Abstract
Rab GTPases are essential regulators of membrane trafficking. We report crystal structures of Rab28 in the active (GppNHp-bound) and inactive (GDP-3′P-bound) forms at 1.5Å and 1.1Å resolution. Rab28 is a distant member of the Rab family. While the overall fold of Rab28 resembles that of other Rab GTPases, it undergoes a larger nucleotide-dependent conformational change than other members of this family. Added flexibility resulting from a double-glycine motif at the beginning of switch 2 might partially account for this observation. The double-glycine motif, which is conserved in the Arf family, only occurs in Rab28 and Rab7B of the Rab family, and may have a profound effect on their catalytic activities.
Keywords: Crystal structures, GDP-3′P, GTP analog GppNHp, Rab GTPases family
1. Introduction
Human cells contain over 170 Ras-related GTPases that control a vast array of signal transduction pathways [1,2]. Based on sequence and functional similarities, the Ras superfamily has been subdivided into five families: Ras, Rho, Rab, Ran and Arf. Sequence similarity among members of the separate families is generally low, but they all share a common fold and a similar nucleotide-dependent switching mechanism [3]. Thus, GTP binding increases the affinities of Ras GTPases for downstream effectors, whereas GTP hydrolysis and phosphate release produce a conformational change that lowers their affinities for effector proteins. Because Ras proteins typically present low intrinsic GTPase activities and slow nucleotide exchange rates, additional factors modulate their activities in cells, including: a) GTPase-activating proteins (GAPs), which accelerate GTP hydrolysis and promote the formation of the inactive state [4], b) guanine-nucleotide exchange factors (GEFs), which promote the exchange of GDP for GTP resulting in activation [5], and c) guanine nucleotide dissociation inhibitors (GDIs), which prolong the inactive state by inhibiting nucleotide exchange and the association of some GTPases with membranes [4].
With ~70 proteins, Rab GTPases constitute the largest family of the Ras superfamily [1,2,6–8]. There is significant sequence variation within this family (Supplementary Fig. 1). Phylogenetic analysis shows that most Rab proteins segregate into six subfamilies, with proteins in each subfamily tending to have similar cellular localization and/or function [9]. Despite this diversity, Rab GTPses present high sequence conservation within five regions known as Rab-family motifs (RabF1-RabF5) [6]. The functions of approximately half the members of the Rab family have been characterized and found to be generally associated with the regulation of vesicular transport and the trafficking of proteins between intracellular organelles [8,10].
The structures of various members of the Rab family have been determined, including in a number of cases the structures of both the active GTP-bound and inactive GDP-bound forms (Supplementary Table 1). These structures show that nucleotide-dependent conformational changes in the Rab family are generally small and affect primarily the switch region near the nucleotide. A question remaining is what factors determine effector specificity among Rab GTPases? Recent evidence suggests that small sequence and structural differences in the switch and interswitch regions are the primary determinants of effector specificity [11]. Such precise information can only be obtained from high-resolution structures of representative family members in complexes with different nucleotides, regulatory proteins and effectors.
We describe crystal structures of Rab28 with bound GppNHp and GDP-3′P at 1.5Å and 1.1Å resolution. Rab28 is expressed predominantly in testis and brain, and to a lesser extent in heart and skeletal muscles [12]. Since its identification Rab28 was recognized as a distant member of the Rab family [12], with lower sequence identity with other members of the family (19–31%) than typically found among Rab GTPases (>40%). Subsequent phylogenetic analyses consistently placed Rab28 at the periphery of the Rab family [1,2,6]. We find that while the overall fold of Rab28 resembles that of other Rab GTPases, it undergoes a larger nucleotide-dependent conformational change than other members of this family.
