Abstract
Incorporation of noncanonical amino acids into cellular proteins often requires engineering new aminoacyl-tRNA synthetase activity into the cell. A screening strategy that relies on cell-surface display of reactive amino acid side-chains was used to identify a diverse set of methionyl-tRNA synthetase (MetRS) mutants that allow efficient incorporation of the methionine (Met) analog azidonorleucine (Anl). We demonstrate that the extent of cell-surface labeling in vivo is a good indicator of the rate of Anl activation by the MetRS variant harbored by the cell. By screening at low Anl concentrations in Met-supplemented media, MetRS variants with improved activities toward Anl and better discrimination against Met were identified.
Keywords: click chemistry, high-throughput screening, protein engineering
Amino acids that carry noncanonical side-chains have become useful tools for protein engineering and analysis. Their uses include stabilization of protein–protein interfaces (1–3), modification of spectral properties of proteins (4, 5), and determination of three-dimensional protein structure (6–8). The value and versatility of noncanonical amino acids has been increased significantly by their recent use in conjugation (9, 10), detection (11, 12), and selective isolation (13) of proteins. Among the reactive groups introduced into biological molecules, azides are especially useful (14). Azides can be ligated under physiological conditions to phosphines through the Staudinger ligation or to terminal alkynes through Cu(I)-catalyzed [3 + 2] azide-alkyne cycloaddition, both in bioorthogonal fashion. Taking advantage of the bioorthogonality of these reactions, azides have been used to selectively label a diverse set of biomolecules including proteins (15), glycans (16), and lipids (17, 18).
Azide functionality can be introduced into proteins in vivo through use of the methionine (Met) surrogate, azidohomoalanine (Aha) (15, 19). Because Aha is activated by the wild-type MetRS (15), it enables convenient cotranslational labeling of proteins in vivo. Dieterich and coworkers recently reported the identification of newly synthesized proteins in mammalian cells through pulse-labeling with Aha (13). In this method, called bioorthogonal noncanonical amino acid tagging (BONCAT), azide-bearing proteins synthesized during the Aha pulse were separated from the pool of preexisting proteins in the cell through a Cu(I)-catalyzed ligation to an alkyne-derivatized biotin tag and subsequent affinity chromatography.
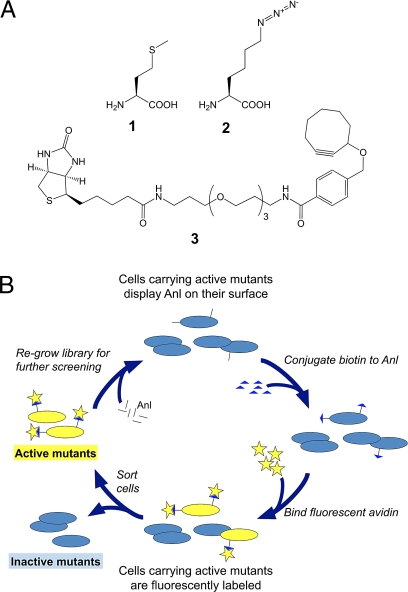
Although Aha is ideal for identification of newly synthesized proteins in cultures containing a single cell type, it is not suitable for directing protein labeling to specific cell types in complex cellular mixtures (e.g., in microbial communities, in tissues or in intact animals) because MetRS is common to all cells. However, we have recently shown that cell-selective metabolic labeling can be achieved by using azidonorleucine (Anl, 2) (Fig. 1A), rather than Aha as the Met surrogate (20). Unlike Aha, Anl is not activated efficiently by any of the wild-type aminoacyl-tRNA synthetases (aaRS), and Anl labeling can be restricted to cells outfitted with mutant forms of MetRS. Link and coworkers recently reported a high-throughput screen for E. coli MetRS mutants that activate Anl (21). Cells carrying such mutants display Anl residues on their surfaces, and can be identified and isolated by fluorescence-activated cell sorting (FACS; Fig. 1B). Although the clones isolated in this way showed only modest levels of Anl incorporation into marker proteins, analysis of the isolated MetRS mutants (all of which shared a Leu-to-Gly mutation at position 13) led to the preparation of the single-site L13G variant (MetRS-L13G). MetRS-L13G allows near-complete replacement of Met by Anl.
Fig. 1.
Reagents and screening method used in this study. (A) Amino acids and tagging reagents: 1, Met; 2, Anl; 3, biotin-PEO-cyclooctyne. (B) Scheme for the library screen for identification of MetRS mutants that allow Anl incorporation into cellular proteins.
Here, we report screening methods that yield MetRS mutants with greatly enhanced activity toward Anl and improved discrimination against Met. We describe the selection and characterization of these MetRS variants, and discuss the factors that we believe to be most important in their selection. Several of the mutants reported here are superior to MetRS-L13G with respect to metabolic protein labeling with Anl in media supplemented with Met.
