-5-iodouracil (FIAU). To identify homologous recombinants, genomic DNA of clones obtained from both positive and negative selection (see below) were examined by Southern blot analysis as described previously [22] using a 533 bp 5′ flanking probe (sequence available upon request).
Generation of the Olig2-GFP knockin reporter line R-Olig2 from BG01
The BG01 hESC line (46, XY) was maintained as described [23]. Briefly, BG01 cells were cultured on a layer of mitomycin C (Sigma) inactivated mouse embryonic fibroblast cells (MEF) in hESC medium containing DMEM-F12, 20% knockout serum replacement, 1% non-essential amino acid, 55 μM 2-mercaptoethanol, 2 mM Lglutamine, supplemented with 4 ng/ml basic FGF (all above from Invitrogen). Cells were passaged using collagenase IV (1 mg/ml, Invitrogen) at a ratio of 1:2 to 4 every 4–5 days. Routine karyotyping examination was done every 10 passages.
To generate the Olig2-GFP knockin reporter R-Olig2, a total of 5×106 to 1×107 BG01 cells were dissociated using accutase (Sigma) and incubated with 30 μg of linearized pWSTK3_hOlig2eGFP. The mixture of DNA and cells was then transferred to a 4 mm cuvette and electroporated using a Bio-Rad Xcell Total system for a single pulse of 250V, 250 μF. Electroporated cells were plated onto MEF layers for recovery. Seventy two hours post-transfection, G418 (50 μg/ml, Invitrogen) and FIAU (125 nM, Maravek Biochmicals) were added to medium everyday. Resistant clones were picked after 21 days of double selection and plated on MEF feeder layers for further expansion. A total of 106 clones were obtained from which genomic DNA was extracted. HindIII-digested genomic DNA from each clone was hybridized to a probe derived from 5′ of the homology arm by Southern blot analysis. Positive clones of Olig2-GFP knockin hESCs were dissociated by TrypLE (Invitrogen) into single cells and transiently transfected using a Cre construct to remove the floxed neo cassette. Single cell clones were isolated and manually picked. Genomic DNA of these clones were examined by PCR to demonstrate the removal of neo cassette and absence of integration of the Cre construct.
In vitro differentiation of R-Olig2
For random differentiation [23], embryoid bodies (EBs) were generated by triturating R-Olig2 ES cells into small clumps and grown in hESC medium (without bFGF) in suspension for 4 days. EBs were then plated onto Geltrex (Invitrogen) coated dishes and continued to differentiate as adherent cultures for an additional 17 days before immunocytochemistry analysis for germ layer specific markers.
Differentiation of R-Olig2 into Olig2+/GFP+ cells
To induce GFP (Olig2) expression, R-Olig2 cells were maintained in hESC medium and detached using collagenase IV. Cells were triturated to make small clumps, which were then transferred to Ultralow attachment dishes (Corning) to form spheres. Spheres were cultured in suspension for 5 days in MDFK5 medium [24, 25] (DMEM-F12 with glutamax, 5% knockout serum replacement, 1x N2, 1% Non-essential amino acid, 55 μM 2-mercaptoethanol, all from Invitrogen). Spheres were further induced to express Olig2-GFP by adding 10 μM all-trans-retinoic acid (Sigma) and 30 ng/ml recombinant sonic hedgehog (Shh, R&D systems) or 1 μM Shh agonist Purmorphamine [26] (CalBiochem) everyday. Spheres generated from these R-Olig2 cells were then examined under fluorescence microscopy for the expression of GFP and subsequent co-labeling of Olig2 by performing immunocytochemistry with an Olig2 antibody. At various time points of differentiation, spheres were harvested for flow cytometric analysis to evaluate the expression of GFP or were fixed with 2% paraformaldehyde and sectioned for direct visualization of GFP and immunocytochemistry of other markers.
To further induce differentiation along the motoneuron or oligodendrocyte lineage, GFP+ spheres were manually picked under fluorescence microscopy. For motoneuron differentiation, GFP+ spheres were seeded onto C2C12 myoblasts (ATCC) and cultured for an additional 5–10 days in MDFK5 medium supplemented with 1xB27, brain-derived neurotrophic factor (BDNF, 10 ng/ml), glial cell line derived neurotrophic factor (GDNF, 10 ng/ml), insulin like growth factor 1 (IGF1, 10 ng/ml), and retinoic acid (1 μM). For oligodendrocyte precursor and oligodendrocyte differentiation, GFP+ spheres were seeded onto fibronectin coated plates and induced in MDFK5 medium supplemented with platelet derived growth factor (PDGF-AA, 10 ng/ml), 3,3,5 triiodothyronine (T3, 30 ng/ml) and Purmorphamine (1 μM) for 5–10 days.
Purification of Olig2+/GFP+ cells by fluorescence activated cell sorting (FACS)
Purification of Olig2+/GFP+ cells was carried out as described previously [27]. Briefly, differentiated R-Olig2 cells were harvested using 0.25% Trypsin and resuspended in 5% FBS in PBS at a density of 5×106 to 1×107 per milliliter. Cell purification was performed using a FACSstarPlus cell sorter (Becton Dickinson Immunocytometry Systems) at 4°C at a rate of 2,500 cells/s. The positively sorted cells were re-examined by FACS and found to have a purity of 91%–95%.
Transplantation of GFP+ cells to adult rat spinal cords
Animal experiments were performed with the approval of the Johns Hopkins University IACUC. Sprague–Dawley rats were purchased from Taconic. Transplantation was performed as described previously [28]. Briefly, GFP+ cells at late stage of differentiation (day 38–39) were dissociated and resuspended in fresh culture media at a concentration of 20,000 or 75,000 cells/μl. Adult male Sprague–Dawley rats (60–70 days old) were anesthetized and two grafts of cells (0.4 × 105 or 1.5 × 105 cells per 2 μl injection) were bilaterally implanted into the dorsal column of white matter at segment C5 of the cervical spinal cord. Cells were delivered using a 10-μl Gastight syringe (Hamilton) with an attached 30-gauge 45° beveled needle (Hamilton). The injection pipette was secured to a manual micromanipulator (World Precision Instruments) attached to an 80° tilting base. The tip was lowered to a depth of 1.25 mm below the surface of the cord and was held in place for 2 min before and after cell injection. Cells were delivered under the control of a microsyringe pump controller (World Precision Instruments) at a rate of 0.5 μl/min. Animals received Buprenex and were immunosuppressed with Cyclosporin A (Sandoz Pharmaceuticals).
