Abstract
The nucleosome has a central role in the compaction of genomic DNA and the control of DNA accessibility for transcription and replication. To help understanding the mechanism of nucleosome opening and closing in these processes, we studied the disassembly of mononucleosomes by quantitative single-molecule FRET with high spatial resolution, using the SELEX-generated “Widom 601” positioning sequence labeled with donor and acceptor fluorophores. Reversible dissociation was induced by increasing NaCl concentration. At least 3 species with different FRET were identified and assigned to structures: (i) the most stable high-FRET species corresponding to the intact nucleosome, (ii) a less stable mid-FRET species that we attribute to a first intermediate with a partially unwrapped DNA and less histones, and (iii) a low-FRET species characterized by a very broad FRET distribution, representing highly unwrapped structures and free DNA formed at the expense of the other 2 species. Selective FCS analysis indicates that even in the low-FRET state, some histones are still bound to the DNA. The interdye distance of 54.0 Å measured for the high-FRET species corresponds to a compact conformation close to the known crystallographic structure. The coexistence and interconversion of these species is first demonstrated under non-invasive conditions. A geometric model of the DNA unwinding predicts the presence of the observed FRET species. The different structures of these species in the disassembly pathway map the energy landscape indicating major barriers for 10-bp and minor ones for 5-bp DNA unwinding steps.
Keywords: chromatin, mononucleosomes, structure
The nucleosome, the basic unit of genome compaction (1), consists of ≈2 turns of double-stranded DNA wound around a histone protein octamer. Its structure is known to a resolution of 1.9 Å (2). DNA sequence, chemical modifications and histone composition modulate gene activity through this structure.
Central to nucleosomal function is its restructuring during processes that act on DNA, e.g., transcription or replication. Biochemical data indicate that nucleosome dissociation is temporary. Mechanisms for nucleosome unfolding, unwrapping or repositioning have been proposed (3), but so far none has been proven by direct physical evidence (e.g., detection of intermediate states). Here, we use single molecule Förster resonance energy transfer (FRET) to obtain quantitative structural information for elucidating such mechanisms.
Bulk solution FRET is a proven tool for measuring average distances; nucleosome dynamics have been quantified by FRET between dyes attached to DNA and/or histone proteins (4–6). Analyzing FRET from single molecules (7, 8) can give detailed information on structural diversity; e.g., FRET on surface-tethered nucleosomes demonstrated spontaneous structure fluctuations (9, 10), whereas confocal spFRET experiments (11, 12) on freely diffusing single nucleosomes detected structural subpopulations under various conditions.
The transit of a single molecule through the laser focus also provides information complementary to simple FRET efficiencies, e.g., fluorescence lifetime and anisotropy. Such multiparameter fluorescence detection (MFD) with appropriate data analysis allows characterizing heterogeneous populations and structural substates in molecules (13, 14). Here, we applied MFD to FRET data from mononucleosomes reconstituted from recombinant histones on a dye-labeled 170-bp DNA fragment. Using population-filtered fluorescence correlation analysis, modeling of possible dye positions, and an analysis of salt-dependent nucleosome dissociation, we could unravel intermediates in the assembly pathway of nucleosomes.
Results and Discussion
Multiparameter Analysis Detects Multiple Nucleosomal Species.
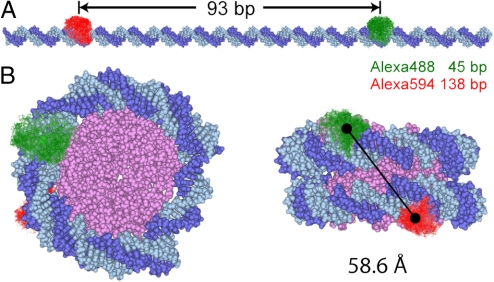
The disassembly studies use the idea that for a fully extended DNA fragment no significant FRET is expected due to the 93-bp separation between the donor Alexa Fluor 488 and the acceptor Alexa Fluor 594 (RDA = 306 Å) as shown in Fig. 1A. Within the intact nucleosome the dyes approach each other to <70 Å (Fig. 1B), enabling efficient FRET. The nucleosomes were reconstituted on one of the strongest positioning sequences (“Widom 601”) (15) to ensure a high sample homogeneity.
Fig. 1.
Internal labeling of nucleosomes. (A) Extended nucleosomal DNA duplex [D strand (dark blue), A strand (light blue)]. The conformational space of the D and A dyes from MD simulations is shown in green and red respectively. The dye positions and the number of separating basepairs of 93 are given for the D strand. (B) Nucleosome viewed from top (Left) and from the side (Right), based on the crystal structure 1KX5 from the RCSB Protein Data Bank (2) and fluorophore positions from (16) (see SI Appendix, section 2.6). Only the core of the histone (magenta) is shown for simplicity. The solid line connects the centers of mass (black circles) of the fluorophores' accessible space.