Results and Discussion
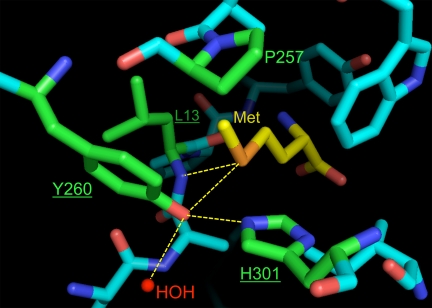
Link and coworkers randomized four positions (L13, P257, Y260, and H301) in the Met-binding pocket of E. coli MetRS (21) (Fig. 2). Screening of the resulting library for in vivo incorporation of Anl into proteins yielded three MetRS mutants: L13G-P257L-Y260T-H301A, L13G-P257S-Y260T-H301L, and L13G-P257L-Y260L-H301V, but not L13G. We wondered whether L13G failed to emerge from the screen because of incomplete (62%) library coverage, or because of other aspects of our experimental design that might have precluded isolation of highly active mutants. To ensure complete coverage of the saturation mutagenesis library, we created a library of lower diversity by reducing the number of randomized sites from four to three, while increasing the number of clones that carry the library by optimizing ligation and transformation conditions. Examination of the crystal structure of Met-bound E. coli MetRS (22) reveals that the sulfur atom of Met is recognized through hydrogen bonds with Oη of Y260 and the backbone amide of L13. (Fig. 2) H301 is also implicated in recognition of the sulfur atom (23). Positions 13, 260, and 301 were therefore selected for randomization. The resulting library is a subset of that screened by Link and coworkers, and includes the L13G mutant. Triple mutants isolated from this library were named according to the single-letter code for each mutation they carry (e.g., the MetRS variant carrying the L13G, Y260M, and H301L mutations is designated “GML”).
Fig. 2.
Residues surrounding the ligand side-chain in the amino acid binding pocket of the wild-type E. coli MetRS. The ligand is displayed in yellow, whereas residues considered for saturation mutagenesis are shown in green. Underlined residues were selected for randomization in this study. The hydrogen-bonding network that recognizes the sulfur atom on the ligand is highlighted by yellow lines. An enzyme-bound water molecule (labeled HOH) also participates in this network. Figure was generated using PyMOL (v1.0, DeLano Scientific) based on coordinates from the Protein Data Bank (PDB ID: 1F4L) (22).
The library was constructed through PCR gene assembly using primers containing degenerate NNK (N = A, C, G, T; K = G, T) codons at residues 13, 260, and 301. The assembled fragments were ligated into plasmid pAJL-20 (24), and the resulting library was transformed into M15MA (19). The number of independent clones was maintained >107 for every transformation step. The library coverage was estimated to be 99.9% using the program GLUE (25), which assumes an unbiased library where each member is equally likely to occur.
OmpC Expression and Cell-Surface Labeling with Anl.
pAJL-20 directs the synthesis of an E. coli OmpC variant carrying six surface-exposed Met residues under control of the isopropyl-β-thiogalactoside (IPTG)-inducible T5 promoter (21). Synthesis of Anl-bearing OmpC was performed in Met-auxotrophic M15MA cells transformed with variants of pAJL-20 carrying MetRS mutants. Minimal medium supplemented with all 20 canonical amino acids was inoculated with an aliquot of transformants and grown to OD600 = 1.0. Cells were washed and shifted into minimal medium lacking Met, but supplemented with Anl. OmpC expression was initiated by addition of IPTG. The azide groups displayed on the surfaces of the resulting cells were ligated to biotin-PEO-cyclooctyne (3) (Fig. 1A) and labeled with avidin conjugated to Alexa Fluor 488.
The screen outlined in Fig. 1B uses the extent of fluorescence labeling as a reporter for Anl incorporation, and in turn, for MetRS activity toward Anl; the highest levels of OmpC expression and fluorescence labeling are expected in cells carrying the MetRS mutants most active toward Anl. However, over-production of membrane proteins such as OmpC can have adverse effects on cell viability (26, 27); efficient synthesis of OmpC may place the most active clones at a disadvantage in the screen. To determine the extent to which OmpC expression affects cell viability and labeling efficiency, OmpC was induced at 1 mM Anl in M15MA cells carrying MetRS-L13G, and changes in cell-surface labeling and cell viability were monitored for 4 h. Significant losses of viable cells were observed over this period (Fig. S1A), but fluorescence labeling was nearly complete after only 30 min of induction (Fig. S1B). To minimize the effects of OmpC toxicity while maintaining high fluorescence levels, OmpC expression was limited to 30–40 min in all subsequent experiments.
Screening and Identification of Active MetRS Mutants.
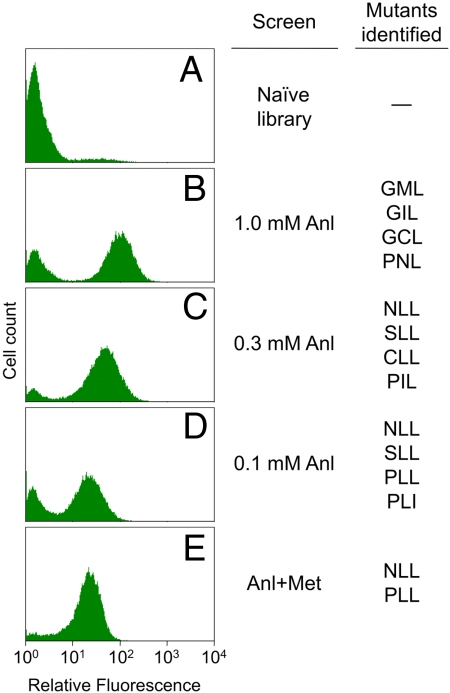
Near-complete replacement of Met by Anl was shown in M15MA cells carrying MetRS-L13G when protein synthesis was induced in Met-free minimal medium supplemented with 1.0 mM Anl (21). In a first step toward isolation of MetRS variants that exceed the activity of L13G, M15MA cells carrying the naïve three-position library (Fig. 3A) were screened under similar conditions. Expression of OmpC was induced at 1.0 mM Anl in minimal medium lacking Met, and Anl residues displayed on the cell surface were fluorescently labeled. The top 1% of fluorescent clones were isolated by FACS and regrown in rich medium for a second round of screening. After a single round of screening, fluorescent clones constituted 65% of the sorted cells. After two rounds of screening, individual clones from the resulting population (Fig. 3B) were sampled. Although the identified clones revealed a highly diverse set of sequences, all clones exhibited high levels of labeling at 1.0 mM Anl. Within this set, four MetRS mutants (GML, GIL, GCL, and PNL) yielded the highest levels of fluorescence.