For post-transplantation analysis, animals were euthanized at 2 or 6 weeks, perfused with 4% paraformaldehyde, and cryoprotected in 30% sucrose–0.1 M phosphate buffer at 4°C for 3 days [28]. Spinal cord fragments centered at the injection site were identified and embedded in O.C.T. (Fisher Scientific). Saggital and transverse sections were cut and processed for direct fluorescence visualization of GFP and immunohistochemical analysis of relevant genes.
Immunocytochemistry
Immunocytochemistry and histology were performed as described [29, 30]. Briefly, hESCs, EBs or tissue sections were fixed using 2% paraformaldehyde and incubated in blocking buffer (5% goat serum, 1% bovine serum albumin, and 0.1% Triton X-100) for 30 min. Cells or sections were then incubated in primary antibodies diluted in blocking buffer at 4°C overnight. For live cell staining (A2B5 and GalC antibodies), primary antibodies were diluted in DMEM and incubated for 30 min without the blocking step. Appropriate secondary antibodies were used for single and double labeling. All secondary antibodies were tested for cross-reactivity and nonspecific immunoreactivity. The following primary antibodies were used: A2B5 (1:20, ATCC), α-fetoprotein (AFP) (1:500, A8452; Sigma), β-III tubulin (1:2000, T8660; Sigma), CNP (1:200, Sigma), GalC (1:5, kind gift of Dr. Rancht), HB9 or MNR2 (1:50, Developmental Studies Hybridoma Bank, DSHB), human nuclei (hNA, 1:200, Chemicon), Nestin (1:500, BD bioscience), NG2 (1:500, Chemicon), Nkx2.2 (1:1, DSHB), Oct4 (1:500, Abcam), Olig2 (1:8000, kind gift of Dr. Rowitch), Pax6 (1:50, DSHB), PDGFRα (1:200, BD), PLP (1:100, Chemicon), SMA (1:200, Sigma), SSEA4 (1:500, Invitrogen), Tra1–60 (1:100, Chemicon). α-Bungarotoxin (BTX) (Alexa Fluor 594 conjugated, 1:500, Invitrogen) was used to label nicotinic acetylcholine receptors. Bis-benzamide (DAPI, 1:1000; Sigma) was used to identify the nuclei. Images were captured using a Zeiss Axiovision microscope with z-stack split view function. For z stacks, images were taken at 1 μm increments and processed using Axiovision software and AdobePhotoshop. Please note that all images involved GFP were captured directly under fluorescence or confocal microscope without immunostaining using a GFP antibody unless indicated otherwise.
Comparison of global gene expression of early and late GFP+ sorted cells by bead based cDNA microarray
Bead based Illumina microarray was performed as described previously [31]. Briefly, RNA was isolated from undifferentiated R-Olig2 or GFP+ sorted cells using TRIzol (Invitrogen) and 100 ng total RNA was used for amplification and hybridization to Illumina HumanRef-8 BeadChip according to the Manufacturer’s instructions (Illumina). Array raw data were processed using Illumina BeadStudio software. Gene expression levels were considered significant only when their detection p-value ≤0.01. Comparison was made between GFP+ sorted cells of early (day 17 of differentiation) and late stage (day 38 of differentiation), and stage specific genes were identified.
RESULTS
Generation of the Olig2-GFP knockin hESC line R-Olig2
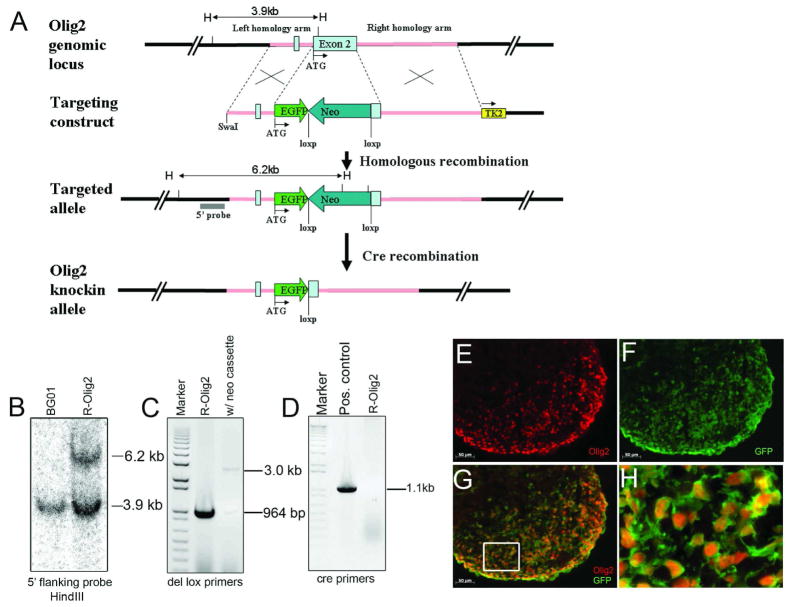
To generate the Olig2-GFP knockin reporter line R-Olig2, we transfected BG01 hESCs with targeting vector pWSTK3_hOlig2eGFP using electroporation (Figure 1). After successful homologous recombination, exon 2 of the Olig2 gene was replaced by EGFP. Among the 106 clones that have been selected through both positive (G418) and negative (2′-Deoxy-2′-fluoro-β-D-arabinofuranosyl-5-iodouracil, FIAU) selection, 6 have been identified to be correctly targeted in one allele while the other allele remained intact as confirmed by Southern blot analysis (Figure 1). The efficiency was 5.7% (6/106). In order to eliminate possible interference on GFP expression, the floxed neo cassette was removed by transient transfection of a supercoiled Cre construct. The removal of neo cassette and the absence of genomic integration of the Cre fragment were confirmed by PCR (Figure 1C, D).
Figure 1.