For single-molecule experiments, mononucleosomes were diluted to a final concentration of <20 pM. After mixing, data were taken for >1 h to analyze the sample by counting single molecules. Labeled molecules generate brief fluorescence bursts during their passage through a confocal detection volume (diffusion time ≈ 3.5 ms). Single-molecule events were selected from the signal by threshold criteria and signal intensities, lifetimes (τ), and anisotropies (r) of the donor (D) and acceptor (A) probes were determined. After correcting for mean background signal, spectral cross-talk, direct acceptor excitation and detection efficiencies we obtained the FRET efficiency E (see SI Appendix, section 2.1).
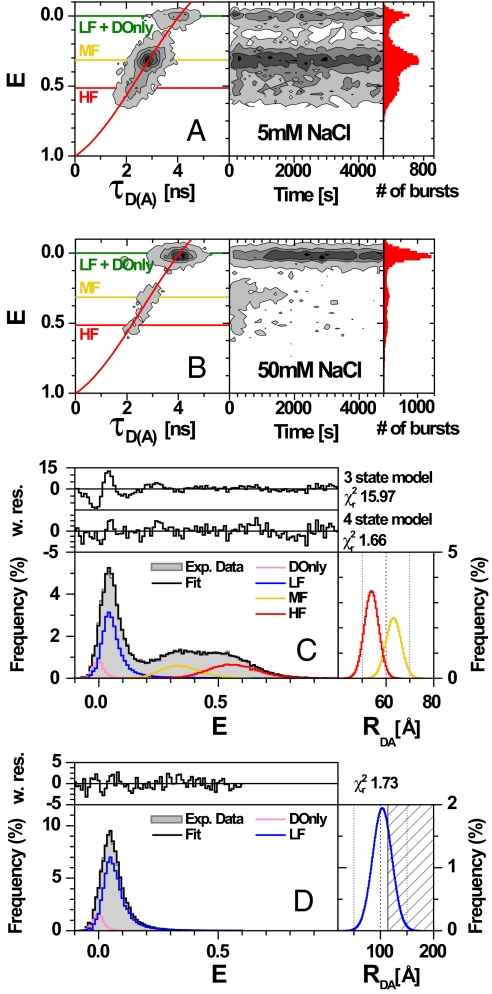
Two-dimensional frequency histograms of E against donor lifetime in the presence of acceptor (τD(A)) are shown in Fig. 2A and B Left. The corresponding 1D parameter histograms are given as projections. To check the stability of the sample, E is plotted against measurement time (Fig. 2 A and B Right).
Fig. 2.
MFD plots of single nucleosomes at different NaCl concentrations. The number of molecules (fluorescent bursts) in each bin is gray scale colored from white (lowest) to black (highest). (A and B) Intensity-based FRET efficiency is plotted versus donor lifetime in the presence of the acceptor, τD(A), (Left) and versus the measurement time (Right). FRET efficiencies were obtained from raw signals S by correcting for green and red background (BG = 2.3 kHz, BR = 1.2 kHz), spectral cross-talk (α = 0.07), detection efficiencies (gG/gR = 0.58), direct A excitation (DE = 1.35 kHz) and the fluorescence quantum yields (ΦFD(0) = 0.70 and ΦFA = 0.70) (see SI Appendix, Eqs. 3 and 4). The red overlaid line (E = 1 − (0.00479 + 0.4813τD(A) + 0.26694τD(A)2 − 0.03435τD(A)3)/4.1) takes linker movements into account (see SI Appendix, Eq. 10). (C and D) PDA of the FRET efficiency of nucleosomes at 25 mM NaCl (C) and free DNA at 5 mM NaCl (D) with the data histogram (gray) for time intervals of 3 ms. The black solid line is the fit to a 4 state model with HF, MF, LF, DOnly (red, yellow, blue, and pink lines, respectively). Weighted residuals are displayed on top of each E histogram. The 1 σ standard deviations were determined by reduced χ2 surfaces (SI Appendix, section 2.4 and Fig. S2E). The fit also includes corrections for multimolecule events. Gaussian distributions for the FRET species yielded mean distances RDA, half-widths HW and fractions xi. The corresponding DA distance distributions of the populations are displayed in the Right Inset. Nucleosomes (C): Four-state model with HF (RDA = 54.0 ± 0.4 Å, HW = 3.0 ± 0.3 Å, xHF = 0.26 ± 0.03), MF (RDA = 63.1 ± 0.4 Å, HW = 3.2 ± 0.3 Å, xMF = 0.19 ± 0.03), LF (RDA = 119 ± 2 Å, HW = 31 ± 2 Å, xLF = 0.47 ± 0.04) and DOnly (E = 0, xDOnly = 0.08 ± 0.01). The residuals of the fit to a 3 and 4 state model are shown for comparison. The corresponding DA distance distributions of the HF and MF populations are displayed in the Right Inset. Free DNA (D): Two-state model with LF (RDA = 103 Å, HW = 18 Å, xLF = 0.89) and DOnly (E = 0, xDOnly = 0.11).
