Abstract
Claudins are tight junction integral membrane proteins that are key regulators of the paracellular pathway. Defects in claudin-16 (CLDN16) and CLDN19 function result in the inherited human renal disorder familial hypomagnesemia with hypercalciuria and nephrocalcinosis (FHHNC). Previous studies showed that siRNA knockdown of CLDN16 in mice results in a mouse model for FHHNC. Here, we show that CLDN19-siRNA mice also developed the FHHNC symptoms of chronic renal wasting of magnesium and calcium together with defective renal salt handling. siRNA knockdown of CLDN19 caused a loss of CLDN16 from tight junctions in the thick ascending limb (TAL) without a decrease in CLDN16 expression level, whereas siRNA knockdown of CLDN16 produced a similar effect on CLDN19. In both mouse lines, CLDN10, CLDN18, occludin, and ZO-1, normal constituents of TAL tight junctions, remained correctly localized. CLDN16- and CLDN19-depleted tight junctions had normal barrier function but defective ion selectivity. These data, together with yeast two-hybrid binding studies, indicate that a heteromeric CLDN16 and CLDN19 interaction was required for assembling them into the tight junction structure and generating cation-selective paracellular channels.
Keywords: hypomagnesemia, transgenic animal, siRNA, paracellular ionic channel, renal calcium wasting
Tight junctions (TJs) play a key role in mediating paracellular ion reabsorption in epithelia. TJs are composed of three transmembrane proteins, occludin, claudins, and junctional adhesion molecule (1). The claudins are a 24-member family of tetraspan proteins that range in molecular mass from 20 to 28 kDa (1, 2). Claudin and occludin are the major components of the branching and anastomosing network of tight junctional strands in the plasma membrane revealed by freeze-fracture microscopy (3, 4). It has been hypothesized that claudin oligomerization occurs before strand assembly on the basis of claudin-4 (CLDN4) expression studies in insect cells that do not form TJs (5) and exhibit ≈10-nm-sized multimers. After trafficking to the cell surface, it is believed that oligomerized claudins then assemble into the TJ strands where they interact with cognate claudins in the adjacent cell (1, 6). Assembly of claudins into TJ strands requires the TJ scaffold proteins ZO-1 or ZO-2, which interact with both claudin PDZ binding domains (7–10) and TJ peripheral proteins such as cingulin, Par-3, and Par-6 (11–13).
Familial hypomagnesemia with hypercalciuria and nephrocalcinosis (FHHNC) is a human hereditary disorder caused by mutations in the TJ proteins CLDN16 (14) and CLDN19 (15). The expression of CLDN16 is restricted to the thick ascending limb (TAL) of the nephron (16). CLDN16-deficient mice exhibit defects in paracellular cation selectivity and develop severe renal wasting of magnesium and calcium (17) similar to that seen in the human disease. In the kidney, CLDN19 is also exclusive to the TAL (15). In vitro, CLDN16 and CLDN19 interact and form a cation-selective TJ paracellular channel (18). On the basis of these observations, we hypothesized that the CLDN19-deficient animals will phenocopy the CLDN16 knockdown (KD) and that CLDN16–CLDN19 interaction is a critical component of paracellular cation selectivity in the TAL.
Here, we find that the renal handling of electrolytes by CLDN19-deficient mice is very similar to that of CLDN16-deficient mice. TAL TJs contain not only CLDN16 and CLDN19 but also CLDN10, CLDN18, and occludin. Depletion of CLDN19 suppressed accumulation of CLDN16 in TAL TJs without a decrease in CLDN16 expression level, whereas knockdown of CLDN16 produced a similar effect on CLDN19. Two-hybrid experiments revealed stronger interactions between CLDN16 and CLDN19 than with the other two claudins in the TAL. These data suggest that the CLDN16–CLDN19 oligomerization occurs before assembly of either claudin into the TJ and that the oligomerization is required for trafficking to the cell surface. Furthermore, neither loss of CLDN16 nor CLDN19 affected the junctional localization of CLDN10, CLDN18, ZO-1, or occludin, suggesting that CLDN10, CLDN18, and/or occludin are largely responsible for the TAL TJ barrier function as assessed by transepithelial resistance, which is unaffected in the CLDN16 KD. Together, these data indicate that CLDN19 oligomerizes with CLDN16, and that this interaction is required for their assembly into the TAL TJs that provide key properties of cation selectivity, underlying the mechanism of FHHNC pathogenesis.