Successful gene targeting of GFP cassette into the Olig2 locus in the hESC line BG01. Homologous recombination was performed using a GFP vector that targets the Olig2 gene in BG01 (A). Pink line represents the homology arms and two light blue boxes represent the two exons of Olig2. EGFP cassette is shown in green. Neo (dark blue box) and Tk2 (yellow box) cassettes are designed for positive and negative selection respectively. Restriction enzyme HindIII (abbreviated as “H” in panel A) was used to digest the genomic DNA of hESC clones for Southern blot analysis. Six clones were identified to be correctly targeted to the Olig2 locus by Southern blot using a 5′ flanking probe. R-Olig2, the representative clone is shown with a 6.2 kb band of targeted allele and a 3.9kb band for the wild type allele, as predicted (B). Floxed neo cassette was further removed by transient transfection of a Cre construct (C) and displayed absence of genomic integration of Cre fragment by PCR (D). To examine GFP expression, R-Olig2 cells were induced by Shh agonist and retinoic acid for 16 days before differentiated spheres were fixed, sectioned and stained with Olig2 antibody (E). All cells that were labeled with Olig2 antibody co-expressed GFP (F, G), which was directly visualized under fluorescence microscope. Panel H shows higher magnification of the boxed area in G. Bar, 50 μm.
R-Olig2 cells are pluripotent
To further evaluate the basic hESC characteristics of R-Olig2, we examined pluripotency markers in R-Olig2 using BG01 as a wild type control. Identical to BG01, R-Olig2 cells were grown on a layer of inactivated MEF or in MEF conditioned medium and were passaged every 4–5 days at a ratio of 1:2 to 4. As shown in Supplementary Figure 1, R-Olig2 uniformly expressed ES cell markers Oct4, SSEA4, and Tra1–60. When differentiated, R-Olig2 gave rise to cells of all three germ layers, including ectodermal cells expressing nestin and β-III tubulin, mesodermal cells expressing smooth muscle actin (SMA) and endodermal cells expressing α-fetoprotein (AFP). R-Olig2 maintained a normal karyotype as the parental line BG01. Gene expression profiles of ROlig2 and BG01 were also indistinguishable. These data combined indicated that R-Olig2 shared all basic properties of a normal hESC line and can be used for further differentiation experiments.
Induction of GFP expression in R-Olig2 and purification of GFP+ cells by FACS
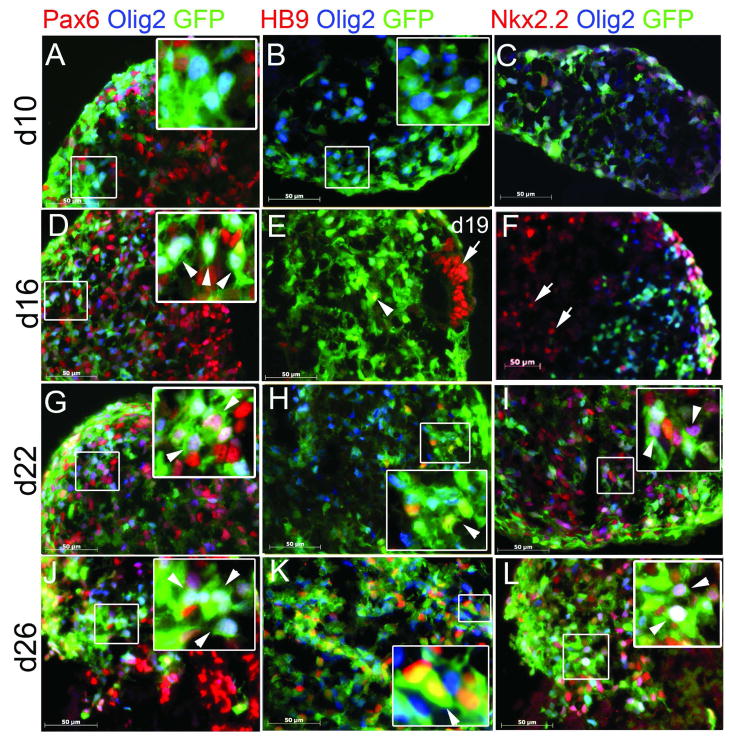
One major goal of this work was to generate a tool which could be used to purify Olig2+ precursors. We therefore differentiated R-Olig2 into Olig2+/GFP+ cells using MDFK5 medium supplemented with sonic hedgehog (Shh) or Shh agonist Purmorphamine and retinoic acid, in suspension culture as spheres [20, 24, 26]. Spheres were then harvested and sectioned, and GFP expression examined using direct fluorescence microscopy. Almost every Olig2-expressing cell, as identified by immunocytochemistry using an Olig2 antibody, expressed GFP (Figure 1E–H), indicating that GFP recapitulated Olig2 expression. Expression of several transcription factors of the neural lineage was also examined along the time course (Figure 2 and Supplementary Figures 2–4). The neural stem cell (NSC) marker Pax6 was first detected at d10 of induction (Figure 2A) and at this time Pax6 did not co-label with Olig2 or GFP. Starting at d16, a subset of Pax6+ cells expressed Olig2 and GFP, indicating early specification of these NSCs (Figure 2D, G, J). Motoneuron lineage marker HB9 was first evident at d19, when HB9 and Olig2 were expressed in different cells. However, a few HB9+ cells coexpressed GFP (but not Olig2), indicating that GFP could serve as a short term tracer for the differentiation process of Olig2+ precursors to mature motoneurons (Figure 2E, H, K), likely due to GFP’s longer half life [32]. Consistent with data obtained in rodents, homeodomain transcription factor Nkx2.2 did not co-label with Olig2 or GFP at very early stages of differentiation (Figure 2C, F), and the initiation of co-expression of Nkx2.2 and Olig2 or GFP indicated a commitment of the cells toward the oligodendrocytic lineage (Figure 2L).
Figure 2.
GFP expression reflects endogenous Olig2 expression along the neural differentiation pathway. R-Olig2 cells were induced with Shh agonist and retinoic acid and the expression of transcription factors Olig2, Pax6 (A, D, G, J), HB9 (B, E, H, K) and Nkx2.2 (C, F, I, L) was examined by immunocytochemistry along with GFP expression (direct visualization under fluorescence microscope) at d10, d16, d22 and d26 of differentiation. Pax6 was first detected at d10 (red, A) and starting at d16, a subset of Pax6+ cells co-expressed Olig2 and GFP (arrowheads, D, G, J). Although Olig2 and GFP expression was detected as early as d10, almost no expression was detected for motoneuron marker HB9 (B), until d19 of differentiation (arrow, E), when the majority of cells did not co-express HB9 and Olig2. Some HB9+ cells co-expressed GFP but not Olig2 (arrowheads, E, H, K). Nkx2.2 did not co-label with Olig2 or GFP at the early stages of differentiation (arrow, C, F), and the initiation of co-expression of Nkx2.2 and Olig2 and GFP could be observed starting from d22 (arrowheads, I, L). Insets show higher magnification images of boxed areas in corresponding panels. Detailed time course images are shown in Supplementary Figures 2–4. Bar, 50 μm.