Three populations with different fluorescence properties are immediately distinguishable in Fig. 2A. One small population is characterized by E and τD(A) similar to those of a free donor dye (E ≈ 0 and τD(A) ≈ 4 ns). It displays little to no FRET and will be referred to as Low-FRET and D-Only (LF + DOnly) (for details see Nucleosome Subpopulations Show Distinct Dissociation Properties at Low Salt Concentration and pM Dilution). The other 2 major populations overlap and display higher FRET, and shortening of the donor lifetime. These species are FRET-active and will be denoted as Mid-FRET (MF) (E ≈ 0.32; τD(A) ≈ 2.9 ns) and High-FRET (HF) (E ≈ 0.51; τD(A) ≈ 2.2 ns). Based on the structure in Fig. 1B we may assign MF and HF to conformations in which the DNA is wrapped around the histones and LF to species where the DNA is more extended.
At low salt concentration (Fig. 2A), most events showed FRET and the populations remained stable over time, demonstrating good sample integrity, negligible photobleaching and insignificant sample adsorption to the coverslip. At 50 mM NaCl (Fig. 2B) a large fraction of the FRET population converted into LF (see below).
A detailed MFD analysis ensured that the changes in FRET efficiency were indeed due to different structures and not due to local quenching interactions or orientation artifacts. (i) Analysis of the 2D E-τD(A) frequency histograms showed that the measured data followed the theoretical relationship between τD(A) and E predicted for purely FRET-related D quenching (red curves in Fig. 2 A and B) (see SI Appendix, section 2.2). (ii) To justify dynamic isotropic averaging of donor and acceptor dipole moments, resulting in a mean orientation factor κ2 = 2/3, we analyzed the anisotropies of the fluorophores. Compared to a free DNA, which shows a mean donor rotational correlation time of ρD = 0.63 ns (16) the observed increase in ρD when attached to the nucleosome is small (ρD = 1.16 ns) (see Fig. S1A). The assumption of κ2 = 2/3 is further supported by modeling the sterically allowed fluorophore positions obtained by MD simulations (Fig. 1B) (16) of free DNA. We estimated that 7.4% of all donor and 39% of all acceptor dye positions—accessible in free DNA—were blocked by the proximity of the histones. With the transition dipole moments of the remaining dye positions we computed κ2 = 0.65, a value close to that of mobile dyes attached to free DNA (κ2 = 0.67 ± 0.01). A corresponding analysis of acceptor anisotropy also revealed a single dye population (see Fig. S1 B and C). To conclude, the multiparameter analysis proves that the differences in E are indeed due to varying DA distances.
Nucleosome Subpopulations Show Distinct Dissociation Properties at Low Salt Concentration and pM Dilution.
Previous work showed dissociation of nucleosomes at higher salt and/or at small nucleosome concentration (9, 11, 17). So far no analysis exists of potential conformational changes and intermediates upon nucleosome destabilization. To address this question we analyzed the FRET data by probability distribution analysis (PDA) of the photon counts as described elsewhere (18–20). The FRET efficiency histogram is affected by the stochastic nature of photon detection (shot noise) and other sources of dynamic or static heterogeneity. Using an exact description for the theoretical shot noise distribution, PDA allows us to separate shot noise from inhomogeneous broadening (see SI Appendix, section 2.3).
Fig. 2C shows a PDA of a nucleosome sample at 25 mM NaCl. To minimize statistical uncertainty we combined all bursts of a 1-hour measurement into a single PDA analysis. Judging the quality of the fit by the reduced χr2 and weighted residuals, PDA showed unambiguously that at least 4 species are required to describe the data. Statistical uncertainties (1σ standard deviation) (see SI Appendix, section 2.4) were calculated from multidimensional (Table S1) and from 2D χr2 surfaces (Fig. S2E). Moreover, a fixed DA distance for each FRET species is not sufficient, and Gaussian distance distributions, each with a mean interdye distance (RDA) and a distribution half-width (HW), had to be used instead. The RDA values and standard deviations were 54.0 ± 0.4 Å and 63.1 ± 0.4 Å for the HF (red) and the MF species (yellow), respectively. The half-widths (HW) and standard deviations of 3.0 ± 0.3 Å are essentially identical for both species.
PDA also shows that the broad population near zero energy transfer consists of 2 species: (i) a very broad distribution of double labeled molecules showing low to very low FRET (referred to as Low-FRET (LF), blue line), and (ii) molecules labeled only with a donor (DOnly, pink line). The two are easily distinguishable, because LF shows direct acceptor excitation and a mean FRET efficiency 〈E〉 = 0.047 whereas DOnly had no acceptor signal. At all salt concentrations PDA gave DOnly fractions of 〈xDOnly〉 = 0.09 ± 0.015 that did not change with time. In contrast, the fraction of LF molecules (blue) increased significantly with time at the expense of the FRET species, especially MF (see Relative Stability of Nucleosome Subspecies Depends on NaCl Concentration). The DA distances did not change with time and NaCl concentration (see Table S2a).
To further understand the nature of the LF species, we compared it to free DNA fragments (shown in Fig. 2D), where PDA also required a broad Gaussian distribution with RDA = 103 Å and HW = 18 Å for an appropriate description. The shaded area represents the region with high uncertainties at distances RDA > 2R0. The only relevant contribution to the mean FRET efficiency 〈E〉 = 0.047 comes from the tail of the Gaussian distribution toward higher E (up to E = 0.5) (see SI Appendix, Eq. 13 and Table S2b).
