Abstract
A strictly host-dependent lifestyle has profound evolutionary consequences for bacterial genomes. Most prominent is a sometimes-dramatic amount of gene loss and genome reduction. Recently, highly reduced genomes from the co-resident intracellular symbionts of sharpshooters were shown to exhibit a striking level of metabolic interdependence. One symbiont, called Sulcia muelleri (Bacteroidetes), can produce eight of the 10 essential amino acids, despite having a genome of only 245 kb. The other, Baumannia cicadellinicola (γ-Proteobacteria), can produce the remaining two essential amino acids as well as many vitamins. Cicadas also contain the symbiont Sulcia, but lack Baumannia and instead contain the co-resident symbiont Hodgkinia cicadicola (α-Proteobacteria). Here we report that, despite at least 200 million years of divergence, the two Sulcia genomes have nearly identical gene content and gene order. Additionally, we show that despite being phylogenetically distant and drastically different in genome size and architecture, Hodgkinia and Baumannia have converged on gene sets conferring similar capabilities for essential amino acid biosynthesis, in both cases precisely complementary to the pathways conserved in Sulcia. In contrast, they have completely divergent capabilities for vitamin biosynthesis. Despite having the smallest gene set known in bacteria, Hodgkinia devotes at least 7% of its proteome to cobalamin (vitamin B12) biosynthesis, a significant metabolic burden. The presence of these genes can be explained by Hodgkinia's retention of the cobalamin-dependent version of methionine synthase instead of the cobalamin-independent version found in Baumannia, a situation that necessitates retention of cobalamin biosynthetic capabilities to make the essential amino acid methionine.
Keywords: cobalamin, genome reduction, genome sequencing, proteomics
The most dramatic example of the massive effects that a strict intracellular lifestyle can have on the evolution of the participating lineages is the eukaryotic mitochondrion, which evolved from a symbiosis of an α-Proteobacteria (1). Upon transitioning from a free-living bacterium to a cellular organelle, most, and in some cases all (2), of the symbiont genes were lost or transferred to the host nucleus, with the result that the mitochondrial proteome is a complex mosaic of products encoded in both genomes and showing different ancestral sources (3). Independent examples of genome reduction that are more evolutionarily recent but nearly as dramatic are found in the nutritional endosymbionts of insects: the four smallest cellular genomes reported to date are all insect symbionts that have formed a mutually obligate relationship with their hosts [Hodgkinia cicadicola, 144 kb (4); Carsonella ruddii PV, 160 kb (5); Sulcia muelleri GWSS, 246 kb (6); and Buchnera aphidicola Cc, 416 kb (7)]. In these cases, it is unclear if some of the lost symbiont genes are transferred to the host nucleus, but their extremely small gene sets suggest that the host plays some major role in the biology of the symbiont.
Insects that develop these intimate symbioses with bacteria usually have restricted diets, and it is has long been predicted that a basis for the symbioses is nutritional (8, 9). Genome sequencing has dramatically confirmed this hypothesis, as the retained bacterial gene sets strongly suggest that the symbionts supply the host with compounds that are lacking in their diet (8, 10). Bacterial symbionts are particularly common in insects that feed exclusively on plant sap, whether it is phloem, which is relatively rich in carbohydrates but poor in amino acids and vitamins, or xylem, which is limited in carbohydrates, amino acids, and vitamins (11). Xylem in particular is an extremely dilute food source and probably the least nutritive of any plant material used by herbivores (11).
Elaborate symbioses with a diversity of bacteria are found in the insect suborder Auchenorrhyncha, a large group of sap-feeding insects including sharpshooters, treehoppers, planthoppers, and spittlebugs (8, 12). The Auchenorrhynchan ancestor established a symbiosis with a Bacteroidetes called Sulcia muelleri approximately 280 million years ago, and during the diversification of this group many lineages have acquired an additional symbiont, usually from one of the classes of Proteobacteria (12). Recently, complete genomes were sequenced from the symbiont pair Sulcia (6) and Baumannia cicadellinicola (13) from the xylem-feeding Glassy-winged sharpshooter (GWSS). These genomes revealed a striking metabolic complementarity: the Sulcia genome encoded nearly complete biosynthetic pathways for eight of the 10 essential amino acids, while Baumannia possessed enzymatic pathways for the remaining two essential amino acids as well as the ability to produce a large number of vitamin cofactors (Fig. 1) (6, 13). Cicadas, also in the Auchenorrhyncha, feed exclusively on xylem sap from plant roots during their long underground juvenile stage (2–17 years, depending on the species) (14–16). Over the course of a few weeks during their last summer, they emerge from the ground, metamorphose into adults, mate, lay eggs, and die.