Fig. 3.
Fluorescence histograms of the naïve library and populations enriched in fluorescent clones. The naïve library (A) is shown labeled at 1.0 mM Anl. Populations enriched through two rounds of screening at 1.0 mM Anl (B), 0.3 mM Anl (C), or 0.1 mM Anl (D) are displayed next to MetRS mutants identified by these screens. The Anl+Met screen was performed at 1.0 mM Anl over three rounds, increasing the Met concentration from 0.01 mM to 0.03 mM to 0.1 mM. The final population from this screen (E) is shown after labeling at equal concentrations (1.0 mM) of Anl and Met.
To identify mutants that can sustain protein synthesis at lower Anl concentrations, the naïve library was screened at 0.3 mM Anl. In addition to increasing the stringency of the screen, a lower Anl concentration further limits OmpC synthesis and the associated toxicity. In two rounds of screening, the top 0.6% of the labeled clones were sorted, and individual clones were identified from the resulting population. (Fig. 3C) The NLL mutant appeared in >50% of the clones sampled. Analysis of isolated clones by FACS identified NLL and three other mutants (SLL, CLL, and PIL) that yield the highest fluorescence levels. Further reduction of the Anl concentration to 0.1 mM resulted in enrichment of clones carrying the NLL, SLL, PLL, and PLI mutants. (Fig. 3D) Since clones carrying the NLL mutant represent the dominant products of both screens, we concluded that the screen had converged to a solution.
Residue-specific incorporation of noncanonical amino acids into recombinant proteins is often carried out under conditions that minimize the intracellular concentration of the canonical amino acid to be replaced (28, 29). However, for applications that require the use of nonauxotrophic cells and/or rich media, it is necessary to engineer aaRS that discriminate against their natural substrates. To identify such mutants, the naïve library was screened against Met for three rounds at 1.0 mM Anl, with increasing concentrations of Met (0.01, 0.03, and 0.1 mM) added to the medium in successive rounds. The final population showed only limited sensitivity to Met, and exhibited substantial labeling even when Met and Anl were both supplied at concentrations of 1.0 mM (Fig. 3E). The NLL and PLL mutants were highly enriched by this screen.
Characterization of MetRS Mutants in Vivo.
Although the NLL, SLL, and PLL mutants emerged from higher stringency screens, these mutants appear to be absent from the population isolated from the 1.0 mM Anl screen. To understand why highly active mutants were not identified through screens of lower stringency, we compared the extent of fluorescence labeling, at Anl concentrations varying between 0.03 and 3.0 mM, of clones isolated from the 1.0 mM Anl (GML, GIL, GCL, and PNL) and 0.3 mM Anl (NLL, CLL, SLL, and PIL) screens. The median values of the fluorescence from labeled cell populations were determined by flow cytometry. The dose-response data were fit to the Hill equation, and an EC50 value was determined for each MetRS mutant (Table S1, Fig. 4A, and Fig. S2). We found similar labeling levels for all eight mutants at 1.0 mM Anl, but only those clones isolated from the more stringent screen maintained high fluorescence levels at 0.3 mM Anl. The absence of highly active mutants such as NLL from the population selected at 1.0 mM Anl might be due simply to undersampling, but increased OmpC expression and toxicity resulting from faster Anl activation might also have hindered the identification of highly active mutants in this screen.
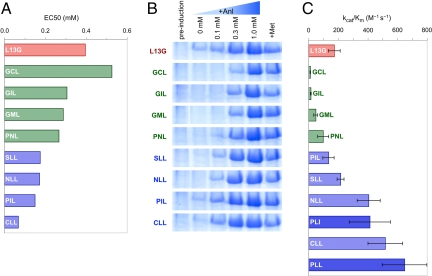
Fig. 4.
Comparison of in vivo and in vitro activities of the identified MetRS mutants. Data shown for mutants identified by screens at 1.0 mM and 0.3 mM Anl, and the L13G mutant are marked in green, blue, and red, respectively. Protein expression, cell-surface labeling in vivo and Anl activation in vitro are consistent in indicating better activity for mutants screened at lower Anl concentrations. (A) EC50 values describing the fluorescence response to Anl availability in the expression media. (B) SDS/PAGE analysis of whole-cell lysates for cells expressing mDHFR under different conditions. Cells grown to midlog phase in medium containing all 20 canonical amino acids were then shifted into 19-amino acid medium (no Met) supplemented with 0.1, 0.3, or 1.0 mM Anl (lanes 3, 4, and 5, respectively), 40 mg/L Met (lane 6), or no 20th amino acid (lane 2) before induction of mDHFR synthesis. Lane 1 shows cells before induction. For clear presentation images were resized and lanes on the GML panel were reordered. Original images can be found in Fig. S3. (C) Specificity constants (kcat/Km) for Anl activation measured in vitro. Average of three measurements is reported with errors bars indicating one standard deviation. Data for mutants identified by the 0.1 mM Anl screen (PLL and PLI) are shown in dark blue.