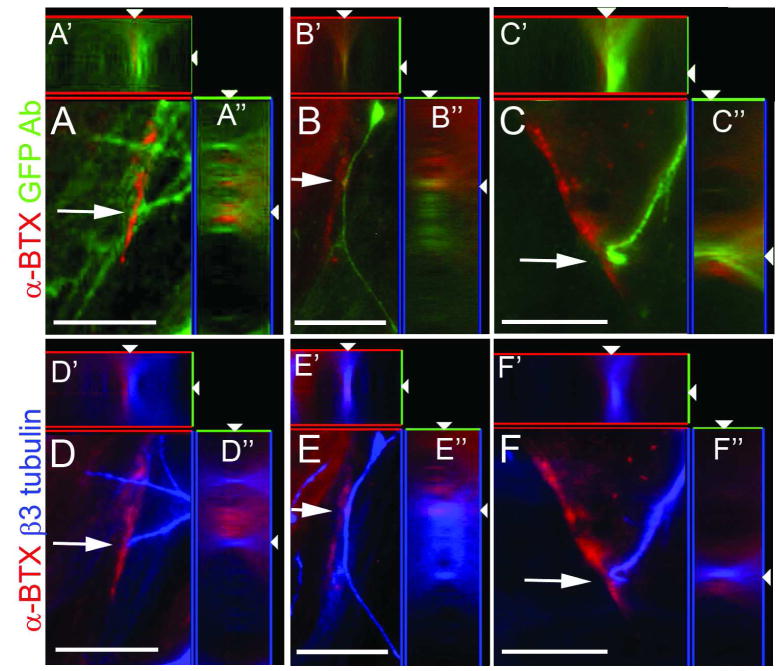
The Olig2-GFP knockin mouse line showed stage-dependent differentiation potential in culture [33]. Bearing this and the in vivo developmental data [34, 35] in mind, we set out to assess the properties of R-Olig2 along the differentiation pathway. When GFP+ cells harvested from early stages (d16-d21) of R-Olig2 differentiation were co-cultured with C2C12 myoblasts for 5–10 days in MDFK5 medium supplemented with BDNF, GDNF, IGF1 and retinoic acid, aggregations of nicotinic acetylcholine receptors on the co-cultured myotubes could be detected by the positive staining of an α-Bungarotoxin (α-BTX) antibody. Immediately adjacent to these innervated myotubes were neuronal processes delineated by GFP and βIII tubulin expression (Figure 3), indicating that the early GFP+ spheres differentiated into motoneurons. When GFP+ cells harvested from late stages (d35-d42) of the induction culture were differentiated under the same condition, however, few or no aggregations of acetylcholine receptors were found (data not shown), indicating that late stage GFP+ cells did not have the potential to give rise to motoneurons.
Figure 3.
Early GFP-expressing cells are likely motoneuron precursors which have the ability to make synapses. Twenty-one-day differentiated GFP+ spheres were harvested and co-cultured with C2C12 myoblasts for 5–10 days. GFP+ cells were able to cause acetylcholine receptor aggregation, as detected by staining using an antibody against α-BTX (arrows, panels A and D, B and E, and C and D show aggregations of three representative fields at the neuromuscular junction). Although cell bodies of the motoneurons were still weakly visible under green fluorescence microscopy, GFP expression in most neuron processes was downregulated as cells matured. Antibodies of GFP (A–C) and βIII tubulin (blue, D–F) were used to delineate the cell bodies and processes of these GFP+ cell-derived motoneurons. Images of z stacks were taken at 1 μm increments and A′–F′ and A″–F″ are xz and yz views processed by Zeiss Axiovision software. GFP Ab: GFP antibody. Bar, 25 μm.
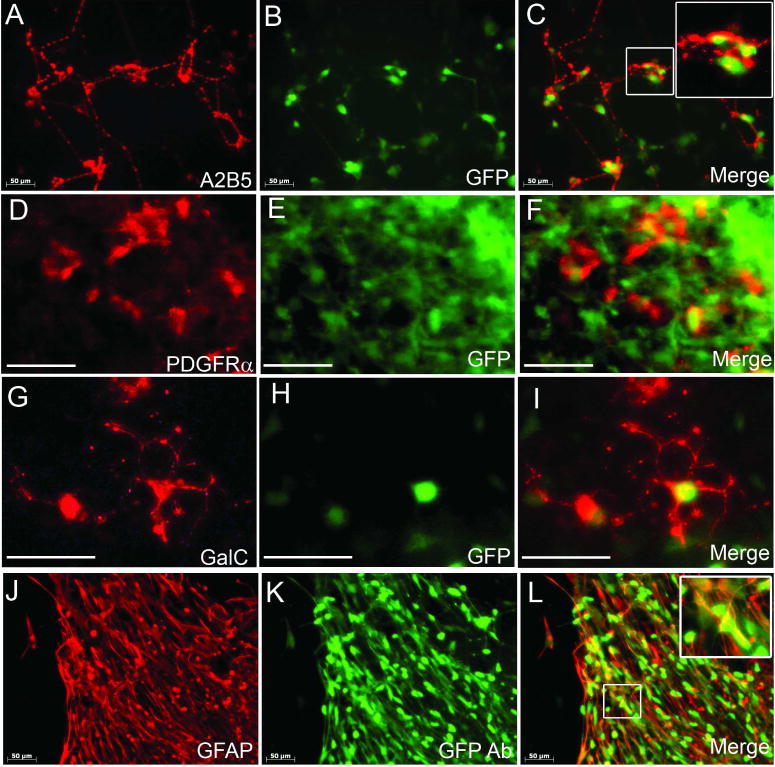
The late stage GFP+ cells were then assessed for their differentiation capacity into the glial cell lineage. After 5–10 days of induction in platelet derived growth factor (PDGFAA), 3,3,5 triiodothyronine (T3) and Shh agonist, the differentiated cells expressed glial and oligodendrocytic markers A2B5 and PDGFRα (Figure 4A–F). When further differentiated, these cells matured into GalC+ oligodendrocytes and GFAP+ astrocytes. Olig2 expression level was maintained in oligodendrocytes (Figure 4G–I) and was downregulated below the detection level of immunocytochemistry in astrocytes, yet weak residual GFP expression remained (Supplementary Figure 5) and was apparent when stained with GFP antibody (Figure 4K, L). These results combined indicated that early and late stage GFP+ cells might have different differentiation potential.