2. Materials and methods
2.1. Expression and purification of Rab28
The cDNA encoding for human Rab28 (NP_004240) was purchased from ATCC. The fragment Rab2811–184 (hereafter referred to as Rab28) was amplified by PCR and inserted between the NdeI and EcoRI sites of vector pET-28a(+) (Novagen). The protein was expressed in E. coli strain BL21 Star (DE3) (Invitrogen) and purified at 4°C on a Ni-NTA affinity column (Qiagen). Cleavage of the His-tag was carried out in the column with addition of 0.75 μg/ml thrombin (HTI) in 500 mM NaCl, 20 mM Tris-HCl pH 7.5 and 0.1 mM MgCl2.
2.2. Nucleotide exchange
E. coli-expressed Rab28 contains GDP-3′P bound in the catalytic site, a fact that became apparent only after determination of the structure (see below). To obtain the complex with GppNHp, Rab28 at 2 mg ml−1 was incubated for 24 h at 4°C with 2 mM GppNHp in 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 2 mM 1,4-dithiothreitol (DTT) and 100 unit of calf intestinal alkaline phosphatase (New England Biolabs) per μmol of Rab28.
2.3. Crystallization and data collection
For crystallization, Rab28 was concentrated to ~20 mg ml−1 in 20 mM HEPES pH 7.5, 100 mM NaCl, 2 mM MgCl2, 2 mM DTT and 5% (v/v) glycerol (and 2 mM GppNHp for the complex of GppNHp-Rab28). Crystals of Rab28 with bound GDP-3′P and GppNHp were obtained using the hanging drop vapor diffusion method at 4°C and under identical conditions: 20% (w/v) polyethylene glycol 3350 and 100 mM potassium acetate. Diffraction datasets were collected at the BioCARS beamlines 14-BM-C (GDP-3′P-Rab28) and 14-BM-D (GppNHp-Rab28) of the Advance Photon Source (Argonne, IL). The datasets were indexed and scaled with the program HKL2000 (HKL Research Inc.) (Table 1).
Table 1.
Data collection and refinement statistics
| GppNHp-Rab28 | GDP-3′P-Rab28 | |
|---|---|---|
| Data Collection | ||
| Space group | P41212 | P212121 |
| Unit cell a, b, c (Å) | 79.75, 79.75, 66.98 | 30.51, 56.62, 85.11 |
| Resolution range (Å) | 50.0–1.50 (1.55–1.50) | 50.0–1.10 (1.14–1.10) |
| Data completeness (%) | 99.9 (99.8) | 98.4 (96.8) |
| Data redundancy | 19.1 | 8.4 |
| Rmerge (%) | 8.0 (32.1) | 7.5 (36.7) |
| I/σ | 47.1 (10.0) | 24.7 (4.3) |
| Refinement | ||
| Resolution range (Å) | 50.0–1.50 (1.54–1.50) | 47.0–1.10 (1.13–1.10) |
| Rfactor (%) | 15.1 (12.0) | 15.0 (17.1) |
| Rfree (%) | 18.1 (17.9) | 18.4 (18.0) |
| Rms deviations from ideal | ||
| Bonds (Å) | 0.007 | 0.024 |
| Angles (°) | 1.202 | 2.184 |
| Average B-factor (Å2) | ||
| PDB Code | 3E5H | 2HXS |
Values in parentheses correspond to highest resolution shell.
Rmerge=Σ(|I-<I>|)/Σ (I), where I and <I> are the observed and mean intensities of all observations of a reflection, including its symmetry-related equivalents.
Rfactor=Σhkl ||Fobs|-|Fcalc||/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors of reflection hkl.
Rfree, Rfactor calculated for a randomly selected subset of the reflections (5%) that were omitted during refinement.
2.4. Structure determination
The solution of the structure of GDP-3′P-Rab28 was obtained by molecular replacement with the program AMoRe [13], using the GDP-bound structure of Rab23 [11] as a search model. The structure was refined anisotropically and semi-automatically with the programs ARP/wARP [14] and Refmac [15], and alternating cycles of manual rebuilding with the program Coot [16]. During the final stages of the refinement, it became apparent that GDP-3′P was bound in the catalytic site, and not GDP as expected (discussed below). The refinement of the structure of GDP-3′P-Rab28 converged to an R-factor of 15% (R-free=18.4%) for all the data to 1.1Å resolution (Table 1). The structure of Rab28-GppNHp was obtained by molecular replacement using the structure of GDP-3′P-Rab28 as a search model. After several rounds of refinement with the program PHENIX [17] and manual rebuilding with the program Coot [16], the R-factor converged to 15.1% (R-free=18.1%) for all the data up to 1.5Å resolution.