To determine how well the isolated MetRS mutants support protein synthesis in Met-depleted media supplemented with Anl, they were transferred to plasmid pAJL-61, which encodes a 6×His-tagged murine dihydrofolate reductase (mDHFR) under control of the IPTG-inducible T5 promoter (21). The resulting plasmids were transformed into M15MA, and protein expression was induced for 3.5 h in minimal media supplemented with 0.1, 0.3, or 1.0 mM Anl, 0.3 mM Met, or no 20th amino acid. mDHFR was purified from cell lysates by Ni-NTA chromatography, and tryptic fragments from purified proteins were analyzed by MALDI-MS. Analysis of whole-cell lysates by SDS/PAGE revealed a pattern of mDHFR expression levels consistent with those inferred from cell-surface fluorescence, as described above: All eight mutants supported similar levels of mDHFR expression at 1.0 mM Anl, but only those mutants isolated from the 0.3 mM Anl screen sustained similar levels of expression at 0.3 mM Anl (Fig. 4B and Fig. S3). Similarly, MALDI-MS analysis of tryptic peptides indicated near-complete replacement of Met by Anl by all eight mutants at 1.0 mM Anl, whereas complete replacement at 0.3 mM Anl was observed only for mutants isolated from the 0.3 mM Anl screen (Fig. S4).
Determination of Activation Kinetics in Vitro.
MetRS mutants were expressed and purified for determination of Anl activation kinetics in vitro (Table 1). In our experience, the relative rates of activation of noncanonical amino acids measured in vitro correlate well with their efficiencies of incorporation in vivo (15, 30). As expected, we observed faster activation of Anl by mutants isolated through screens performed at lower concentrations of Anl (Fig. 4C). Cell-surface display is a good predictor of MetRS activity.
Table 1.
Kinetic parameters for the activation of Anl and Met by the MetRS variants identified in this study and the L13G mutant
| MetRS | Anl activation kcat/Km, M−1 s−1 | Met activation kcat/Km, M−1 s−1 | Selectivity* |
|---|---|---|---|
| wt† | - | 550,000 | - |
| L13G | 170 ± 40 | 5,600 ± 1,500 | 0.03 |
| PLL | 650 ± 150 | 200 ± 50 | 3.2 |
| NLL | 410 ± 80 | 350 ± 70 | 1.2 |
| SLL | 220 ± 20 | 900 ± 80 | 0.24 |
| CLL | 520 ± 120 | 7,100 ± 600 | 0.07 |
| PLI | 410 ± 140 | - | - |
| PIL | 130 ± 40 | - | - |
| PNL | 98 ± 37 | - | - |
| GML | 47 ± 13 | - | - |
| GIL | 14 ± 3 | - | - |
| GCL | 11 ± 2 | - | - |
Clones carrying the NLL and PLL mutants exhibited fluorescence labeling in screens performed in Met-supplemented media. The rates of activation of Met by NLL, PLL, and two similar mutants (SLL and CLL, both obtained from the noncompetitive high-stringency screen) were determined and their substrate preferences compared. Only the NLL and PLL mutants activate Anl faster than Met (Table 1); only the competitive screen yielded mutants that prefer the noncanonical substrate. The results of competitive screens depend directly on the relative rates of activation of Anl and Met by each clone. In contrast, noncompetitive screens are insensitive to the rates of activation of Met, and can yield clones in which Anl is activated rapidly yet cannot compete with the natural substrate.
Comparison of MetRS-L13G and Selected MetRS Mutants.
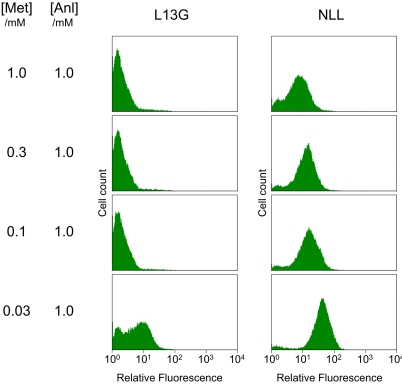
We previously reported comparable specificity constants (kcat/Km) for activation of Anl and Met by MetRS-L13G (21). Careful reexamination of the activation kinetics suggests that our earlier conclusions were incorrect, and that MetRS-L13G exhibits a strong preference for Met (Table 1). Consistent with these observations, fluorescence labeling of cells harboring the L13G mutant shows significant sensitivity to the addition of Met to the expression medium; fluorescence labeling is not detected above a Met concentration of 0.03 mM (Fig. 5). In contrast, cells bearing MetRS-NLL show much lower sensitivity to the presence of Met in the medium and produce a distinct fluorescent population even when Anl and Met are both provided at 1.0 mM, in agreement with the activation kinetics obtained for the NLL mutant. Furthermore, while the incorporation of a natural amino acid was detected when Anl was presented to cells bearing the L13G mutant at low concentrations in Met-free medium, MetRS-NLL supports only minimal levels protein synthesis under similar conditions (Fig. 4B and Fig. S4). These results suggest that mutants such as NLL and PLL may be suitable for in vivo labeling applications at physiological amino acid concentrations.
Fig. 5.
Cell-surface labeling in the presence of Met. Labeling levels were evaluated for cells carrying the L13G and NLL mutants after OmpC expression at 1.0 mM Anl and varying concentrations of Met. Fluorescence histograms show that the NLL mutant is less susceptible than L13G to the presence of Met in the medium, in agreement with substrate preferences of these mutants determined in vitro (Table 1).
Significance of Selected Mutations.