Figure 4.
Late stage GFP+ cells are likely glial precursors and give rise to oligodendrocytes and astrocytes. Thirty-five-day differentiated GFP+ spheres were harvested and seeded on fibronectin and further induced by PDGFAA, T3 and Shh agonist for 5 days. Cells expressed GFP (B, E, direct fluorescence) and glial progenitor markers A2B5 (A) and PDGFRα (D). Merged images (C, F) show co-expression. When further induced, GFP+ cells (H, direct fluorescence) gave rise to GalC+ oligodendrocytes (G-I) and GFAP+ astrocytes (J-L). While a small subpopulation of GFAP+ cells still expressed GFP, which were visualized directly by fluorescence microscopy (Supplementary Figure 5), in most GFAP+ astrocytes, GFP expression was revealed by antibody staining (K, L). GFP Ab: GFP antibody. Insets in C and L show higher magnification images of the boxed area. Bar, 50 μm.
Distinct gene expression profile for early and late GFP+ cells
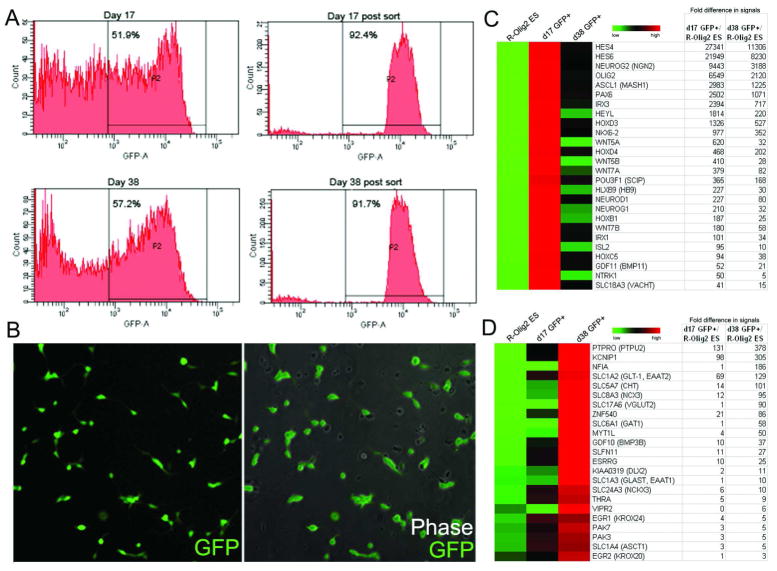
To further characterize the early and late stage GFP+ cells, we harvested differentiated spheres at both early (day 17) and late (day 38) stages, and sorted for GFP+ cells (Figure 5A, B) for gene expression analysis using Illumina bead array. As expected, Olig2 was highly expressed in both populations as compared to undifferentiated R-Olig2 ES cells (Figure 5C, D, 6549 and 2120 folds, respectively). Early stage GFP+ sorted cells differentially expressed several Hox proteins, indicating the initiation of the caudalization process. Genes that are important in motoneuron specification and maturation were also highly expressed [36], including HB9, Isl2, Mash1, GDF11, SCIP, and NeuroD (Figure 5C). By contrast, genes that are involved in the early commitment of glia and oligodendrocyte differentiation [37, 38] were upregulated in late stage sorted GFP+ cells, among them PTPU2, Dlx2, GLT-1 (EAAT2), GLAST (EAAT1), GDF10, THRA (thyroid hormone receptor α), and Nkx6.2. Some myelination associated genes such as MYT1L were also upregulated (Figure 5D).
Figure 5.
Early and late stage GFP+ cells show distinct gene expression profile. GFP+ cells derived from R-Olig2 at early (day 17) and late (day 38) stages were purified by FACS (A) for gene expression profile analysis. Cells maintained GFP expression after sorting (B, direct fluorescence) and showed distinct global gene expression profiles. When compared to undifferentiated R-Olig2 ES cells, GFP+ cells from early stage showed high expression of genes of the motoneuron lineage (C), while those from a later stage showed an upregulation of genes of the oligodendrocyte lineage (D). In the heatmaps, high expression levels relative to mean values (average signals of ES, d17 and d38 of the corresponding genes) are colored red. Low expression levels are colored green. Black represents no significant change in the expression level between mean and sample. Fold increase of gene expression level of d17 and d38 GFP+ cells compared to undifferentiated R-Olig2 ES cells is listed in the tables in C and D.
GFP+ cells gave rise to oligodendrocytes in vivo after transplantation
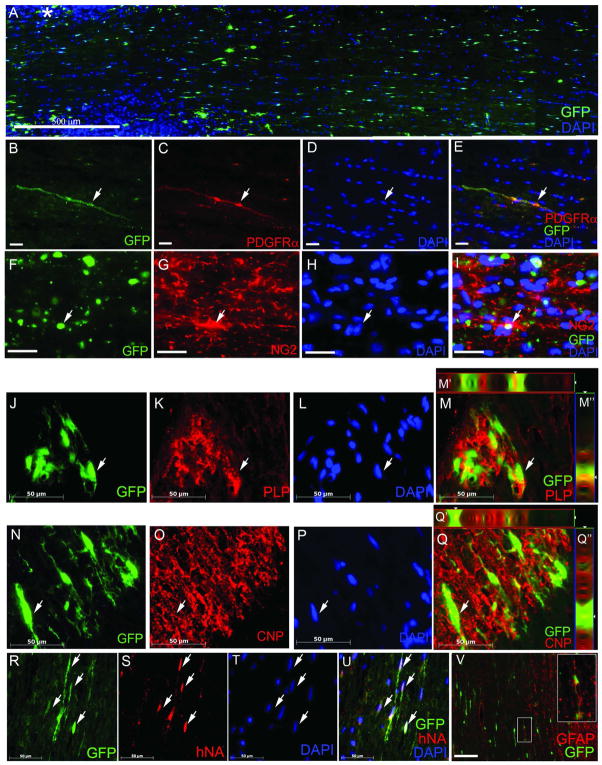
To evaluate R-Olig2 derived GFP+ cells in vivo, we transplanted day 38 differentiated GFP+ cells into the dorsal column white matter of the spinal cord (cervical segment C5) of adult male Sprague-Dawley rats. The engrafted cells were identified by GFP expression (direct fluorescence) and by immunostaining using an antibody against the human nuclei (hNA, Figure 6R–U). Two weeks posttransplantation, GFP+ cells survived in both white and gray matter regions of the cervical spinal cord and coexpressed PDGFRα and NG2, two oligodendrocyte progenitor makers (Figure 6B–I). By six weeks post engraftment, the majority of GFP+ cells migrated from the initial injection site and robustly integrated into the host spinal cord (Figure 6A). In addition, many GFP+ cells differentiated into oligodendrocytes that produced myelin, as indicted by the coexpression of GFP and proteolipid protein (PLP, Figure 6J-M″) or 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNP, Figure 6N-Q″). A subset of transplanted cells differentiated into GFAP-expressing astrocytes (Figure 6V).