Results
Loss of CLDN19 Resulted in Renal Wasting of Magnesium and Calcium.
We previously described a CLDN16 KD animal model that recapitulates human FHHNC phenotypes (17). Human mutations in CLDN19 also result in FHHNC (15). By using identical strategies to developing our CLDN16 KD mouse lines, we first tested siRNA sequences within the CLDN19 coding region in cultured mouse TAL cells (see Methods and Fig. S1) to determine optimal siRNA constructs for knocking down intracellular CLDN19 mRNA. Sequences 438 (GAAGTGCACTCGGGTTGGA) and 517 (GCAGGTCTCTGTACTTTGA) were highly effective in vitro (>100-fold knockdown) (Fig. S2). These sequences were then cloned into the pUG-U6 vector (17), upstream of the red fluorescent protein (“cherry”) driven by the ubiquitin-C promoter. After testing of the lentivirus in cultured mouse TAL cells, transgenic mouse lines were generated by perivitelline injection of virus to fertilized mouse embryos. Transgenic F0 pups were directly selected on the basis of fluorescent skin (Fig. S3). Fluorescent mice were then outbred with wild-type (WT) animals to expand the colony. Immunohistochemical analysis of kidneys from the transgenic mice revealed a loss of the CLDN19 protein from the TAL cells (Fig. S4). Preliminary analyses of plasma and urine electrolytes found hypomagnesemia and hypercalciuria in both pUG-U6-#438 and pUG-U6-#517 transgenic mouse lines (Table S1).
To rigorously analyze the renal handling of Mg2+ and Ca2+, we performed renal clearance measurements on CLDN19 KD mice (pUG-U6-#517) and their littermate WT controls. Age (9–12 weeks old)- and sex (female)-matched animals from each group were analyzed and elaborated below. The plasma Mg2+ level in CLDN19 KD mice was significantly lower (≈22%) than in WT, whereas the Ca2+ level was not significantly altered (Table 1). Urinalysis showed that CLDN19 KD mice were losing both Mg2+ and Ca2+ more rapidly than WT mice. The absolute excretion of Mg2+ and Ca2+ in KD mice (EMg and ECa) was 1.6- and 3.0-fold higher, respectively, than in WT, whereas the fractional excretion (FEMg% and FECa%) was increased by 2.3- and 3.0-fold, respectively. These phenotypes closely resembled those of CLDN16 KD animals (17).
Table 1.
Analyses of plasma and urine electrolytes in WT and CLDN19 KD mice
| WT | CLDN19 KD | Significance | |
|---|---|---|---|
| UV, μL/min/100 g | 9.29 ± 0.85 | 10.03 ± 0.78 | NS |
| GFR, mL/min/100 g | 0.87 ± 0.06 | 0.79 ± 0.08 | NS |
| Osm, mOsmo/kg | 994.9 ± 54.6 | 1,073.6 ± 82.3 | NS |
| PMg, mM | 0.83 ± 0.03 | 0.65 ± 0.04 | P < 0.05 |
| EMg, nmol/min/100 g | 138.56 ± 15.45 | 220.37 ± 16.72 | P < 0.05 |
| FEMg, % | 19.83 ± 2.14 | 46.09 ± 5.22 | P < 0.01 |
| PCa, mM | 2.13 ± 0.07 | 2.25 ± 0.08 | NS |
| ECa, nmol/min/100 g | 11.94 ± 1.66 | 35.83 ± 3.73 | P < 0.01 |
| FECa, % | 0.69 ± 0.13 | 2.10 ± 0.17 | P < 0.01 |
| PNa, mM | 137.2 ± 1.4 | 136.0 ± 1.2 | NS |
| ENa, μmol/min/100 g | 1.68 ± 0.20 | 2.06 ± 0.25 | NS |
| FENa, % | 1.40 ± 0.11 | 1.91 ± 0.16 | NS |
| PK, mM | 3.89 ± 0.13 | 4.13 ± 0.16 | NS |
| EK, nmol/min/100 g | 309.27 ± 84.66 | 610.58 ± 68.93 | P < 0.01 |
| FEK, % | 9.04 ± 2.13 | 18.93 ± 1.21 | P < 0.01 |
Values are expressed as means ± SE; n = 8, the number of animals; sex, female; age, 9–12 weeks.UV, urine volume; NS, not significant; GFR, glomerular filtration rate; Osm, urine osmolality; PMg, PCa, PNa, PK, plasma Mg2+, Ca2+, Na+, and K+ concentrations, respectively; EMg, ECa, ENa, EK, absolute excretion of Mg2+, Ca2+, Na+, and K+, respectively; FEMg, FECa, FENa, FEK, fractional excretion of Mg2+, Ca2+, Na+, and K+, respectively.