We identified 41 different MetRS mutants that allow Anl incorporation in Met-depleted media supplemented with 1.0 mM Anl (Table S2). Examination of the distribution of mutations across the randomized sites shows strong selection for Leu and other aliphatic side-chains at position 301, and a diverse set of mutations at positions 13 and 260 (Fig. S5A). In mutants isolated from screens of higher stringency (0.3 mM Anl or lower; 13 mutants), positions 13 and 260 are dominated by Pro and Leu, respectively (Fig. S5B). Alternatives to Leu at position 260 are mostly hydrophobic, whereas position 13 is commonly mutated to small and/or polar residues and to Pro. Although the L13P mutation is unique in eliminating the backbone hydrogen bond to the native ligand, small and/or polar mutations might have a role in modulating solvation, and thus the availability of the backbone hydrogen bond to Met at this site. The overall result of these mutations is the elimination of hydrophilic contacts to the Met side-chain, thereby reducing competition with Met and polar amino acids and creating a hydrophobic pocket for Anl.
A survey of small molecule crystal structures indicates that organic azides rarely participate in hydrogen bonds, in some cases forming weak hydrogen bonds through the terminal nitrogen (31). The high-resolution (1.5 Å) X-ray crystal structures of free and Anl-bound MetRS-SLL reveal that such a hydrogen bond facilitates Anl binding. The terminal azide on the ligand is recognized by an enzyme-bound water molecule, which is well conserved in wild-type MetRS (22, 32) (HOH) (Fig. 2). Moreover, the SLL mutant does not exhibit the ligand-induced conformational change that leads to formation of a “closed” binding pocket around the ligand observed in the wild-type enzyme (22). Instead, the SLL mutant is locked in the “closed” conformation even in the absence of ligand. The crystal structure shows the orientation of the L301 side-chain to be incompatible with the “open” conformation of F300 (22), and suggests that formation of the closed pocket before Anl binding might lead to faster Anl activation and contribute to the strong bias for the H301L mutation observed in the screens.
Most selected mutations remove from the binding site the hydrophilic elements that ensure specificity toward Met, but do not introduce specific contacts for Anl recognition. Such substitutions might create enzymes that are relatively indiscriminant with respect to hydrophobic ligands. Recently, a leucyl-tRNA synthetase variant evolved for incorporation of 2-amino-6-(methylthio)hexanoic acid was reported to show promiscuity toward other long-chain hydrophobic ligands (33). Similarly, the SLL mutant discussed above was identified by library screening to enable incorporation of 6,6,6-trifluoronorleucine into recombinant proteins (34). Thus, the mutants discovered in this study may permit incorporation of other long-chain hydrophobic ligands, or may be used as design platforms for incorporation of longer side-chains.
Conclusions
We have successfully identified a diverse set of MetRS mutants that enable efficient in vivo incorporation of the azide-bearing amino acid Anl into proteins, by using a fast, high-throughput screening strategy based on cell-surface display of reactive side-chains. Through characterization of identified mutants, we established in vivo fluorescence labeling of displayed azides as an accurate predictor of both protein synthesis levels in vivo and Anl activation rates in vitro. The results of the library screens point to the importance of the H301L mutation, and structural analysis of MetRS-SLL mutant suggests a role for this mutation in locking the enzyme in the closed conformation.
Despite the small size of the library explored, we were successful in shifting the specificity of MetRS in favor of the unnatural ligand through screens against the natural substrate. The identified MetRS variants provide faster activation of Anl and significantly lower sensitivity to Met than any previously known mutant. These properties are essential for the use of these enzymes in proteome-wide labeling of live cells in complex environments.
Materials and Methods
Chemical Reagents.
Anl (2) (24) was a gift from A. J. Link and M. B. van Eldijk (California Institute of Technology, Pasadena, CA), and biotin-PEO-cyclooctyne (3) (35) was a gift from J. D. Fisk (California Institute of Technology, Pasadena, CA).
Plasmids, Cell Strains, and Cloning Reagents.
All restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. PfuUltra HighFidelity polymerase (Stratagene) was used in all PCR steps. E. coli strain XL-1 Blue (Stratagene) was used in all recombinant DNA manipulations, while the Met-auxotroph M15MA (19) carrying the plasmid pREP4 (Qiagen) was used as the expression host, unless noted otherwise. All DNA sequencing was performed by Laragen Inc. or by the Caltech DNA sequencing facility. Plasmids pAJL-20 and pAJL-61 (21) were used in cell-surface labeling and mDHFR expression, respectively. Both plasmids carry a cassette encoding a monomeric version of E. coli MetRS truncated at residue 548 (36) under control of its natural promoter. In addition, pAJL-20 encodes a variant of E. coli OmpC that carries six surface-exposed Met residues (19), while pAJL-61 directs the synthesis of an N-terminally 6×His-tagged mDHFR, both under control of the IPTG-inducible T5 promoter. Plasmid pMTY21 (34) encodes an N-terminally 6×His-tagged MetRS regulated by the T5 promoter for expression and purification of MetRS mutants and was constructed by ligation of BamHI/SalI-digested pQE-80L (Qiagen) with a similarly digested MetRS gene PCR-amplified from pAJL-20 or pAJL-61 using the primers MRS_BamHI and MRS_SalI-r. Sequences for all primers used in this study are listed in Table S3. Nomenclature for plasmids modified to carry MetRS mutants is explained in the SI Text.
MetRS Library Construction.