3. Results and discussion
3.1. Structure of Rab28 with bound GDP-3′P
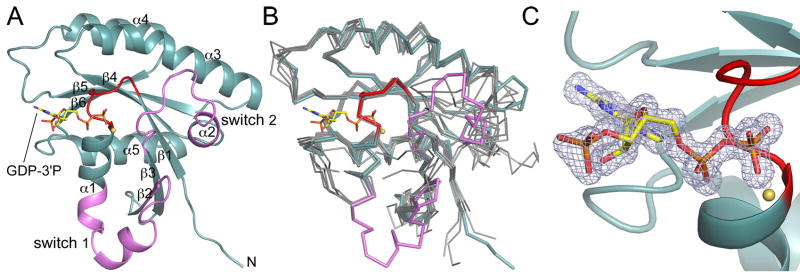
The structure of Rab28 was determined at 1.1Å resolution (Table 1). Like other Ras-related GTPases, Rab28 presents an α/β-fold, consisting of a central six-stranded β-sheet surrounded by five α-helices (Fig. 1A). The β-sheet contains five parallel strands and one antiparallel strand and is twisted, so that it forms an incomplete β-barrel. Two of the α-helices lie along the inside of the incomplete β-barrel, whereas the other three are on the outside. This core structure superimposes well with other Rab GTPases (Fig. 1B). However, the structure revealed two unexpected observations.
Fig. 1.
Structure of Rab28 with bound GDP-3′P. (A) Ribbon representation of the structure of GDP-3′P-Rab28, showing the switch region (defined as the region that changes conformation between the two structures determined here) in pink and the P-loop in red. The β-strands and α-helices are numbered according to their order in the sequence. (B) Superimposition of the structure of GDP-3′P-Rab28 (colored as in part A) with a representative group of inactive structures of Rab GTPases (gray) for which the active structures are also known (m-Rab5C, h-Rab7A, r-Rab7A, h-Rab11A, h-Rab11B, h-Rab21, Sc-Sec4, and Sc-Ypt7). The references and PDB codes of these structures are listed in Supplementary Table 1. (C) Close-up view of the catalytic site, showing a 2Fo-Fc omit electron density map (contoured at 1.4 σ) around the GDP-3′P nucleotide.
First, inspection of Fourier difference maps during the final stages of the refinement revealed a strong electron density peak (~ 10 σ) connected to the O3′ atom the nucleotide. The additional density was consistent with the presence of a molecule of guanosine-3′-monophosphate-5′-diphosphate (GDP-3′P or G3D) in the nucleotide site (Fig. 1C), and not GDP as originally expected. Because of the high resolution of this structure, we were able to refine the individual occupancies of the GDP-3′P atoms, which converged to values near 1, suggesting that the substitution of GDP for GDP-3′P was nearly 100%. GTPases purified from mammalian tissues or bacteria typically present GDP bound in the catalytic site, which could be due to either higher affinity for GDP than GTP [18–21] or nucleotide hydrolysis during purification. The latter possibility is supported by the fact that Rab6A, which is the slowest GTPase of the Rab family, is isolated as a mixture of GTP- and GDP-bound forms [22]. A search of the protein data bank (http://www.rcsb.org/) reveals that in addition to Rab28 three other proteins contain GDP-3′P bond in the catalytic site. Interestingly, all three proteins are GTPases of the Arf family [23–25] (Supplementary Table 2). A common trait of these structures is their high resolutions, which raises the possibility that the modified nucleotide might have been overlooked in some lower resolution structures. GDP-3′P is produced in E. coli during the stringent response [26], and no biological significance has yet been ascribed to its presence in the inactive structures of certain GTPases. From the viewpoint of protein-nucleotide contacts, GDP-3′P appears indistinguishable from GDP. This is because in the current structure, as well as those of the three Arf GTPases where it has been observed, the phosphate group on the O3′ atom of the nucleotide is solvent-exposed and does not make direct contacts with protein atoms (Fig. 1C). Therefore, it appears reasonable to suggest that the structure of Rab28 with bound GDP-3′P corresponds to that of its inactive GDP-bound state.