Approximating DNA by a worm-like chain model (see SI Appendix, section 3.2, Eq. 17) a mean FRET efficiency of only 〈E〉 ≅ 0.01 would be expected due to fast bending motions in the microsecond to millisecond range (21). Our FRET analysis, however, yielded a broadly distributed species, which is stable for at least the diffusion time of some milliseconds. We attribute this fraction with DA distances RDA < 100 Å to preexisting sequence-dependent bending or higher bendability in the DNA (22). The fact that the same LF species are also formed by disassembly of nucleosomes indicates extensive histone dissociation at pM concentrations and higher salt concentration (see Nucleosome Dissociation Does Not Completely Proceed to Free DNA).
Finally we checked whether the FRET distribution was broadened due to dynamical changes between substates during the diffusion time. PDA with time windows of 1 and 3 ms (see Fig. S2 C and D), using a model consisting of only static distributions, gave the same fit results independently of the time window used. This implies that the 4 populations did not change during the millisecond dwell time.
Relative Stability of Nucleosome Subspecies Depends on NaCl Concentration.
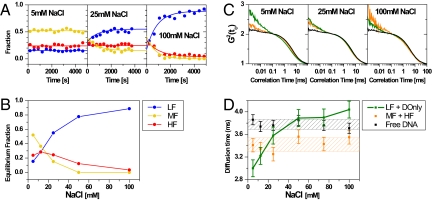
The time evolution of the DA populations at different NaCl concentrations is plotted in Fig. 3A. At 5 mM NaCl both HF and MF populations remained stable over time, whereas at higher salt their proportions decreased significantly, most pronounced for the MF species (e.g., Fig. S2 A and B). This implies that the HF species is considerably more stable. The solid lines describe the interconversion using a model with first order decays for the HF and MF species linked to a growth term for the LF species. A global fit to all time points yields apparent relaxation times for HF and MF of the order of 700 to 1,000 s for all conditions (for details see SI Appendix, section 3.3, Eq. 18 and Table 4 a and b). NaCl concentrations >100 mM rapidly destabilized the samples such that single molecule experiments became impossible. A similar dissociation occurred when we added 2 mM MgCl2 to the buffer.
Fig. 3.
Stability of HF and MF species (A) Time evolution of the 3 different DA species (HF, MF, LF) obtained by PDA for 5, 25 (e.g., Fig. S2 A and B) and 100 mM NaCl. The solid lines are fits using Eq. 18 (SI Appendix, section 3.3) with the results listed in Table S4 a and b. (B) Equilibrium fractions of HF, MF and LF (DOnly not shown) as a function of NaCl concentration. (C) Subensemble-filtered FCS curves for free DNA (black), LF+DOnly (green) and HF+MF (orange) species at 5, 25 and 100 mM NaCl. (D) Characteristic FCS diffusion times of the subspecies as a function of NaCl concentration (for all curves see Fig. S4). The data were obtained by fitting the curves with Eq. 14 (SI Appendix, section 2.5). Shaded areas indicate the 1-σ standard deviation.
The constant initial fractions at time 0 (0.29 ± 0.05, 0.42 ± 0.09, 0.21 ± 0.07 and 0.08 ± 0.02, for the HF, MF, LF and DOnly species respectively) demonstrate that the destabilization is not caused by sample handling but indeed by the salt.
Notably, at moderate salt the HF, MF—populations did not drop to zero, suggesting that these species approached a reversible equilibrium with LF. Fig. 3B summarizes the equilibrium fractions for all 3 species (obtained from a global fit) as a function of NaCl concentration. Although low salt preferentially destabilizes the MF species, at >25 mM NaCl the HF species also starts to dissociate.
High FRET Species Represents the Intact Nucleosome.
To obtain a structural model of the FRET subpopulations, we determined—based on the crystal structure 1KX5—all possible positions that the fluorophores may occupy when they are attached to the nucleosomal DNA via a flexible C6-linker. Only the histone core without the protruding histone tails was considered. Sterically allowed positions of the dyes on DNA were from ref. 16. and aligned to the nucleosome as described in SI Appendix, section 2.6. Views of the nucleosome with aligned donor and acceptor dye clouds defining their accessible space are shown in Fig. 1B. The distance between the 2 mean dye positions, Rmp = 58.6 Å, translates to a mean FRET efficiency of Emp = 0.42. However, experiments do not allow one to measure Emp but rather an isotropic average FRET efficiency 〈E〉iso (see SI Appendix, Eq. 6) with an apparent distance 〈RDA〉isoexp. To compare the experimental results with the model we used all allowed dye coordinates from the model and calculated 〈E〉isomodel = 0.41 and the theoretical apparent distance 〈RDA〉isomodel = 59 Å. For this special case this is very close to Rmp.
For the HF species the PDA derived FRET efficiency is 〈E〉isoexp = 0.55 giving an experimental mean distance of 〈RDA〉isoexp = 54.0 ± 0.4 Å. We assign this species to the intact nucleosome, because it has the most compact structure and the highest stability. The MF species, due to its larger interdye distance of 63.1 ± 0.4 Å (〈E〉isoexp = 0.31) and its reduced stability is probably less compact. Further evidence for this assignment is presented in Consequences of DNA Unwrapping: The Incomplete and Broken Nucleosome.