Oligonucleotides encoding degenerate NNK (N = A, T, G, C; K = G, T) codons at sites corresponding to Leu-13, Tyr-260, and His-301 in E. coli MetRS were obtained from Operon. Four separate PCR reactions were performed using PfuUltra HighFidelity polymerase (Stratagene), pAJL-20 as a template and the following primer pairs: lib_fwd2/L13_lib2-r, L13_lib3/Y260_lib3-r, Y260_lib3/H301_lib3-r, and H301_lib2/lib_rev2. The DNA fragments obtained were isolated by electrophoresis and desalted on Zymo-spin columns (Zymo Research Corp.). Approximately equimolar quantities of the fragments were mixed and subjected to 20–30 rounds of PCR (95 °C for 30 s, 58 °C for 30 s, 72 °C for 2 min). To amplify the assembled product, the primers lib_fwd3 and lib_rev3 were added to the reaction mixture and 20–30 more rounds of PCR were performed using the same conditions. The resulting 2.0 kb PCR product was digested with NotI and BsrGI to reveal a 1.35-kb insert, which was subsequently ligated using an insert/vector molar ratio of 3:2 into pAJL-20 digested with the same enzymes. Minimizing UV-exposure during DNA manipulations was found to be critical for high ligation efficiency. Resulting plasmids were electroporated into ElectroTen (Stratagene) cells yielding 4.5 × 107 independent transformants. The plasmids from pooled transformants were isolated on Miniprep columns (Qiagen) and electroporated into M15MA[pREP4] to yield 1.3 × 107 independent clones. Aliquots of the cultures containing the library (1 ml, OD600 = 1.0) were stored at −80 °C in 50% glycerol until protein expression.
OmpC Expression and Cell-Surface Labeling.
Expression of the OmpC mutant carrying Anl was performed as described in ref. 21 with minor modifications. M15MA[pREP4/pAJL-20] cells were inoculated into M9 medium (M9 salts, 0.2% glucose, 1 mM MgSO4, 25 mg/L thiamine) supplemented with 40 mg/L of each of the 20 canonical amino acids, 200 mg/L ampicillin, and 35 mg/L kanamycin (referred to as M9 + 20aa) either 1:100 from overnight cultures or 1:30 from frozen stocks carrying the MetRS library. After reaching OD600 = 0.8 to 1.0, cells were pelleted at 5,000 × g for 7 min and resuspended in an equal volume of M9 medium supplemented with 19 amino acids (no Met) and antibiotics (referred to as M9 + 19aa). After cells were incubated while shaking for 15 min at 37 °C to deplete any residual Met, they were moved into fresh M9 + 19aa medium, divided into multiple vessels, and supplemented with Met or Anl (commonly introduced at 0.3 mM and 1.0 mM, respectively), or no Met surrogate. Expression of OmpC was initiated by addition of IPTG (1 mM); after 30–40 min a 1-mL aliquot of each culture adjusted to OD600 = 1.0 was washed with PBS (PBS, pH 7.4) in preparation for cell-surface labeling. After OmpC expression, cells were treated with 100 μM biotin-PEO-cyclooctyne with agitation for 16 h at 37 °C. Biotin-labeled cells were cooled to 4 °C, washed twice with 1-mL PBS, and treated with 2.5 μL of a 1-mg/mL solution of avidin-Alexa Fluor 488 conjugate (Molecular Probes) for 2 h at 4 °C with agitation. Cells were washed three more times with PBS to remove excess fluorophore and labeling was confirmed on a Safire II (Tecan) plate reader or by flow cytometry.
To determine the number of viable cells after OmpC expression and labeling, the cell density was adjusted to OD600 = 0.2. Cells were further diluted 1:104 and 1:2.5 × 105 into PBS, and 100 μL from each dilution was plated onto 2×YT agar supplemented with 200-mg/L ampicillin and 35-mg/L kanamycin. The number of colonies on each plate was determined after overnight incubation at 37 °C.
Flow Cytometry and Cell Sorting.
All flow cytometric analyses were performed on a MoFlo (DakoCytomation) FACS equipped with an argon ion laser emitting at 488 nm. The naïve library was sorted in the “sort purify” mode, whereas for all subsequent sorts the “sort single” mode was used. Sort gates were set and data analysis was performed with Summit software (DakoCytomation). After cell-surface labeling, highly fluorescent cells were isolated by setting gates in the fluorescence channel for the top 0.8% to 1.2% of the population. Additional gates were set in the forward- and side-scatter channels to avoid events of unusual size. The gate on the fluorescence channel was set especially high (0.5%) for libraries expressing OmpC at low concentrations of Anl. In a typical experiment, 108 events were interrogated, of which 5 × 105 were taken to the following round. Sorted cells were rescued in 15 mL of SOC medium for 30 min before antibiotics were added (200 mg/L ampicillin, 35 mg/L kanamycin). An aliquot of rescued cells was plated on selective 2×YT agar plates for analysis of individual clones. Cultures adjusted to OD600 = 1.0 were either stored at −80 °C in 50% glycerol or immediately carried through to the next round of selection.
The dose–response relationship between Anl concentration in expression media and fluorescence observed on FACS after cell-surface labeling was determined on M15MA[pREP4/pAJL-20] cells as described above. OmpC expression was induced in M9 + 19aa medium supplemented with 0.03, 0.1, 0.3, 1.0, and 3.0 mM Anl. After cell-surface labeling, the median population fluorescence was determined based on FACS data. EC50 values were obtained by performing a least-squares fit to the Hill equation using KaleidaGraph (v3.6; Synergy Software).
Recombinant DHFR Expression, Purification, and Analysis.