We next examined the effects of CLDN19 KD on renal salt handling and again found values comparable with those published for the CLDN16 KD line (17, 19). Specifically, we analyzed CLDN19 KD mice for changes in the circulating and urinary levels of Na+ and K+. Neither the plasma level of Na+ nor its urinary excretion was significantly affected in the CLDN19 KD (Table 1). However, the plasma aldosterone levels were significantly elevated in CLDN19 KD mice (573.1 ± 80.4 pg/mL), compared with WT (297.5 ± 41.3 pg/mL; n = 8; P < 0.05). A possible explanation would be that increases in TAL NaCl concentration at the macula densa were activating the renin–angiotensin–aldosterone system. Higher aldosterone levels would elicit compensatory Na+ reabsorption accompanied by kaliuresis. Indeed, the fractional excretion of K+ (Table 1, FEK) in CLDN19 KD was increased 2.1-fold over WT. There was no significant difference in glomerular filtration rate (GFR), urinary volume (UV), and osmolality (Osm) between WT and KD (Table 1); thus, loss of CLDN19 did not result in a urine-concentrating defect.
In summary, CLDN19 KD mice showed chronic renal wasting of Mg2+ and Ca2+ and perturbed renal handling of Na+ and K+, both of which would result from a defective cation permselectivity in the TAL. The plasma and urine electrolyte abnormalities in CLDN19 KD mice were highly similar to those reported in CLDN16 KD mice (17), indicating these two claudins functioned in the same pathway.
Assembly of CLDN16 and CLDN19 into TJ Strands Required CLDN16–CLDN19 Interaction.
In vitro data suggest that interaction between CLDN16 and CLDN19 may be necessary for cation permselectivity of TAL TJs. The two claudins can be coimmunoprecipitated from doubly transfected renal epithelial cells, and coexpression of CLDN16 and CLDN19 in renal epithelial cells generates paracellular cation selectivity, whereas expression of either claudin alone does not. Furthermore, mutations that interfere with their ability to interact also abolish the cationic permselectivity (18). Taken together, these data suggest that cooligomerization of CLDN16 with CLDN19 is required for TAL paracellular channel function.
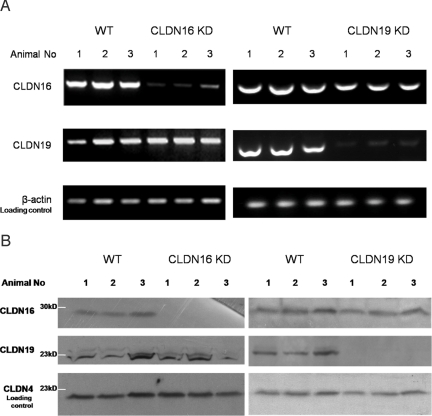
To test this hypothesis in vivo, we looked for changes in the TJ localization of CLDN16 in the CLDN19 KD kidney. We used the characteristic interdigitated appearance of TAL TJs to distinguish TAL from proximal tubules and the collecting duct (Fig. S5). In WT mouse kidneys, CLDN16 was found exclusively in the TJ of the TAL (Fig. S6 and Fig. S1A), consistent with previous reports (16, 17). In CLDN19 KD mouse kidneys, CLDN16 staining completely disappeared from TJs of the TAL (Fig. 1B). However, the mRNA (see Fig. 2A and Table S2) levels of CLDN16 were not affected in the CLDN19 KD, ruling out the possibility that CLDN19 was involved in regulation of CLDN16 transcription. The CLDN16 protein levels slightly decreased in CLDN19 KD (see Fig. 2B) [area densitometry (AD), ADCLDN16/ADloading control: WT, 0.74 ± 0.02, versus CLDN19 KD, 0.57 ± 0.03; P < 0.01; n = 3]. Although CLDN19 may have a role in CLDN16 protein biosynthesis or degradation, our data indicate that CLDN19 was required for CLDN16 assembly into TAL TJs.