Nucleosome Dissociation Does Not Completely Proceed to Free DNA.
Transcription through a nucleosome is postulated to proceed without complete histone dissociation (23). FRET alone cannot easily distinguish complete from partial dissociation, the dye separation exceeding the FRET range (> 120 Å). As an additional parameter for the analysis, we computed FCS autocorrelation functions on the data from the nucleosome experiments and compared them to pure DNA fragments. To separate the LF+DOnly population from contributions by scattered light and by the FRET species (MF and HF), we extended the theory of Enderlein (24) and applied spectrally resolved fluorescence lifetime filters (SI Appendix, section 2.5). Subensemble-filtered FCS curves for the nucleosome sample are compared to free DNA in Fig. 3C. At 25 mM NaCl, the diffusion times increased in the order FRET (HF+MF), LF+DOnly and free DNA. Unlike free DNA, all nucleosomal species displayed a distinct correlation term on the μs-timescale. Although we cannot assign this term to any particular process, we took it as indication that the LF species was distinct from free DNA, probably still associated with histones.
The NaCl dependence of the diffusion times in Fig. 3D allows further insight into the different species. The FRET species (HF, MF) had constant diffusion times at all [NaCl], suggesting that their shape did not change significantly before disassembly. Because the diffusion times did not depend on the HF/MF ratio (see Fig. 3B), MF likely has a hydrodynamic shape similar to a complete nucleosome (HF) with the DNA more loosely bound (lower FRET). We assign MF to an incomplete nucleosome.
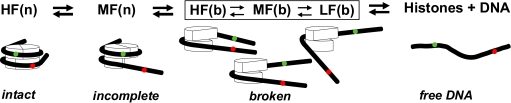
At 5 mM NaCl the LF+DOnly subensemble diffuses fastest. This finding makes an extended DNA structure unlikely: A very open 170-bp DNA should diffuse more slowly than the nucleosome. Because LF is dominant at higher salt (Fig. 3B), we conclude that the fast-diffusing contribution at low salt is caused by minor DOnly-like contaminations that are masked by the dissociated nucleosomal DNA at higher salt. Because the diffusion time of the LF+DOnly subensemble approaches that of free DNA for medium salt and slightly exceeds it at high salt, we propose a subspecies lacking histones referred to as broken nucleosome (see Fig. 6). At high salt the LF species likely includes a lot of free DNA because the amplitude of the μs bunching term in the autocorrelation function drops continuously.
Fig. 6.
Diagram of the possible nucleosomal species. Nucleosomes lacking the contact between DNA and one H2A/H2B dimer are indicated as Incomplete, whereas nucleosomes lacking the contact with >1 H2A/H2B dimer are indicated as Broken. Depending on the progress of disassembly the width of the E-distributions of the species LF, MF and HF can be either narrow (n) or broad (b).
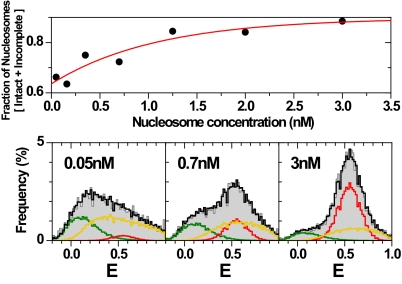
Nucleosome dissociation at higher salt can be suppressed by an excess of unlabeled mononucleosomes (Fig. 4Upper), shifting the equilibrium toward the compact nucleosome (HF) (Fig. 4 Lower). Dilution causes an increase of the MF species without changing the DA distances. A similar redistribution between closed and opened nucleosome structures upon dilution was observed earlier through the distances between dyes attached to the ends of the nucleosomal DNA (11). We estimate the dissociation constant for the transition between HF and MF to be in the nM range at 100 mM NaCl.
Fig. 4.
Unfolding of nucleosomes. (Upper) Concentration-dependent stability of nucleosomes at 100 mM NaCl as a function of total nucleosome concentration. For each data point 50 pM labeled nucleosomes were mixed with unlabeled 601 nucleosomes to obtain the final concentration and measured for 60 min (11). The fractions of the different species were computed through PDA as shown below with the results listed in Table S4c. The red line is a single exponential fit: y = 0.988–0.262 × exp(−x/1.083). (Lower) Three representative E-distributions are shown for 0.05, 0.7 and 3 nM nucleosomes. The fit (black line) of the E-distributions (gray histograms) is decomposed by PDA into 3 populations: HF (red line), MF (yellow line) and combined LF+DOnly (green line). The following parameters were fixed in the PDA fits: HF (RHF = 54.5 Å, HWHF = 3.02 Å; obtained at 3 nM), LF (RLF = 75.1 Å, HWLF = 10.3 Å; obtained from free DNA (data not shown)) and DOnly, (single distance RDA → ∞). Only the properties of MF (RMF, HWMF) and the fractions x of all species were varied. The fit results are: (0.05 nM) RMF = 57.3 Å, HWMF = 12 Å, xMF = 0.61, xHF = 0.05, xLF = 0.24, xDOnly = 0.10; (0.7 nM) RMF = 53.6 Å, HWMF = 9.4 Å, xMF = 0.47, xHF = 0.25, xLF = 0.20, xDOnly = 0.08; (3 nM) RMF = 52.5 Å, HWMF = 8.7 Å, xMF = 0.27, xHF = 0.61, xLF = 0.07, xDOnly = 0.04.