Expression of Anl-bearing mDHFR was performed in M15MA[pREP4/pAJL-61] cells, using the same protocol for OmpC expression, but inducing expression for 3.5 h with IPTG. Variants of the pAJL-61 plasmid carrying different MetRS mutants were generated using QuikChange site-directed mutagenesis (primers listed in Table S3). Integrity of all constructs was verified by DNA sequencing. Protein was expressed in 5–10 mL cultures, aliquots standardized for OD600 were saved for SDS/PAGE analysis, and the remainder was lysed in 8 M urea. Purification was performed under denaturing conditions on Ni-NTA spin columns (Qiagen) according to the manufacturer's suggestions. For MALDI-MS analysis, 15-μL purified mDHFR was diluted with 95 μL of 75 mM NH4HCO3 and digested for 4 h at 37 °C with 0.2 μg of porcine trypsin (Promega). The resulting peptide mixtures were desalted on C18 ZipTip (Millipore) columns. Eluents were diluted 3-fold with matrix solution [saturated α-cyano hydroxycinnamic acid (Fluka) in 1:1 water/acetonitrile and 0.1% trifluoroacetic acid] and analyzed on a Voyager MALDI-TOF mass spectrometer (Applied Biosystems).
MetRS Expression, Purification, and in Vitro Activation Assays.
Variants of pMTY21 carrying different MetRS mutants were constructed either by the PCR amplification and ligation into pMTY21 of MetRS from pAJL-20 or pAJL-61 or by QuikChange site-directed mutagenesis as discussed above. E. coli XL-1 Blue cells transformed with pMTY21 were inoculated 1:100 from an overnight culture into 100 mL of Terrific Broth. Cells were grown to OD600 = 1.0 at 37 °C and were induced with IPTG (1 mM) to produce MetRS for 18 h at 25 °C. Cells were harvested by centrifugation (6,000 × g for 15 min). 6×His-tagged MetRS mutants were isolated under native conditions on Ni-NTA agarose resin (Qiagen) according to the manufacturer's instructions. Eluted product was exchanged into 100 mM Tris buffer (pH 7.5 with 2 mM DTT) on PD-10 columns (GE Healthcare). Enzyme stocks were prepared in 50% (wt/wt) glycerol and stored at −80 °C until needed. Enzyme concentrations were determined based on the absorbance at 280 nm of the enzyme stock diluted 1:9 in 9 M urea, and assuming an extinction coefficient of 93,280 M−1 cm−1 for MetRS mutants as calculated by the ProtParam tool (http://expasy.org/tools/protparam.html). Amino acid activation assays were carried out as described in ref. 37. Radiolabeled sodium 32P-pyrophosphate was purchased from Perkin-Elmer Life Sciences. An enzyme concentration in the range 1–4 μM was used in each reaction, and activation rates were measured at 0.031–16 mM Met and 0.25–64 mM Anl. Kinetic parameters were determined by fitting the initial activation rates to the Michaelis-Menten model using KaleidaGraph (v3.6; Synergy Software).
Supplementary Material
Acknowledgments.
We thank Mona Shahgholi for expert assistance with mass spectrometry, Haick Issaian for assistance with radioisotope use, and Professor Jared Leadbetter for use of his scintillation counter. We thank I. Kwon, A. J. Link, T. H. Yoo, J. D. Fisk, R. E. Connor, M. L. Mock, E. J. Choi and J. T. Ngo for helpful discussions and Dr. J. A. Champion for reviewing the manuscript. This work was supported by National Institutes of Health Grant GM62523 and by the Army Research Office through the Institute for Collaborative Biotechnologies.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905735106/DCSupplemental.
References
- 1.Bilgicer B, Xing X, Kumar K. Programmed self-sorting of coiled coils with leucine and hexafluoroleucine cores. J Am Chem Soc. 2001;123:11815–11816. doi: 10.1021/ja016767o. [DOI] [PubMed] [Google Scholar]
- 2.Tang Y, Tirrell DA. Biosynthesis of a highly stable coiled-coil protein containing hexafluoroleucine in an engineered bacterial host. J Am Chem Soc. 2001;123:11089–11090. doi: 10.1021/ja016652k. [DOI] [PubMed] [Google Scholar]
- 3.Son S, Tanrikulu IC, Tirrell DA. Stabilization of bzip peptides through incorporation of fluorinated aliphatic residues. ChemBioChem. 2006;7:1251–1257. doi: 10.1002/cbic.200500420. [DOI] [PubMed] [Google Scholar]
- 4.Bae JH, et al. Expansion of the genetic code enables design of a novel “gold” class of green fluorescent proteins. J Mol Biol. 2003;328:1071–1081. doi: 10.1016/s0022-2836(03)00364-4. [DOI] [PubMed] [Google Scholar]
- 5.Budisa N, et al. Global replacement of tryptophan with aminotryptophans generates noninvasive protein-based optical pH sensors. Angew Chem Int Ed. 2002;41:4066–4069. doi: 10.1002/1521-3773(20021104)41:21<4066::AID-ANIE4066>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 6.Eichler JF, Cramer JC, Kirk KL, Bann JG. Biosynthetic incorporation of fluorohistidine into proteins in E. coli: A new probe of macromolecular structure. Chembiochem. 2005;6:2170–2173. doi: 10.1002/cbic.200500249. [DOI] [PubMed] [Google Scholar]
- 7.Bae JH, et al. Incorporation of beta-selenolo[3,2-b]pyrrolyl-alanine into proteins for phase determination in protein X-ray crystallography. J Mol Biol. 2001;309:925–936. doi: 10.1006/jmbi.2001.4699. [DOI] [PubMed] [Google Scholar]
- 8.Hendrickson WA, Horton JR, LeMaster DM. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): A vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang K, Diehl MR, Tirrell DA. Artificial polypeptide scaffold for protein immobilization. J Am Chem Soc. 2005;127:10136–10137. doi: 10.1021/ja051457h. [DOI] [PubMed] [Google Scholar]