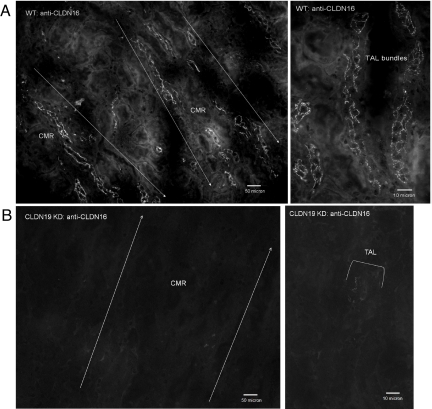
Fig. 1.
Immunofluorescence analysis of CLDN16 protein localization in the kidney. Cryostat sagittal sections (10 μm) from WT mouse kidneys (A) (Right, high magnification) show CLDN16 localization in the TJs of TAL tubules in the corticomedullary rays (CMR). In CLDN19 KD mouse kidneys, CLDN16 staining completely disappears from the TJs of the TAL (B).
Fig. 2.
Analysis of mRNA and protein levels of CLDN16 and CLDN19 in the kidney. (A) RT-PCR measurements showing CLDN16 and CLDN19 mRNA transcript levels in mouse kidneys of WT, CLDN16 KD, and CLDN19 KD (n = 3). (B) Western immunoblots showing CLDN16 and CLDN19 protein levels in the kidneys of WT, CLDN16 KD, and CLDN19 KD mice (n = 3).
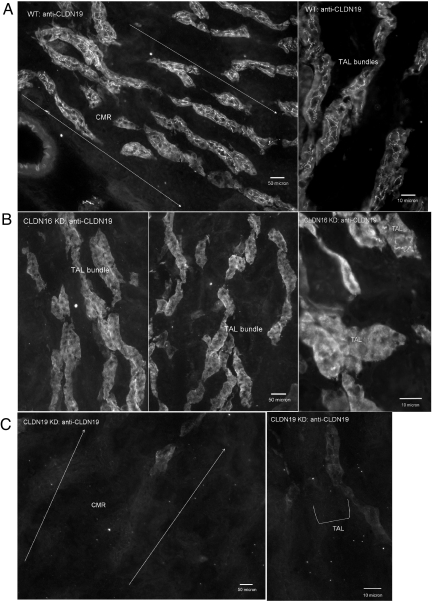
CLDN19 localization in WT mouse kidneys was also restricted to TJs in the TAL (Fig. 3A), consistent with previous findings (15). Strikingly, we found that the majority of CLDN19 TJ immunostaining was lost in CLDN16 KD kidneys (Fig. 3B), with occasional staining spotted in isolated cells. This is consistent with our earlier findings that transgenic expression variegation resulted in a small number of epithelial cells (<5%) escaping siRNA knockdown in the kidney (17). CLDN19 intracellular staining was seen in both WT and CLDN16 KD, but not in CLDN19 KD (Fig. 3C), suggesting a large reservoir of unassembled CLDN19 protein. The mRNA (Fig. 2A and Table S2) and protein (Fig. 2B) levels of CLDN19 in the CLDN16 KD were equal to WT.
Fig. 3.
Immunofluorescence analysis of CLDN19 protein localization in the kidney. Cryostat sagittal sections (10 μm) from WT mouse kidneys (A) (Right, high magnification) show CLDN19 localization in the TJs of TAL tubules in the corticomedullary rays (CMR). In CLDN16 KD mouse kidneys, CLDN19 staining is completely lost in TJs of the TAL (B). A strong intracellular background is seen in both WT and CLDN16 KD specimens. In CLDN19 KD mouse kidneys, CLDN19 staining disappears from both intracellular and junctional areas in the TAL (C).
In summary, deletion of CLDN19 prohibited assembly of CLDN16 into TAL TJs, whereas deletion of CLDN16 has a similar effect on CLDN19. These data provide a clear explanation as to why the phenotypes of CLDN16 KD and CLDN19 KD mice were similar: removal of either claudin created a double deletion at the level of the TJ.
TJ Barrier Function in the Absence of CLDN16 and CLDN19 Is Supplied by CLDN10, CLDN18, and Occludin.