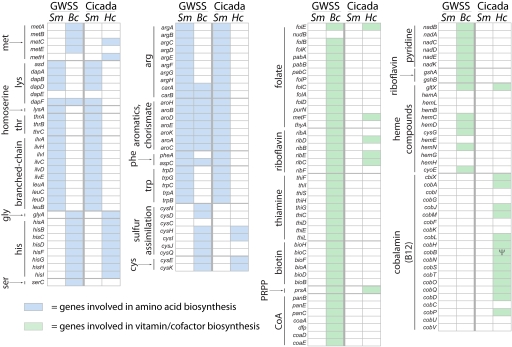
Fig. 1.
Amino acid and vitamin-related gene contents of cicada and GWSS symbiont genomes. Abbreviations are Sm, Sulcia muelleri; Bc, Baumannia cicadellinicola; Hc, Hodgkinia cicadicola; and Ψ, pseudogene. Blue boxes represent amino acid biosynthesis genes that are present in a given genome, and green boxes represent vitamin biosynthesis genes. Note the pattern of conservation evident in amino acid biosynthesis genes but lacking in vitamin biosynthesis genes.
The symbionts of some cicadas are Sulcia and H. cicadicola (4, 12). Hodgkinia has the smallest cellular genome known (144 kb), a high guanine + cytosine (GC) content and an alternative genetic code (UGA Stop→Trp), an unprecedented combination of genomic features (4). Here we describe the complete genome of Sulcia from the cicada Diceroprocta semicincta (17), detail the metabolic contributions of the co-resident symbionts, Sulcia and Hodgkinia, and compare the putative nutritional contributions of these bacteria to their cicada host with the contributions of the GWSS symbionts.
Results
Sulcia from GWSS and Cicada Are Very Similar.
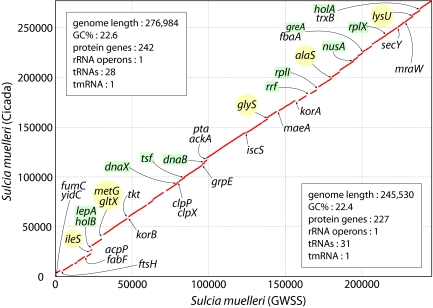
Whole-genome alignments of Sulcia from cicada and GWSS show that despite diverging at least 200 million years ago (18), there are no rearrangements between the two genomes, only differential gene loss and retention (Fig. 2). This perfect colinearity has been observed previously in genomes from symbiont clades within the the γ-Proteobacteria (19, 20), and our observation of the same phenomenon in the distantly related Bacteroidetes phylum indicates that this unusual level of conservation of gene order is a general trend in symbiont genomes, regardless of their phylogenetic origin. The most striking difference in gene content between the two genomes is the retention in the Sulcia of cicada, but not of GWSS, of six aminoacyl tRNA synthetase genes (ileS, metG, gltX, glyS, alaS, and lysU), as well as many other apparent homologs of genes involved in replication (the DNA polymerase III holoenzyme components encoded by holA, holB, and dnaX and the replicative DNA helicase encoded by dnaB), transcription (the transcription elongation and termination factors encoded by greA and nusA), and translation (the ribosomal proteins encoded by rplI and rplX and the ribosomal elongation and recycling factors encoded by tsf, lepA, and rrf) (Fig. 2).
Fig. 2.
Genome conservation in cicada and GWSS Sulcia genomes. The cicada Sulcia genome is represented on the y axis and described in the top left box, and the GWSS Sulcia genome is represented on the x axis and described in the bottom right box. The red line indicates shared synteny between the two genomes. Gene names in the lower triangle are found in the GWSS Sulcia genome but not in the cicada Sulcia genome; gene names in the upper triangle are present in the cicada Sulcia genome but not in GWSS. Genes involved in replication, transcription, and translation are highlighted; aminoacyl tRNA synthetase genes are shown in yellow circles, and all others are shown in green boxes. Only named genes with clear functions were included in the figure for clarity; hypothetical genes, conserved hypothetical genes, and genes with ambiguous functions were omitted.
The Roles of Sulcia and Its Coprimary Symbiont Are Conserved in Amino Acid Biosynthesis.