Geometric Model for Unwrapping of Nucleosomal DNA Explains the Broad Distribution of FRET Species.
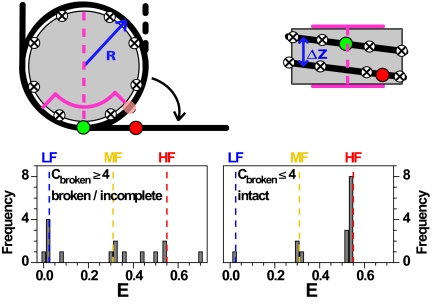
To describe the FRET species during DNA dissociation, we computed E histograms using a geometric model based on the crystal structure and dye positions in Fig. 1B (Fig. 5 and SI Appendix, section 2.7). We considered several dissociation modes (Fig. S3): (i) quasi-continuous with step size 1 bp; (ii) stepwise at defined “contact points” (crossed circles in Fig. 5) from either end in 10 to 11 bp steps. The contact points C were identified from the X-ray structure (summarized in Table S3); and (iii) stepwise in 5- to 6-bp steps as observed recently for mechanical unzipping of DNA (25). All combinations of dissociated contacts of the donor and acceptor arm were considered to be equally probable. In reality this assumption may be violated, but this will change only amplitudes, and not the peak positions in the FRET histograms.
Fig. 5.
Geometric model for nucleosome unfolding. Nucleosomal DNA (black line) is wrapped around a cylindrical histone core (gray). Dyad (magenta dashed line), region with strong interactions (magenta line), contact points (circled crosses), donor and acceptor dyes (green and red spheres). In the top view, the semitransparent red circle represents the position of the acceptor in the fully wrapped nucleosome. The detached DNA segments were straight and tangential to the nucleosome core at the detachment point(s). Model parameters were estimated from the X-ray structure: Effective nucleosome radius R = 40 Å, rise per turn Δz = 45 Å, 80 bp per nucleosome turn, DNA length of 170 bp, donor dye position at 46.5 bp, acceptor dye position 136.5 bp, scaled Förster radius ßR0 = 61.2 Å with ß = 1.1 (for details see SI Appendix, section 2.7). (Lower) Possible FRET efficiency values from the geometrical model with contact points 10 bp apart, showing values obtained for the loss of ≤4 contact points (Right) and ≥4 contact points (Left).
Using this model, we computed FRET histograms for nucleosome intermediates with partly dissociated DNA at the large step size of 10 bp. For 1–4 broken contact points, mainly 2 narrow distributions of FRET species are expected (Right), which nicely agree with the characteristic values measured for the MF and HF species. Four to eight lost contact points result in a broad E distribution ranging from LF to HF (Left). Using only 5-bp dissociation steps would result in additional FRET states between the MF and HF species (Fig. S3 G–I). Even smaller step sizes agree less well with the experiment. A quasi-continuous dissociation in 1-bp steps cannot reproduce the characteristic E peaks of LF and MF (see Fig. S3 A–C).
To uncover eventual intermediate FRET states hidden in the broad MF-HF-distribution as expected for a 5-bp step size, we applied PDA with a model-free approach based on the maximum entropy method (MEM) as an alternative to the Gaussian distance distributions with half-widths of 3.0 Å to describe the MF and HF species in Fig. 2C. MEM-analysis (Fig. S2 F and G) indeed indicates distinct populations of intermediate FRET states, which are less separated at higher salt concentrations. It seems that the loss of the crystallographic contact points is the most important barrier for the dissociation of ds DNA (10-bp step), but intermediate smaller barriers also exist (5-bp step), which are especially visible at low electrostatic shielding (Fig. S2G).
Our result, obtained without external forces, nicely agrees with recent data obtained by mechanical unzipping of nucleosomal DNA, which revealed 3 broad structured barriers in the course of dissociation of DNA from the octamer: two lower ones ranging from +30 bp to +50 bp and −30 to −50 bp from the dyad axis (magenta quadrant in Fig. 5) and a higher one in the vicinity of the dyad (25). Our proposed MF species corresponds to states with the outer bases already released but none of these barriers completely overcome, whereas in the LF states the lower barriers are already open. Moreover, the disassembly intermediates map the energy landscape indicating major barriers for 10-bp and minor ones for 5-bp DNA unwinding steps.
Consequences of DNA Unwrapping: The Incomplete and Broken Nucleosome.
Fig. 6 sketches possible disassembly intermediates. Besides FRET efficiency, the width of the individual distributions is also characteristic. Depending on the progress of disassembly the width of the LF, MF and HF distributions can be either narrow (n) or broad (b), i.e., an HF signal is not per se indicative of an intact nucleosome. Thus, the FRET peak width, presence of other FRET species and measurement conditions are essential to identify the nucleosome species. Besides the prediction of the geometric model there is other evidence that the MF species (RDA ∼ 63 Å, 〈E〉 = 0.31) represents a dissociation intermediate: (i) its accelerated dissociation at higher salt indicates a less stable conformation (Fig. 3 A and B); (ii) its stability depends on the absolute nucleosome concentration—under bulk conditions this population is only a minor species compared to the intact nucleosome, whereas at pM concentrations it comprises the dominant species (Fig. 4, Lower); and (iii) the structure is still quite compact with hydrodynamic properties similar to the intact nucleosome (Fig. 3D).