Physiological study of isolated perfused tubules from CLDN16 KD kidneys revealed that, although permselectivity is altered, there is not a loss of TJ barrier function as the mean transepithelial resistance (Rte) is not significantly different (17). Because we have shown here that CLDN16 KD animals also lacked CLDN19 in the TAL TJs, it was clear that the loss of both CLDN16 and CLDN19 does not affect barrier function. Therefore, we looked to see which other components might be present at TAL TJs. Kiuchi-Saishin et al. (16) examined the expression pattern of claudins in the kidney and found CLDN3, -10, -11, and -16 in the TAL. Our antibodies failed to detect any renal expression of CLDN11 and found CLDN3 exclusively in the collecting duct (Fig. S7, CLDN3 staining). We also screened for TAL localization of claudins 1, 2, 4, 5, 7, 8, 10, 12, 13, 14, 15, and 18. Of these, we found only CLDN10 and CLDN18 in the TAL (Fig. S8), although their expression was not restricted to this portion of the nephron. CLDN10 and CLDN18 showed strong TJ staining in the TAL in WT, CLDN16 KD, and CLDN19 KD mice (Fig. S9 C and D), suggesting CLDN10 and CLDN18 were not associated with the CLDN16–CLDN19 complex. The protein expression levels of CLDN10 and CLDN18 in the kidney were not affected by the loss of CLDN16 or CLDN19 (Fig. S10). In addition, the localizations of ZO-1 and occludin were grossly normal in CLDN16 KD and CLDN19 KD mice (Fig. S9 A and B).
CLDN16 Interacts Strongly only with CLDN19, Whereas CLDN19 Can Interact with Other TAL Claudins.
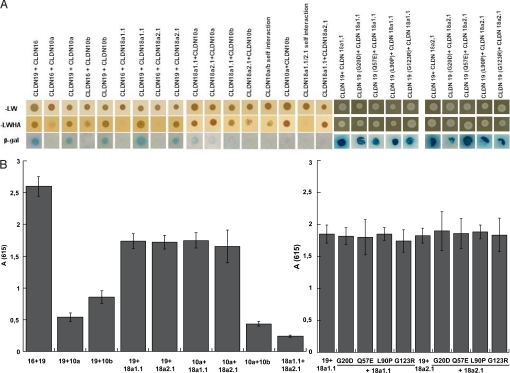
To determine the ability of TAL claudins to physically interact, we used the split-ubiquitin yeast two-hybrid (Y2H) assay as described in ref. 18. CLDN10 has two splice variants (a and b) encoding proteins that differ in the N terminus and the first extracellular loop (ECL) (20). CLDN18 has four splice variants, a1.1, a1.2, a2.1, and a2.2 (21). CLDN18 a1.2 and a2.2 encode C-terminal truncations. Because CLDN18 antibodies used in our study were raised against C-terminal sequences, we were unable to assay CLDN18 a1.2 and a2.2. However, we were able to detect CLDN10a and CLDN10b and CLDN18a1.1 and CLDN18a2.1. Y2H assays revealed no interaction between CLDN16 and CLDN10a, CLDN18a1.1 and CLDN18a2.1 by using three different reporter assays (HIS3, lacZ, and ADE2) (Fig. 4A). HIS3 and ADE2 reported weak interaction between CLDN16 and CLDN10b, but no signal was evident in the lacZ assay (Fig. 4A). However, lacZ positives were obtained when testing for interactions between CLDN19 and CLDN10a, CLDN10b, CLDN18a1.1, and CLDN18a2.1 (Fig. 4B). The level of lacZ signal from CLDN19 and CLDN10a or CLDN10b interactions was ≈35% of that measured for CLDN19–CLDN16, and CLDN19–CLDN18a1.1 or CLDN18a2.1 was ≈65%. These in vitro data may suggest a higher affinity of CLDN16 and CLDN19 for each other than for either CLDN10 or CLDN18 and that CLDN16 does not functionally interact with CLDN10 or CLDN18. Moreover, the interactions of CLDN19 with CLDN18a1.1 or CLDN18a2.1 were not affected by the point mutations (G20D, Q57E, L90P, and G123R) in CLDN19 (Fig. 4) that are known to disrupt the CLDN19–CLDN16 interaction (18). All four mutations in CLDN19 caused FHHNC and abolished the synergistic function of the CLDN19–CLDN16 heteromeric channel (18). CLDN18 is unlikely a functional binding partner for the CLDN19–CLDN16 channel complex or involved in the FHHNC pathogenesis. Although significantly weaker than the interaction between CLDN16 and CLDN19, considerable lacZ signals were found from interaction of CLDN10a with CLDN18a1.1 or CLDN18a2.1 (Fig. 4B), which likely held the TJ together in the absence of CLDN16 and CLDN19.