The second unexpected observation resulting from the structure of GDP-3′P-Rab28 is an unusually large conformational change in the switch region as compared to the inactive structures of other Rab GTPases (Fig 1B). To better understand the meaning and magnitude of this conformational change, we set out to study the structure of the active state of Rab28.
3.2. Structure of Rab28 with bound GppNHp
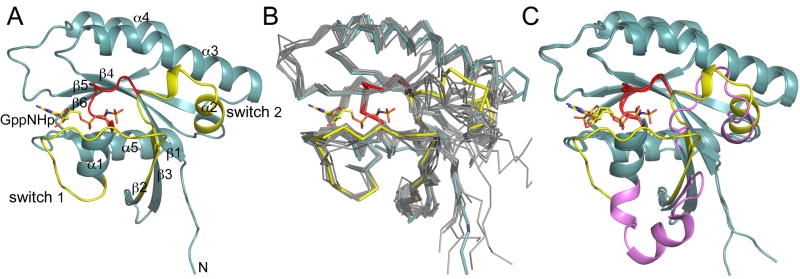
The GDP3′P-bound nucleotide was exchanged for the non-hydrolyzable GTP analog GppNHp (Materials and methods). GppNHp-Rab28 was crystallized under conditions identical to those used for GDP-3′P-Rab28, allowing for a direct comparison of the structures. The structure of GppNHp-Rab28 (Fig. 2A) is similar to those of other GTP-bound Rab GTPases (Fig. 2B). However, compared with the GDP-3′P-bound structure (Fig. 2C), switch 1 from Ala33 to Leu51 adopts a thoroughly different conformation, with some regions of the loop moving ~25Å (Supplementary Movie 1). Residues Gly38-Gly46 in the middle of switch 1, present an extended conformation in the active state, but form two helical turns in the inactive structure. In switch 2 the conformational change affects residues Asp68-Tyr82. Within this region, residues Gln72-Gly75 form a helical turn in the active structure, whereas in the inactive structure their conformation is extended. Differences also occur in the P-loop (Gly21-Lys25) that surrounds the phosphate groups of the nucleotide, but these differences are nearly identical for any active/inactive GTPase structure pair. Among the existing structures of Rab GTPases (Supplementary Table 1) only Saccharomyces sereviceae Sec4 [27] displays a conformational change of a comparable magnitude (albeit different). However, in this case the switch region is involved in interactions with the GEF domain of Sec2 and there is either phosphate (2EQB) or no nucleotide (2OCY) bound in the catalytic site.
Fig. 2.
Structure of Rab28 with bound GppNHp. (A) Ribbon representation of the structure of GppNHp-Rab28 showing the switch region in yellow and the P-loop in red. (B) Superimposition of the structure of GppNHp-Rab28 (colored as in part A) with the active structures of the same group of Rab GTPases shown in figure 1B. (C) Superimposition of the structure of Rab28 with bound GppNHp and GDP-3′P. The switch region was colored according to figures 1A and 2A.
3.3. What causes the conformational change in Rab28 and what are its functional implications?
Although the two nucleotide states of Rab28 crystallize under identical conditions, crystal symmetry and contacts are different in the two structures (Table 1). In particular, switch 1 is involved in crystal contacts in the GDP-3′P-bound structure (Supplementary Fig. 2), but not in the GppNHp-bound structure. Switch 1 is flexible, especially in the GDP state, and its structure is not always resolved in GTPase crystal structures. It is therefore possible that crystal contacts help stabilize the conformation of switch 1 in the inactive structure of Rab28. However, there is sufficient room around switch 1 in the GppNHp-bound structure that a conformational change of the magnitude typically observed in other Rab GTPases could have been accommodated within the same space group. Indeed, some of the evidence suggests that the switch region of Rab28 is particularly flexible compared to other members of the Rab family.