Our data support the idea that nucleosome dissociation is initiated by H2A/H2B dimer release, probably facilitated by transient unwrapping of one DNA arm. Unwrapping was shown to occur in nucleosomes with apparent lifetimes of the unwrapped state of 100–250 ms (9, 26). During this time the dimer might partially dissociate from the histone core and prevent DNA rewrapping, leaving several tens of base pairs exposed. Transient opening is also suggested by recent simulations of nucleosome dynamics (27). Assuming that the DNA stays fully attached to the remaining hexamer, the geometric model predicts an average FRET efficiency 〈E〉 ≅ 0.32, in very good agreement with the experimental 〈E〉 ≅ 0.31.
One possible broken nucleosome species may be a tetrasome where both H2A/H2B are dissociated; the (H3H4)2 tetramer remains attached to the DNA, most likely at the innermost 20–30 bp near the dyad axis. Crystallographic data (2) and force rupture experiments (28) showed these sites to harbor the most intense DNA-histone interactions.
Conclusion
Confocal single molecule FRET analysis of mononucleosome populations shows at least 4 separable molecular subspecies differing in FRET efficiency (HF, MF, LF and DOnly). Analysis of the salt-dependent stability identified these with intermediates in nucleosome disassembly, possibly through partial dissociation starting from one of the linker DNA ends. A stepwise dissociation at salt concentrations exceeding physiological ionic strength has also been suggested in recent bulk FRET fluorimetry (29) and a related histone-histone FRET study (6). Although the latter suggested a local disruption of dimer-tetramer contacts, it did not provide evidence for complete dimer release. Further bulk in vivo and in vitro data on potential subnucleosomal structures has been reviewed recently (30). Neither of these studies quantified the coexistence of subspecies under non-perturbative conditions (zero forces, free diffusion, physiological salt and no enzymes involved) although. Our single molecule approach enabled us to follow the interconversion of nucleosomal subspecies and to provide yet unknown structural details on nucleosomes in free solution. Experiments like these have just opened the route for a much more detailed mapping of the energy landscape of nucleosome and chromatin structures, using DNA-histone FRET and systematic variation of label positions.
Materials and Methods
Preparation of Labeled Nucleosomes.
Mononucleosomes were reconstituted by salt dialysis from a 170-bp DNA fragment containing a positioning sequence (Selex “Widom 601”, sequence; see ref. 31) and recombinant X. laevis histones. The DNA was labeled via thymine with C6-linker at 33 bp (Alexa Fluor 594) and 45 bp (Alexa Fluor 488) from each end. Only samples yielding >80% nucleosomes were accepted. For details see refs. 11 and 31 and SI Appendix, section 1.1.
Single-Molecule Fluorescence Spectroscopy.
The experiments were carried out with a confocal epi-illuminated set-up (13) with spectral detection windows for donor (520/66 nm) and acceptor (630/60 nm). The fluorescently labeled complexes were excited by a linearly polarized, active-mode-locked Argon-ion laser (476 nm, 73 MHz, 150 ps). Fluorescence bursts are distinguished from the background of 3–3.5 kHz by applying threshold intensity criteria defined by 0.1-ms interphoton time and 150 photons minimum per burst. All measurements were done at 20 °C in TE buffer (10 mM Tris, 0.1 mM EDTA pH = 7.5) supplemented with 0.1 g/L BSA, 1 mM ascorbic acid and NaCl as stated. Experiments were essentially done as in refs. 13 and 14 (for details, see SI Appendix, section 1.2).
Supplementary Material
Acknowledgments.
We thank Paul Rothwell and Evangelos Sisamakis for helpful discussions and suggestions. This work was supported by German Science foundation (DFG) in the priority program “Optical analysis of the structure and dynamics of supramolecular biological complexes” Grant SE 843/9-3 and SFB 590 (to J.L. and C.A.M.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.L.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903005106/DCSupplemental.