Fig. 4.
Y2H assays of heteromeric claudin interactions. CLDN10 and CLDN18 are with weak binding affinity to CLDN16 and CLDN19, determined by using three reporter genes (HIS3, lacZ, and ADE2) in the yeast NMY51 strain. Shown are plates with selective medium lacking leucine and tryptophan (A) (SD-LW), indicating the transforming of both bait and prey vectors; with SD-LWHA (A), indicating the expression of reporter genes HIS3 and ADE2; and the β-galactosidase assay (B) [A(615) values] for quantification of interaction strength by using reporter gene LacZ.
Discussion
In this study, we make the demonstration that CLDN16 and CLDN19 interaction is required for their assembly into TAL TJs. We have also shown that CLDN16–CLDN19 interaction underlies the molecular mechanism of FHHNC pathogenesis. Transgenic mouse models deficient in either claudin developed renal wasting of magnesium and calcium, which has its origin in the defective cation permselectivity of the TAL.
Examination of TJ formation in cultured cells involving other claudins suggests a three-stage hypothesis of TJ assembly (6, 22). First, claudins cis associate within the plane of the membrane into dimers, or higher oligomeric state. Second, trans interactions between claudins in adjacent cells take place. Third, additional cis interactions occur elaborating the TJ strands. Our data show that cis interactions between CLDN16 and CLDN19 were required for stable integration into the TJ but do not address the intracellular location of their initial interaction. Integral membrane proteins generally oligomerize in the endoplasmic reticulum (23) or Golgi (24), and it is likely that CLDN16 and CLDN19 follow this pattern. An alternative one-stage hypothesis remains possible that each claudin trafficks independently to the TJ, where it is stabilized by their interaction with each other. Loss of CLDN16 and CLDN19 interaction may increase the rate of their removal from the TJ into the endosomes and lysosomes for degradation, which is supported by one line of evidence that CLDN16 protein levels were down-regulated in CLDN19 KD animals.
There is an apparent conflict between some of the Y2H data and the distributions of the claudins we observed in vivo. In the Y2H system, interactions between CLDN19 and CLDN18 were significant, consistent with heteromeric oligomerization. However, two lines of evidence suggest CLDN18 is not part of the CLDN16–CLDN19 functional complex (1). In the absence of CLDN19, CLDN18 accumulated normally in the TJ (2). The CLDN19 mutations (G20D, Q57E, L90P, and G123R) known to disrupt the CLDN19–CLDN16 interaction had no effects on the CLDN19–CLDN18 interaction. One explanation is that the interactions experienced in the yeast membrane have some levels of nonspecificity that are not present in the mammalian cells, mindful that the lipid compositions of the plasma membrane are significantly different between two model systems. Another explanation is that cellular factors present in TAL epithelial cells but not yeast may influence claudin behavior. For example, interaction between CLDN16 and CLDN19 may create a recognition site required for transport to the plasma membrane absent in CLDN19 and CLDN18 heteromers. These cellular factors may be unique to the TAL epithelial cells. Freeze-fracture replicas revealed the assembly of TJ strands in mammalian fibroblast L cells that only expressed CLDN19, suggesting strong homomeric CLDN19 interactions (18). Nevertheless, in the absence of CLDN16, CLDN19 was not able to assemble in the TJ of the TAL. Third, it is possible that the measured affinities reflect interactions normally experienced only during the third stage of the TJ assembly process envisioned by Piontek et al. (6).
The CLDN16–CLDN19 complex participates in forming a cation-selective paracellular channel in the TAL. It is generally accepted that the charges on the ECLs of claudins line the channel pore and electrostatically influence passage of soluble ions (25, 26). Mutational analysis identified a locus of negatively charged amino acids in the first ECL of CLDN16 critical for its selectivity (27). However, CLDN19 shares limited homology with CLDN16 in this domain. In any case, the mutational analysis was performed on cells in culture in the absence of CLDN19 expression. The coordinated interaction of CLDN16 and CLDN19 in vivo suggests that meaningful conclusions about the molecular basis of charge selectivity will require studies on cells expressing both claudins. It is less likely that CLDN10 and CLDN18 participate in the production of cation selectivity in the TAL, because in the absence of CLDN16 and CLDN19, cation selectivity is lost, although barrier function is preserved. One of the unanswered questions regarding TAL function is the nature of the paracellular cation channel allowing reabsorption of Mg2+. Indeed, it was proposed originally that CLDN16 might serve this function (14). A reasonable hypothesis is that CLDN10 and/or CLDN18 comprise the Mg2+ paracellular channel.