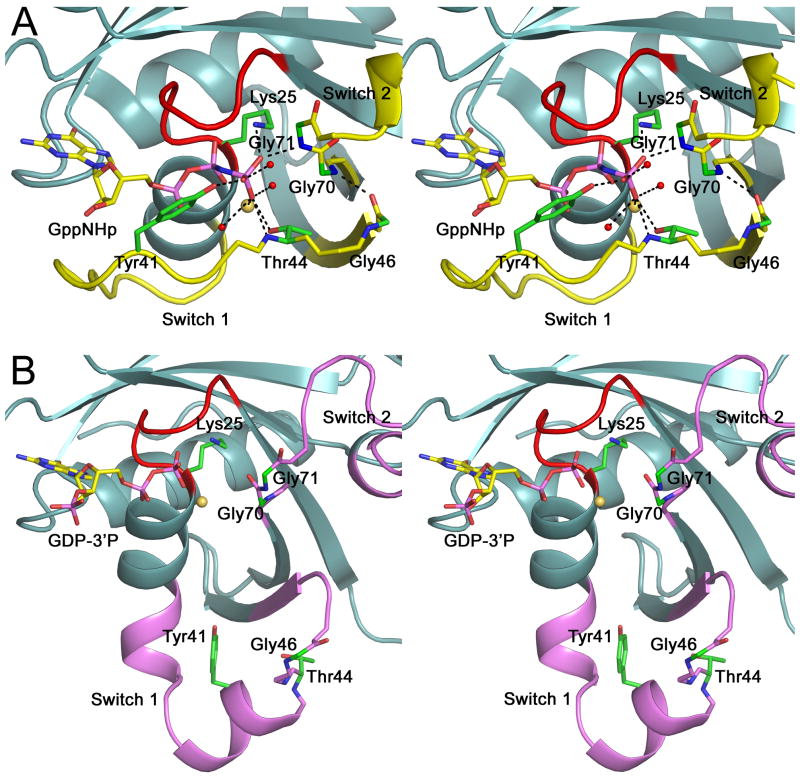
Thus, the sequence motif WDTAGQE is more than 90% conserved among Rab GTPases (Supplementary Fig. 1). This sequence encompasses the C-terminal end of β-strand-3 and the N-terminal portion of switch 2 (Fig 3). Structurally, this sequence is extremely important because it contacts the γ-phosphate of the nucleotide and ‘senses’ its departure by changing conformation in the inactive state, which causes the conformational change in switch 2. Interestingly, this sequence also influences the conformation of adjacent β-strand-2 and, as a result, that of switch 1. Indeed, switch 1 ends with the conserved motif TiGxdF (where uppercase and lowercase letters indicate >90% and >65% conservation), which also contacts the γ-phosphate of the nucleotide and coincides with the beginning of β-strand-2. β-Strands 2 and 3 run antiparallel and are hydrogen bonded to each other (Fig 1A). This network of hydrogen bonding contacts around the γ-phosphate of the nucleotide comprises some of the most highly conserved residues in the Rab family (Supplementary Fig. 1) and determines the conformation of the switch region. However, Rab28 presents three mutations within the WDTAGQE motif (67WDIGGQT73). Most notably, the occurrence of a glycine residue at position 70, together with the canonical glycine at position 71, adds conformational flexibility at the end of β-strand-3 and the beginning of switch 2. The added flexibility also affects lateral contacts with β-strand-2, which becomes shorter in the inactive structure, allowing switch 1 to swing away from the nucleotide-binding site (Fig. 2C).
Fig. 3.