Figure 6.
Engrafted late stage GFP+ cells migrated and differentiated into oligodendrocyte progenitors and oligodendrocytes after transplantation into the rat spinal cords. Engrafted GFP+ cells survived, migrated and integrated into the host white matter regions of the cervical spinal cord robustly as identified by GFP (A) and an antibody against human nuclear antigen (hNA) (arrows, R-U). Specifically, at 2 weeks posttransplantation, engrafted cells co-expressed GFP and oligodendrocyte precursor markers PDGFRα (arrow, B-E) and NG2 (arrow, F-I). By 6 weeks posttransplantation, cells continued to express GFP, migrated further from the injection site (*, A) and started to myelinate the host tissue, as evidenced by co-expression of GFP and PLP (arrow, J-M″), and GFP and CNP (arrow, N-Q″). Some transplanted cells differentiated into astrocytes as detected by GFAP staining (V, inset shows higher magnification of the boxed area). For panels J-Q″, z stack images were taken at 1 μm increments. M′ and Q′ and M″-Q″ are images of xz and yz views processed by Zeiss Axiovision software. GFP was directly visualized under fluorescence microscopy. Bar in A, 500 μm, B–E and J–V, 50 μm, F–I, 20 μm.
DISCUSSION
We reported here the successful generation of R-Olig2, a GFP knockin reporter targeted to one allele of Olig2 gene in hESCs. R-Olig2 retained its normal parental karyotype, was pluripotent, and continued to propagate in culture without showing any changes in proliferation or self-renewal. Upon differentiation, GFP expression faithfully replicated the endogenous Olig2 expression and could be used as a surrogate marker to purify Olig2+ cells, which, depending on what stage they were harvested, further gave rise to motoneurons, oligodendrocyte progenitors and myelinating oligodendrocytes both in vitro and in vivo after transplantation.
The targeting efficiency of Olig2 in BG01 was 5.7%, which is higher than those reported in hESCs by homologous recombination so far (Oct4: 3.9%; Hprt1: 2%, Fezf2: 1.5%, and the human Rosa locus: 2.3%) [3–9]. While the difference could be due to the accessibility of different genomic loci, we believe that the novel method we used to make the targeting construct directly from Olig2-containing human BAC clones played a critical role. Although a non-isogenic DNA vector was used [39], this protocol was more efficient and precise than conventional methods, and provided more flexibility when designing the homology arms for desired genes or gene fragments. We noted that an efficiency of as high as 40% was reported for Oct4 targeting [6], however, this targeting strategy of using endogenous Oct4 promoter to drive the positive selection cassette is only suitable for genes that are highly expressed in the undifferentiated ES cells, and cannot be adapted to lineage-specific genes.
We further found that floxing out the neo cassette in homologous recombinants is essential to ensure appropriate GFP expression. When compared between clones prior to and after the removal of neo cassette, we found that with the presence of neo cassette, GFP expression was very low and could not be used for direct detection by fluorescence microscopy or FACS purification. In mice, elegant systems have been designed to overcome this problem in vivo [40]. In cultured hESCs, a more efficient and preferably a “zero-footprint” system is worth developing although currently this can be done by transiently transfecting a Cre construct (Ref. [3, 4] and the current study) or a cell permeant Cre protein [41].
A useful lineage hESC reporter line needs to meet the following criteria: (1) expression of the reporter recapitulates the targeted endogenous gene, and (2) the transgene does not disrupt or interfere with the function of the gene of interest or any other genes especially the essential genes in the genome. Several genetic engineering strategies have been used to make reporter lines for hESCs, including random integration, BAC transgenesis, and viral vectors [42–44]. These strategies while efficient, are random in nature when integrated to the genome, thus the possibility of bringing undesired effects at molecular or cellular level cannot be completely excluded. Moreover, often times the promoter fragments that drive reporter cassettes are not regulated in the same manner as the endogenous gene and a faithful replication of expression is difficult to achieve [45]. A more reliable and predictable strategy is gene targeting, as reported in this manuscript and by others [3–10] (reviewed in [46, 47]).
In R-Olig2, one copy of Olig2 gene was replaced by an EGFP cassette, while the other allele remained intact. Disruption of one copy of Olig2 gene was shown not to affect characteristics of the Olig2 knockin mESC line [21], and heterozygous Olig2+/− mice did not show any abnormal phenotype either during gestation or after birth [12, 15, 19], In order to ensure that the properties of R-Olig2 was unimpaired relative to the parental line, we conducted a careful parallel comparison of R-Olig2 and BG01 at both pluripotent and differentiated stages. R-Olig2, same as BG01, expressed pluripotency markers at ES stage, and differentiated into motoneurons, oligodendrocyte precursors and myelin-producing oligodendrocytes when appropriately induced in vitro and in vivo after transplantation (Figures 2–6, Supplementary Figures 1–4). Therefore missing one copy of Olig2 gene did not affect R-Olig2’s function as an otherwise normal hESC line. GFP+ cells differentiated and purified from R-Olig2 showed predicted differentiation and gene expression profiles and should reflect the characteristics of the corresponding cell types derived from the non-engineered BG01.