Methods
Antibodies, Cell Lines, and Animals.
The following antibodies were used in this study: rabbit polyclonal anti-CLDN1, anti-CLDN2, anti-CLDN3, anti-CLDN5, anti-CLDN7, anti-CLDN8, anti-CLDN11, anti-CLDN12, anti-CLDN13, anti-CLDN14, anti-CLDN16, and anti-CLDN18 (Zymed Laboratories); rabbit polyclonal anti-CLDN19 (a kind gift from Mikio Furuse, Kobe University, Kobe, Japan); mouse monoclonal anti-CLDN4, anti-CLDN10, anti-CLDN15, and anti-occludin antibodies (Zymed Laboratories); rat monoclonal ZO-1 (28); rabbit polyclonal anti-THP (Biomedical Technologies); mouse monoclonal anti-NKCC2 (Novus Biologicals); goat polyclonal anti-AQP2 (Santa Cruz Biotechnology); fluorescein isothiocyanate-labeled goat anti-rabbit IgG and rhodamine-labeled goat anti-mouse IgG (Chemicon); fluorescein isothiocyanate-labeled Lotus tetragonolobus lectin (Vector Laboratories); and horseradish peroxidase-labeled donkey anti-rabbit and anti-mouse IgG (Amersham Pharmacia Biotech). HEK293T cells (a kind gift from Joan Brugge, Harvard Medical School, Cambridge, MA) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS, penicillin/streptomycin, and 1 mM sodium pyruvate. Animals (strain: C57BL/6, B6D2F1, CD-1 female foster mice, and CD-1 vasectomized male stud mice) were from Charles River Laboratory.
siRNA Screening and Lentivirus Production.
The siRNA hairpin oligonucleotides [complementary to the mouse CLDN19 mRNA sequence (GenBank NM_153105)] were synthesized by Integrated DNA Technologies (IDT), annealed, and cloned into the pUG-U6 lentivirus backbone (17) downstream of the human small nuclear ribonucleoprotein U6 promoter to create the CLDN19 siRNA constructs. A set of 10 short hairpin oligonucleotides (shRNAs) were tested in vitro for the ability to efficiently deplete CLDN19 mRNA in primary cultures of mouse TAL cells. We used an immunomagnetic separation method to isolate the TAL cells from the mouse kidney [modified from Pizzonia et al. (29)]. Antibodies against the TAL cell-specific surface antigen, Tamm-Horsfall protein (THP), were coated onto the paramagnetic polystyrene beads (Dynabeads M-280; Dynal), allowing immunoprecipitation of the TAL cells from collagenase-digested mouse kidneys (Fig. S1A). The isolated cells were viable and express the TAL-specific genes, THP (Fig. S1B) and CLDN16 (Fig. S1C). VSV-G pseudotyped lentivirus were produced in HEK293T cells and used to inject the single cell mouse embryos at a titer of 1 × 106 units/μL, as described in ref. 17.
Animal Protocols, Surgical Protocols and Renal Clearance, Real-Time Quantitative PCR, Protein Electrophoresis and Immunoblotting, Immunolabeling and Fluorescence Microscopy, Y2H Membrane Protein Interaction Assay, and Statistical Analyses.
For additional information on these topics, see SI Methods.
Supplementary Material
Acknowledgments.
We thank the Nikon Imaging Centre of Harvard Medical School for its excellent assistance on fluorescence microscopy. We thank Dr. Daniel Martin and the Washington University George M. O'Brien Center for Kidney Disease Research Core for analyzing plasma and urinary ion concentrations. This work was supported by National Institutes of Health Grants EY02430 and GM37751, by American Heart Association Grant 0930050N (to J.H.), and by The P. E. Kempkes Foundation Grants 15/06 and 10/08 (to A.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907724106/DCSupplemental.