The double-glycine motif and the conformational change. Close-up stereo views of the nucleotide, the double-glycine motif and the switch region in the structures of GppNHp-Rab28 (A) and GDP-3′P-Rab28 (B). The hydrogen-bonding network around the γ-phosphate of the nucleotide is broken after hydrolysis and phosphate release, resulting in destabilization of the double-glycine motif and the conformational change in the switch region. Colors are according to figures 1A and 2A.
In the Rab family, only Rab7B presents a double-glycine motif equivalent to that of Rab28 (Supplementary Fig. 1), but its structure has not been determined. However, the double-glycine motif is strictly conserved among all the members of the Arf GTPase family [1,2], where it has been linked to the observed retraction of the interswitch region in the inactive state [28]. Examination of active/inactive pairs of structures of Arf GTPases (Supplementary Table 2) shows that the members of this family typically undergo more extensive structural transitions on phosphate release than Rab GTPases. Clearly, the differences between these two GTPase families cannot be ascribed solely to the presence/absence of the double-glycine motif, but because of its critical position with respect to the γ-phosphate of the nucleotide and the switch region, this motif most likely plays a pivotal role in the nucleotide-dependent structural transition. The uniqueness and importance of the double-glycine motif has been pointed out in the Arf family [28], and among the heterotrimeric G protein α (Gα) subunits [29], which are more closely related to the Arfs than to other GTPases. Arf GTPases present extremely low intrinsic catalytic activities [30]. Future research should address whether Rab28 also shares this feature.
Interestingly, there is more conformational diversity among the GDP-bound structures of Rab GTPases than among their GTP-bound structures (Figs. 1B and 2B), which is the state relevant to effector recognition. Rab28 is not an exception to this rule; its GTP-bound structure superimposes well with those of other Rab GTPases, with relatively minor differences in the switch region. This observation supports the notion that the interactions of Rab GTPases with specific effectors is based on the exquisitely detailed recognition of minor structural differences in the switch and interswitch regions [7,11,31,32]. Yet, the larger differences among the GDP-bound structures, which set Rab28 apart, are probably significant for nucleotide exchange and hydrolysis and the specific recognition of GEF and GDI cofactors.
Supplementary Material
Supplementary Fig. 1. Alignment of human Rab GTPases.
Supplementary Fig. 2. Crystal packing contacts near switch 1 in the structure of ADP-3 ′P-Rab28.
Supplementary Movie 1. Nucleotide-dependent conformational change in Rab28.
Supplementary Table 1. Structures of Rab GTPases.
Supplementary Table 2. Structures of Raf GTPases.
Acknowledgments
This work was supported by National Institute of Health grant GM073791. Use of the Advanced Photon Source and BioCARS facilities were supported respectively by Department of Energy Contract W-31-109-Eng-38 and National Institute of Health grant RR007707.
References
- 1.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–6. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 3.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 4.Bernards A, Settleman J. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 2004;14:377–85. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 6.Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol. 2000;301:1077–87. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- 7.Pfeffer SR. Structural clues to Rab GTPase functional diversity. J Biol Chem. 2005;280:15485–8. doi: 10.1074/jbc.R500003200. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz SL, Cao C, Pylypenko O, Rak A, Wandinger-Ness A. Rab GTPases at a glance. J Cell Sci. 2007;120:3905–10. doi: 10.1242/jcs.015909. [DOI] [PubMed] [Google Scholar]
- 9.Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 10.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–17. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 11.Eathiraj S, Pan X, Ritacco C, Lambright DG. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature. 2005;436:415–9. doi: 10.1038/nature03798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brauers A, Schurmann A, Massmann S, Muhl-Zurbes P, Becker W, Kainulainen H, Lie C, Joost HG. Alternative mRNA splicing of the novel GTPase Rab28 generates isoforms with different C-termini. Eur J Biochem. 1996;237:833–40. doi: 10.1111/j.1432-1033.1996.0833p.x. [DOI] [PubMed] [Google Scholar]