Olig2 has been shown to have a transcription repressor function [11, 48, 49] and is a key gene in promoting oligodendrocyte and suppressing astrocyte differentiation [11– 13, 15, 18, 19]. In mice, Olig2 interacted with several transcription factors, such as Ngn2, Zfp488 and NFIA/NFIB [11, 50, 51]. In humans, Olig2 is expressed in several types of malignant gliomas and is critical for maintaining proliferation of neural stem cells [17, 52], yet detailed mechanism of the Olig2 function remains elusive. The present work provides a powerful tool to study the role of the Olig2 gene. Stage-dependent Olig2+ populations can be purified by GFP+ sorting, and GFP intensity is high enough to serve as a readout to screen for growth factors or small molecule compounds that promote or inhibit the expression of Olig2. Using R-Olig2 as a genetic engineering platform we can further discover key pathways and transcriptional regulatory networks for oligodendrocyte and/or motoneuron proliferation and differentiation in humans. In addition, R-Olig2 may also be valuable for cross species comparisons to elucidate evolutionarily conserved and divergent pathways regulating development and tissue repair [53, 54].
Transplantation studies provide important clues for clinical trials of cell based therapy. However, silencing of randomly integrated ectopic genes after transplantation has been reported, which has inhibited the use of cell labels for in vivo tracking. We reasoned that such silencing would be less likely to occur in a homologous recombination targeted gene. As expected, after being transplanted into the rat spinal cord, GFP expression was maintained in R-Olig2 derived oligodendrocyte precursors and mature oligodendrocytes, and was readily visualized by fluorescence microscopy without the need of any enhancement (such as immunostaining), indicating that R-Olig2 can be reliably used for short-term lineage tracing experiments in vivo or in transplantation studies of animal disease models.
The pattern of GFP expression suggests that Olig2 is expressed in either two lineages or in a single lineage that changes its developmental potential. Only the early Olig2+ precursors have the ability to make motoneurons and we as yet do not know the fate of the very early Olig2-expressing cells. Our current data do not allow us to distinguish between them, we have however, provided a platform for future study of this topic.
CONCLUSION
We present a genetically engineered hESC reporter line generated by homologous recombination targeting Olig2, a gene specific to the neural lineage. This in vitro model system serves as a source of purified lineage specific population derived from engineered hESCs. The targeting efficiency in the current work is high enough for extending this technique to other genes of both neural and non-neural lineages. It is also feasible to apply similar methods to generate knockout, knockin or mutant lines for functional studies in hESCs or induced pluripotent stem cells (iPSCs).
Supplementary Material
Acknowledgments
This work was supported by Robert Packard Center for ALS Research award to YL, National Institute on Aging (NIA, NIH) Intramural Research Program (MSR and MPM), Intramural Research Training Awards to YL, California Institute for Regenerative Medicine Grant CL1-00501-1 and the Larry L Hillblom Foundation to XZ, and Life Technologies Corporation. We thank Dr. Y. Han for technical assistance, and all members of our laboratories for constant stimulating discussions. MSR acknowledges the contributions of Dr. S. Rao that made undertaking this project possible.
Footnotes
Author contributions:
HX: Experimental design, collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
SW: Experimental design, collection and/or assembly of data, data analysis and interpretation, manuscript editing, final approval of manuscript
STP: Experimental design, collection and/or assembly of data, data analysis and interpretation, manuscript editing, final approval of manuscript
SS: Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
AMS: Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
CCM: Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
MPM: Manuscript editing, financial support, final approval of manuscript
NJM: Experimental design, data analysis and interpretation, manuscript editing, financial support, final approval of manuscript
MRC: Conception and design, manuscript editing, financial support, final approval of manuscript
MSR: Conception and design, data analysis and interpretation, manuscript editing, financial support, final approval of manuscript
XZ: Conception and design, data analysis and interpretation, manuscript editing, financial support, final approval of manuscript
YL: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, financial support, final approval of manuscript
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 3.Davis RP, Ng ES, Costa M, et al. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- 4.Ruby KM, Zheng B. Gene Targeting in a HUES Line of Human Embryonic Stem Cells via Electroporation. Stem Cells. 2009 doi: 10.1002/stem.73. [DOI] [PubMed] [Google Scholar]
- 5.Urbach A, Schuldiner M, Benvenisty N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells. 2004;22:635–641. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- 6.Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 7.Irion S, Luche H, Gadue P, et al. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- 8.Lombardo A, Genovese P, Beausejour CM, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki K, Mitsui K, Aizawa E, et al. Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc Natl Acad Sci U S A. 2008;105:13781–13786. doi: 10.1073/pnas.0806976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa M, Dottori M, Sourris K, et al. A method for genetic modification of human embryonic stem cells using electroporation. Nat Protoc. 2007;2:792–796. doi: 10.1038/nprot.2007.105. [DOI] [PubMed] [Google Scholar]
- 11.Deneen B, Ho R, Lukaszewicz A, et al. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Lu QR, Sun T, Zhu Z, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75– 86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 13.Marshall CA, Novitch BG, Goldman JE. Olig2 directs astrocyte and oligodendrocyte formation in postnatal subventricular zone cells. J Neurosci. 2005;25:7289–7298. doi: 10.1523/JNEUROSCI.1924-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takebayashi H, Nabeshima Y, Yoshida S, et al. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 16.Rowitch DH, Lu QR, Kessaris N, et al. An ‘oligarchy’ rules neural development. Trends Neurosci. 2002;25:417–422. doi: 10.1016/s0166-2236(02)02201-4. [DOI] [PubMed] [Google Scholar]
- 17.Ligon KL, Huillard E, Mehta S, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Rao MS. Olig genes are expressed in a heterogeneous population of precursor cells in the developing spinal cord. Glia. 2004;45:67–74. doi: 10.1002/glia.10303. [DOI] [PubMed] [Google Scholar]