References
- 1.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 2.Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodenough DA, Revel JP. A fine structural analysis of intercellular junctions in the mouse liver. J Cell Biol. 1970;45:272–290. doi: 10.1083/jcb.45.2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Staehelin LA. Structure and function of intercellular junctions. Int Rev Cytol. 1974;39:191–283. doi: 10.1016/s0074-7696(08)60940-7. [DOI] [PubMed] [Google Scholar]
- 5.Mitic LL, Unger VM, Anderson JM. Expression, solubilization, and biochemical characterization of the tight junction transmembrane protein claudin-4. Protein Sci. 2003;12:218–227. doi: 10.1110/ps.0233903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piontek J, et al. Formation of tight junction: Determinants of homophilic interaction between classic claudins. FASEB J. 2008;22:146–158. doi: 10.1096/fj.07-8319com. [DOI] [PubMed] [Google Scholar]
- 7.Furuse M, et al. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh M, et al. Direct binding of three tight junction associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol. 1999;147:1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bazzoni G, et al. Interaction of junctional adhesion molecule with the tight junction components ZO-1, cingulin, and occludin. J Biol Chem. 2000;275:20520–20526. doi: 10.1074/jbc.M905251199. [DOI] [PubMed] [Google Scholar]
- 10.Umeda K, et al. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126:741–754. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- 12.Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 13.Cordenonsi M, et al. Cingulin contains globular and coiled-coil domains and interacts with ZO-1, ZO-2, ZO-3, and myosin. J Cell Biol. 1999;147:1569–1582. doi: 10.1083/jcb.147.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon DB, et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- 15.Konrad M, et al. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet. 2006;79:949–957. doi: 10.1086/508617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiuchi-Saishin Y, et al. Differential expression patterns of claudins, tight junction membrane proteins, in mouse nephron segments. J Am Soc Nephrol. 2002;13:875–886. doi: 10.1681/ASN.V134875. [DOI] [PubMed] [Google Scholar]
- 17.Hou J, et al. Transgenic RNAi depletion of claudin-16 and the renal handling of magnesium. J Biol Chem. 2007;282:17114–17122. doi: 10.1074/jbc.M700632200. [DOI] [PubMed] [Google Scholar]
- 18.Hou J, et al. Claudin-16 and claudin-19 interact and form a cation-selective tight junction complex. J Clin Invest. 2008;118:619–628. doi: 10.1172/JCI33970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Himmerkus N, et al. Salt- and acid/base metabolism in claudin-16 knockdown mice—impact for the pathophysiology of FHHNC patients. Am J Physiol Renal Physiol. 2008;295:F1641–F1647. doi: 10.1152/ajprenal.90388.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Itallie CM, et al. Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. Am J Physiol Renal Physiol. 2006;291:F1288–F1299. doi: 10.1152/ajprenal.00138.2006. [DOI] [PubMed] [Google Scholar]
- 21.Niimi T, et al. Claudin-18, a novel downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor, encodes lung- and stomach-specific isoforms through alternative splicing. Mol Cell Biol. 2001;21:7380–7390. doi: 10.1128/MCB.21.21.7380-7390.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blasig IE, et al. On the self-association potential of transmembrane tight junction proteins. Cell Mol Life Sci. 2006;63:505–514. doi: 10.1007/s00018-005-5472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurtley SM, Helenius A. Protein oligomerization in the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- 24.Musil LS, Goodenough DA. Multisubunit assembly of an integral plasma membrane channel protein, gap junction connexin43, occurs after exit from the ER. Cell. 1993;74:1065–1077. doi: 10.1016/0092-8674(93)90728-9. [DOI] [PubMed] [Google Scholar]
- 25.Colegio OR, Van Itallie C, Rahner C, Anderson JM. Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am J Physiol Cell Physiol. 2003;284:C1246–C1254. doi: 10.1152/ajpcell.00547.2002. [DOI] [PubMed] [Google Scholar]
- 26.Van Itallie CM, Fanning AS, Anderson JM. Reversal of charge selectivity in cation or anion selective epithelial lines by expression of different claudins. Am J Physiol Renal Physiol. 2003;285:F1078–F1084. doi: 10.1152/ajprenal.00116.2003. [DOI] [PubMed] [Google Scholar]
- 27.Hou J, Paul DL, Goodenough DA. Paracellin-1 and the modulation of ion selectivity of tight junctions. J Cell Sci. 2005;118:5109–5118. doi: 10.1242/jcs.02631. [DOI] [PubMed] [Google Scholar]
- 28.Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. Identification of ZO-1: A high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pizzonia JH, et al. Immunomagnetic separation, primary culture, and characterization of cortical thick ascending limb plus distal convoluted tubule cells from mouse kidney. In Vitro Cell Dev Biol. 1991;27:409–416. doi: 10.1007/BF02630961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.