References
- 1.Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 2.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 3.Kulic IM, Schiessel H. Nucleosome repositioning via loop formation. Biophys J. 2003;84:3197–3211. doi: 10.1016/S0006-3495(03)70044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lovullo D, Daniel D, Yodh J, Lohr D, Woodbury NW. A fluorescence resonance energy transfer-based probe to monitor nucleosome structure. Anal Biochem. 2005;341:165–172. doi: 10.1016/j.ab.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 5.Tóth K, Brun N, Langowski J. Chromatin compaction at the mononucleosome level. Biochemistry. 2006;45:1591–1598. doi: 10.1021/bi052110u. [DOI] [PubMed] [Google Scholar]
- 6.Hoch DA, Stratton JJ, Gloss LM. Protein–protein Förster resonance energy transfer analysis of nucleosome core particles containing H2A and H2A.Z. J Mol Biol. 2007;371:971–988. doi: 10.1016/j.jmb.2007.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ha T, et al. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuler B, Lipman EA, Eaton WA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 9.Koopmans WJ, Brehm A, Logie C, Schmidt T, van Noort J. Single-pair FRET microscopy reveals mononucleosome dynamics. J Fluoresc. 2007;17:785–795. doi: 10.1007/s10895-007-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomschik M, Zheng H, van Holde K, Zlatanova J, Leuba SH. Fast, long-range, reversible conformational fluctuations in nucleosomes revealed by single-pair fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2005;102:3278–3283. doi: 10.1073/pnas.0500189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gansen A, Tóth K, Schwarz N, Langowski J. Structural variability of nucleosomes detected by single-pair Förster resonance energy transfer: Histone acetylation, sequence variation, and salt effects. J Phys Chem B. 2009;113:2604–2613. doi: 10.1021/jp7114737. [DOI] [PubMed] [Google Scholar]
- 12.Kelbauskas L, et al. Sequence-dependent variations associated with H2A/H2B depletion of nucleosomes. Biophys J. 2008;94:147–158. doi: 10.1529/biophysj.107.111906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Widengren J, et al. Single-molecule detection and identification of multiple species by multiparameter fluorescence detection. Anal Chem. 2006;78:2039–2050. doi: 10.1021/ac0522759. [DOI] [PubMed] [Google Scholar]
- 14.Rothwell PJ, et al. Multi-parameter single-molecule fluorescence spectroscopy reveals heterogeneity of HIV-1 reverse transcriptase:primer/template complexes. Proc Natl Acad Sci USA. 2003;100:1655–1660. doi: 10.1073/pnas.0434003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thastrom A, et al. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 16.Woźniak AK, Schröder G, Grubmüller H, Seidel CAM, Oesterhelt F. Single molecule FRET measures bends and kinks in DNA. Proc Natl Acad Sci USA. 2008;105:18337–18342. doi: 10.1073/pnas.0800977105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claudet C, Angelov D, Bouvet P, Dimitrov S, Bednar J. Histone octamer instability under single molecule experiment conditions. J Biol Chem. 2005;280:19958–19965. doi: 10.1074/jbc.M500121200. [DOI] [PubMed] [Google Scholar]
- 18.Antonik M, Felekyan S, Gaiduk A, Seidel CAM. Separating structural heterogeneities from stochastic variations in fluorescence resonance energy transfer distributions via photon distribution analysis. J Phys Chem B. 2006;110:6970–6978. doi: 10.1021/jp057257+. [DOI] [PubMed] [Google Scholar]
- 19.Kalinin S, Felekyan S, Antonik M, Seidel CAM. Probability distribution analysis of single-molecule fluorescence anisotropy and resonance energy transfer. J Phys Chem B. 2007;111:10253–10262. doi: 10.1021/jp072293p. [DOI] [PubMed] [Google Scholar]
- 20.Kalinin S, Felekyan S, Valeri A, Seidel CAM. Characterizing multiple molecular states in single-molecule multiparameter fluorescence detection by probability distribution analysis. J Phys Chem B. 2008;112:8361–8374. doi: 10.1021/jp711942q. [DOI] [PubMed] [Google Scholar]
- 21.Merlitz H, Rippe K, Klenin KV, Langowski J. Looping dynamics of linear DNA molecules and the effect of DNA curvature: A study by Brownian dynamics simulation. Biophys J. 1998;74:773–779. doi: 10.1016/S0006-3495(98)74002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruscio JZ, Onufriev A. A computational study of nucleosomal DNA flexibility. Biophys J. 2006;91:4121–4132. doi: 10.1529/biophysj.106.082099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark DJ, Felsenfeld G. A nucleosome core is transferred out of the path of a transcribing polymerase. Cell. 1992;71:11–22. doi: 10.1016/0092-8674(92)90262-b. [DOI] [PubMed] [Google Scholar]
- 24.Enderlein J, Gregor I. Using fluorescence lifetime for discriminating detector afterpulsing in fluorescence-correlation spectroscopy. Rev Sci Instrum. 2005;76 033102. [Google Scholar]
- 25.Hall MA, et al. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 27.Voltz K, et al. Coarse-grained force field for the nucleosome from self-consistent multiscaling. J Comput Chem. 2008;29:1429–1439. doi: 10.1002/jcc.20902. [DOI] [PubMed] [Google Scholar]
- 28.Brower-Toland B, et al. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J Mol Biol. 2005;346:135–146. doi: 10.1016/j.jmb.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 29.Park YJ, Chodaparambil JV, Bao YH, McBryant SJ, Luger K. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J Biol Chem. 2005;280:1817–1825. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- 30.Zlatanova J, Bishop TC, Victor JM, Jackson V, van Holde K. The nucleosome family: Dynamic and growing. Structure. 2009;17:160–171. doi: 10.1016/j.str.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 31.Gansen A, Hauger F, Tóth K, Langowski J. Single-pair fluorescence resonance energy transfer of nucleosomes in free diffusion: Optimizing stability and resolution of subpopulations. Anal Biochem. 2007;368:193–204. doi: 10.1016/j.ab.2007.04.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.