- 10.Carrico IS, et al. Lithographic patterning of photoreactive cell-adhesive proteins. J Am Chem Soc. 2007;129:4874–4875. doi: 10.1021/ja070200b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beatty KE, et al. Fluorescence visualization of newly synthesized proteins in mammalian cells. Angew Chem Int Ed. 2006;45:7364–7367. doi: 10.1002/anie.200602114. [DOI] [PubMed] [Google Scholar]
- 12.Beatty KE, Xie F, Wang Q, Tirrell DA. Selective dye-labeling of newly synthesized proteins in bacterial cells. J Am Chem Soc. 2005;127:14150–14151. doi: 10.1021/ja054643w. [DOI] [PubMed] [Google Scholar]
- 13.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci USA. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baskin JM, Bertozzi CR. Bioorthogonal click chemistry: Covalent labeling in living systems. Qsar Comb Sci. 2007;26:1211–1219. [Google Scholar]
- 15.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc Natl Acad Sci USA. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 17.Kho Y, et al. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc Natl Acad Sci USA. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hang HC, et al. Chemical probes for the rapid detection of fatty-acylated proteins in mammalian cells. J Am Chem Soc. 2007;129:2744–2745. doi: 10.1021/ja0685001. [DOI] [PubMed] [Google Scholar]
- 19.Link AJ, Tirrell DA. Cell surface labeling of Escherichia coli via copper(I)-catalyzed [3+2] cycloaddition. J Am Chem Soc. 2003;125:11164–11165. doi: 10.1021/ja036765z. [DOI] [PubMed] [Google Scholar]
- 20.Ngo JT, et al. Cell-selective metabolic labeling of proteins. Nat Chem Biol. 2009 doi: 10.1038/nchembio.200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Link AJ, et al. Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids. Proc Natl Acad Sci USA. 2006;103:10180–10185. doi: 10.1073/pnas.0601167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serre L, et al. How methionyl-tRNA synthetase creates its amino acid recognition pocket upon L-methionine binding. J Mol Biol. 2001;306:863–876. doi: 10.1006/jmbi.2001.4408. [DOI] [PubMed] [Google Scholar]
- 23.Crepin T, et al. Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase. J Mol Biol. 2003;332:59–72. doi: 10.1016/s0022-2836(03)00917-3. [DOI] [PubMed] [Google Scholar]
- 24.Link AJ, Vink MK, Tirrell DA. Presentation and detection of azide functionality in bacterial cell surface proteins. J Am Chem Soc. 2004;126:10598–10602. doi: 10.1021/ja047629c. [DOI] [PubMed] [Google Scholar]
- 25.Firth AE, Patrick WM. Statistics of protein library construction. Bioinformatics. 2005;21:3314–3315. doi: 10.1093/bioinformatics/bti516. [DOI] [PubMed] [Google Scholar]
- 26.Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 27.Wagner S, Bader ML, Drew D, de Gier JW. Rationalizing membrane protein overexpression. Trends Biotechnol. 2006;24:364–371. doi: 10.1016/j.tibtech.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Link AJ, Tirrell DA. Reassignment of sense codons in vivo. Methods. 2005;36:291–298. doi: 10.1016/j.ymeth.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Budisa N. Prolegomena to future experimental efforts on genetic code engineering by expanding its amino acid repertoire. Angew Chem Int Ed. 2004;43:6426–6463. doi: 10.1002/anie.200300646. [DOI] [PubMed] [Google Scholar]
- 30.Kiick KL, Tirrell DA. Protein engineering by in vivo incorporation of non-natural amino acids: Control of incorporation of methionine analogues by methionyl-tRNA synthetase. Tetrahedron. 2000;56:9487–9493. [Google Scholar]
- 31.Tchertanov L. Structural metrics relationships in covalently bonded organic azides. Acta Crystallogr B. 1999;55:807–809. doi: 10.1107/s0108768199003936. [DOI] [PubMed] [Google Scholar]
- 32.Mechulam Y, et al. Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features. J Mol Biol. 1999;294:1287–1297. doi: 10.1006/jmbi.1999.3339. [DOI] [PubMed] [Google Scholar]
- 33.Brustad E, Bushey ML, Brock A, Chittuluru J, Schultz PG. A promiscuous aminoacyl-tRNA synthetase that incorporates cysteine, methionine, and alanine homologs into proteins. Bioorg Med Chem Lett. 2008;18:6004–6006. doi: 10.1016/j.bmcl.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 34.Yoo TH, Tirrell DA. High-throughput screening for methionyl-tRNA synthetases that enable residue-specific incorporation of noncanonical amino acids into recombinant proteins in bacterial cells. Angew Chem Int Ed. 2007;46:5340–5343. doi: 10.1002/anie.200700779. [DOI] [PubMed] [Google Scholar]
- 35.Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3+2] azide-alkyne cycloaddition for covalent modification of blomolecules in living systems. J Am Chem Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh G, Brunie S, Schulman LH. Transition state stabilization by a phylogenetically conserved tyrosine residue in methionyl-tRNA synthetase. J Biol Chem. 1991;266:17136–17141. [PubMed] [Google Scholar]
- 37.Kiick KL, Weberskirch R, Tirrell DA. Identification of an expanded set of translationally active methionine analogues in Escherichia coli. FEBS Lett. 2001;502:25–30. doi: 10.1016/s0014-5793(01)02657-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.