- 13.Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr D Biol Crystallogr. 2001;57:1367–72. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 14.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–9. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55:247–55. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 16.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 17.Zwart PH, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–35. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 18.Zhang B, Zhang Y, Shacter E, Zheng Y. Mechanism of the guanine nucleotide exchange reaction of Ras GTPase--evidence for a GTP/GDP displacement model. Biochemistry. 2005;44:2566–76. doi: 10.1021/bi048755w. [DOI] [PubMed] [Google Scholar]
- 19.Itzen A, Pylypenko O, Goody RS, Alexandrov K, Rak A. Nucleotide exchange via local protein unfolding--structure of Rab8 in complex with MSS4. Embo J. 2006;25:1445–55. doi: 10.1038/sj.emboj.7601044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delprato A, Lambright DG. Structural basis for Rab GTPase activation by VPS9 domain exchange factors. Nat Struct Mol Biol. 2007;14:406–12. doi: 10.1038/nsmb1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz SL, et al. Flow cytometry for real-time measurement of guanine nucleotide binding and exchange by Ras-like GTPases. Anal Biochem. 2008 doi: 10.1016/j.ab.2008.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergbrede T, Pylypenko O, Rak A, Alexandrov K. Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8A resolution. J Struct Biol. 2005;152:235–8. doi: 10.1016/j.jsb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Renault L, Guibert B, Cherfils J. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature. 2003;426:525–30. doi: 10.1038/nature02197. [DOI] [PubMed] [Google Scholar]
- 24.Amor JC, Horton JR, Zhu X, Wang Y, Sullards C, Ringe D, Cheng X, Kahn RA. Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases. J Biol Chem. 2001;276:42477–84. doi: 10.1074/jbc.M106660200. [DOI] [PubMed] [Google Scholar]
- 25.Wang ZX, Shi L, Liu JF, An XM, Chang WR, Liang DC. 2.0 A crystal structure of human ARL5-GDP3′P, a novel member of the small GTP-binding proteins. Biochem Biophys Res Commun. 2005;332:640–5. doi: 10.1016/j.bbrc.2005.04.168. [DOI] [PubMed] [Google Scholar]
- 26.Pao CC, Gallant J. A new nucleotide involved in the stringent response in Escherichia coli. Guanosine 5′-diphosphate-3′-monophosphate. J Biol Chem. 1979;254:688–92. [PubMed] [Google Scholar]
- 27.Sato Y, Fukai S, Ishitani R, Nureki O. Crystal structure of the Sec4p.Sec2p complex in the nucleotide exchanging intermediate state. Proc Natl Acad Sci U S A. 2007;104:8305–10. doi: 10.1073/pnas.0701550104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasqualato S, Renault L, Cherfils J. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for ‘front-back’ communication. EMBO Rep. 2002;3:1035–41. doi: 10.1093/embo-reports/kvf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuwald AF. Galpha Gbetagamma dissociation may be due to retraction of a buried lysine and disruption of an aromatic cluster by a GTP-sensing Arg Trp pair. Protein Sci. 2007;16:2570–7. doi: 10.1110/ps.073098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randazzo PA, Kahn RA. GTP hydrolysis by ADP-ribosylation factor is dependent on both an ADP-ribosylation factor GTPase-activating protein and acid phospholipids. J Biol Chem. 1994;269:10758–63. [PubMed] [Google Scholar]
- 31.Ostermeier C, Brunger AT. Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell. 1999;96:363–74. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- 32.Zhu G, Zhai P, Liu J, Terzyan S, Li G, Zhang XC. Structural basis of Rab5-Rabaptin5 interaction in endocytosis. Nat Struct Mol Biol. 2004;11:975–83. doi: 10.1038/nsmb832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Alignment of human Rab GTPases.
Supplementary Fig. 2. Crystal packing contacts near switch 1 in the structure of ADP-3 ′P-Rab28.
Supplementary Movie 1. Nucleotide-dependent conformational change in Rab28.
Supplementary Table 1. Structures of Rab GTPases.
Supplementary Table 2. Structures of Raf GTPases.