- 19.Masahira N, Takebayashi H, Ono K, et al. Olig2-positive progenitors in the embryonic spinal cord give rise not only to motoneurons and oligodendrocytes, but also to a subset of astrocytes and ependymal cells. Dev Biol. 2006;293:358– 369. doi: 10.1016/j.ydbio.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 20.Xian H, Gottlieb DI. Dividing Olig2-expressing progenitor cells derived from ES cells. Glia. 2004;47:88–101. doi: 10.1002/glia.20010. [DOI] [PubMed] [Google Scholar]
- 21.Xian HQ, McNichols E, St Clair A, et al. A subset of ES-cell-derived neural cells marked by gene targeting. Stem Cells. 2003;21:41–49. doi: 10.1634/stemcells.21-1-41. [DOI] [PubMed] [Google Scholar]
- 22.Wu S, Ying G, Wu Q, et al. A protocol for constructing gene targeting vectors: generating knockout mice for the cadherin family and beyond. Nat Protoc. 2008;3:1056–1076. doi: 10.1038/nprot.2008.70. [DOI] [PubMed] [Google Scholar]
- 23.Zeng X, Miura T, Luo Y, et al. Properties of pluripotent human embryonic stem cells BG01 and BG02. Stem Cells. 2004;22:292–312. doi: 10.1634/stemcells.22-3-292. [DOI] [PubMed] [Google Scholar]
- 24.Wichterle H, Lieberam I, Porter JA, et al. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 25.Xian HQ, Werth K, Gottlieb DI. Promoter analysis in ES cell-derived neural cells. Biochem Biophys Res Commun. 2005;327:155–162. doi: 10.1016/j.bbrc.2004.11.149. [DOI] [PubMed] [Google Scholar]
- 26.Li XJ, Hu BY, Jones SA, et al. Directed differentiation of ventral spinal progenitors and motor neurons from human embryonic stem cells by small molecules. Stem Cells. 2008;26:886–893. doi: 10.1634/stemcells.2007-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Han SS, Wu Y, et al. CD44 expression identifies astrocyte-restricted precursor cells. Dev Biol. 2004;276:31–46. doi: 10.1016/j.ydbio.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 28.Lepore AC, Rauck B, Dejea C, et al. Focal transplantation-based astrocyte replacement is neuroprotective in a model of motor neuron disease. Nat Neurosci. 2008;11:1294–1301. doi: 10.1038/nn.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng X, Chen J, Sanchez JF, et al. Stable expression of hrGFP by mouse embryonic stem cells: promoter activity in the undifferentiated state and during dopaminergic neural differentiation. Stem Cells. 2003;21:647–653. doi: 10.1634/stemcells.21-6-647. [DOI] [PubMed] [Google Scholar]
- 30.Cai J, Chen J, Liu Y, et al. Assessing self-renewal and differentiation in human embryonic stem cell lines. Stem Cells. 2006;24:516–530. doi: 10.1634/stemcells.2005-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campanelli JT, Sandrock RW, Wheatley W, et al. Expression profiling of human glial precursors. BMC Dev Biol. 2008;8:102. doi: 10.1186/1471-213X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- 33.Shin S, Xue H, Mattson MP, et al. Stage-dependent Olig2 expression in motor neurons and oligodendrocytes differentiated from embryonic stem cells. Stem Cells Dev. 2007;16:131–141. doi: 10.1089/scd.2006.0023. [DOI] [PubMed] [Google Scholar]
- 34.Wu S, Wu Y, Capecchi MR. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineagerestricted progenitors in vivo. Development. 2006;133:581–590. doi: 10.1242/dev.02236. [DOI] [PubMed] [Google Scholar]
- 35.Mukouyama YS, Deneen B, Lukaszewicz A, et al. Olig2+ neuroepithelial motoneuron progenitors are not multipotent stem cells in vivo. Proc Natl Acad Sci U S A. 2006;103:1551–1556. doi: 10.1073/pnas.0510658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalla Torre di Sanguinetto SA, Dasen JS, Arber S. Transcriptional mechanisms controlling motor neuron diversity and connectivity. Curr Opin Neurobiol. 2008;18:36–43. doi: 10.1016/j.conb.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Pescini Gobert R, Joubert L, Curchod ML, et al. Convergent Functional Genomics of Oligodendrocyte Differentiation Identifies Multiple Autoinhibitory Signaling Circuits. Mol Cell Biol. 2009 doi: 10.1128/MCB.01375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J Mol Neurosci. 2008;35:3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- 39.Sedivy JM, Vogelstein B, Liber HL, et al. Gene Targeting in Human Cells Without Isogenic DNA. Science. 1999;283:9. [Google Scholar]
- 40.Bunting M, Bernstein KE, Greer JM, et al. Targeting genes for self-excision in the germ line. Genes Dev. 1999;13:1524–1528. doi: 10.1101/gad.13.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nolden L, Edenhofer F, Haupt S, et al. Site-specific recombination in human embryonic stem cells induced by cell-permeant Cre recombinase. Nat Methods. 2006;3:461–467. doi: 10.1038/nmeth884. [DOI] [PubMed] [Google Scholar]
- 42.Placantonakis DG, Tomishima MJ, Lafaille F, et al. Bac Transgenesis in Human Es Cells as a Novel Tool to Define the Human Neural Lineage. Stem Cells. 2008 doi: 10.1634/stemcells.2008-0884. [DOI] [PubMed] [Google Scholar]
- 43.Koch P, Siemen H, Biegler A, et al. Transduction of human embryonic stem cells by ecotropic retroviral vectors. Nucleic Acids Res. 2006;34:e120. doi: 10.1093/nar/gkl674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren C, Zhao M, Yang X, et al. Establishment and applications of epstein-barr virus-based episomal vectors in human embryonic stem cells. Stem Cells. 2006;24:1338–1347. doi: 10.1634/stemcells.2005-0338. [DOI] [PubMed] [Google Scholar]
- 45.Liew CG, Draper JS, Walsh J, et al. Transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007 doi: 10.1634/stemcells.2006-0634. [DOI] [PubMed] [Google Scholar]
- 46.Giudice A, Trounson A. Genetic modification of human embryonic stem cells for derivation of target cells. Cell Stem Cell. 2008;2:422–433. doi: 10.1016/j.stem.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Yates F, Daley GQ. Progress and prospects: gene transfer into embryonic stem cells. Gene Ther. 2006;13:1431–1439. doi: 10.1038/sj.gt.3302854. [DOI] [PubMed] [Google Scholar]
- 48.Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]
- 50.Lee SK, Lee B, Ruiz EC, et al. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang SZ, Dulin J, Wu H, et al. An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. Development. 2006;133:3389–3398. doi: 10.1242/dev.02522. [DOI] [PubMed] [Google Scholar]
- 52.Lu QR, Park JK, Noll E, et al. Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. Proc Natl Acad Sci U S A. 2001;98:10851–10856. doi: 10.1073/pnas.181340798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ginis I, Luo Y, Miura T, et al. Differences between human and mouse embryonic stem cells. Dev Biol. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 54.Sun Y, Li H, Liu Y, et al. Cross-species transcriptional profiles establish a functional portrait of embryonic stem cells. Genomics. 2007;89:22–35. doi: 10.1016/j.ygeno.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.