In both the GWSS and cicada systems, genomic sequences support the production of eight of the 10 essential amino acids by Sulcia (arginine, phenylalanine, tryptophan, lysine, threonine, isoleucine, leucine, and valine), and the remaining two (methionine and histidine) by Baumannia in GWSS and by Hodgkinia in cicada (Fig. 1, blue boxes). Thus, although arising from very different bacterial groups, Hodgkinia and Baumannia have converged upon functionally similar gene sets with respect to amino acid synthesis. The most important differences in the amino acid metabolisms of Baumannia and Hodgkinia are in the methionine biosynthetic pathway. In Hodgkinia, no homologs for homoserine O-succinyltransferase (metA) and O-succinylhomoserine(thiol) lyase (metB) can be found by sequence comparisons, so it is unclear how cystathionine is synthesized in this system. [Cystathionine is present in plant root exudate (21, 22), possibly obviating the need for metA and metB in Hodgkinia since cicadas feed primarily on plant roots.] Both Baumannia and Hodgkinia have a homolog of MetC, which converts cystathionine into homocysteine. It is in the last step of the pathway, the conversion of homocysteine into methionine, where Baumannia and Hodgkinia diverge: Baumannia uses the cobalamin (vitamin B12)-independent version of methionine synthase (MetE), whereas Hodgkinia uses the cobalamin-dependent version of the enzyme (MetH).
The vitamin and cofactor biosynthetic capabilities of Baumannia and Hodgkinia are extremely different. In contrast to the convergence of gene sets related to amino acid biosynthetic capabilities in Baumannia and Hodgkinia, there is very little overlap in vitamin and cofactor biosynthetic capabilities of these organisms (Fig. 1, green boxes). The only significant overlap is in genes devoted to riboflavin synthesis, where three of five genes in the pathway can be found in Hodgkinia. What is striking, especially considering the extremely small size of the Hodgkinia genome, is the presence of 13 identifiable genes with intact ORFs (plus one apparent pseudogene), homologous to genes devoted to the synthesis of cobalamin.
Cobalamin is a complex small molecule which requires approximately 20–25 enzymatic steps for its biosynthesis (23, 24), although there is a great deal of diversity in cobalamin-related genes in different organisms (25). Some free-living α-Proteobacteria such as Mesorhizobium loti have a complete or nearly complete set of cobalamin synthesis genes, while others such as Caulobacter crescentus have a highly fragmentary set of only two genes (26). The Hodgkinia genome contains homologs of the the cobN, cobS, and cobT cobalt insertion genes, indicating that it uses the aerobic, or late cobalt insertion, version of the pathway, although a copy of cbiX is present in the genome. It is unclear what the final structure of this cobalamin-like molecule would be in Hodgkinia: homologs of genes involved in cobalt insertion (cobNST) and adenosylation (cobO) are present, but genes involved in the synthesis and addition of the dimethylbenzimidazole moiety (cobU and cobV), among others, are apparently missing.
Discussion
A tightly integrated metabolic complementarity between Sulcia and Baumannia from GWSS was inferred from genome sequencing of the symbionts: Sulcia produces eight of 10 essential amino acids, and Baumannia produces the remaining two essential amino acids as well as a number of vitamins (6, 13). Evidence from phylogenetics (12) and the fossil record (18) indicates that a symbiosis with Sulcia evolved by 270 million years ago in an ancestor of Auchenorrhyncha (12) and that the divergence of GWSS and cicada occurred at least 200 million years ago, during the early Jurassic (18). Since their divergence, GWSS and cicada have each acquired one additional symbiont, Baumannia (γ-Proteobacteria) and Hodgkinia (α-Proteobacteria), respectively. By sequencing the genomes of both Sulcia and Hodgkinia from cicada, we sought to uncover the genomic changes that had occurred in the two Sulcia strains over hundreds of millions of years as well as to delineate how this system distributed the metabolic roles of Sulcia and its partner symbiont when compared to the case of GWSS.
The Sulcia genomes from GWSS and cicada are very similar (Fig. 2). Genes that are retained in both members of the genome pair are perfectly co-linear, despite at least 200 million years of divergence. Similar levels of genomic stability have been observed for other highly reduced symbiont genomes for which there are at least two complete genomes from the same clade, that is, the four Buchnera genomes reflecting divergence over about 150 million years (7, 19, 27, 28) and the two genomes from the ant symbiont Blochmannia reflecting divergence over about 20 million years (20, 29). This conserved genomic organization reflects a lack of recombination and lateral gene transfer, possibly resulting from a loss of pathways involved in DNA uptake and recombination (19, 30). Differential gene loss is therefore the only remaining process shaping genome organization in Sulcia. Both Buchnera and Blochmannia arose from the same group of γ-Proteobacteria; our observation of extreme genome stability in Sulcia extends this finding to a distantly related Bacteroidetes symbiont.
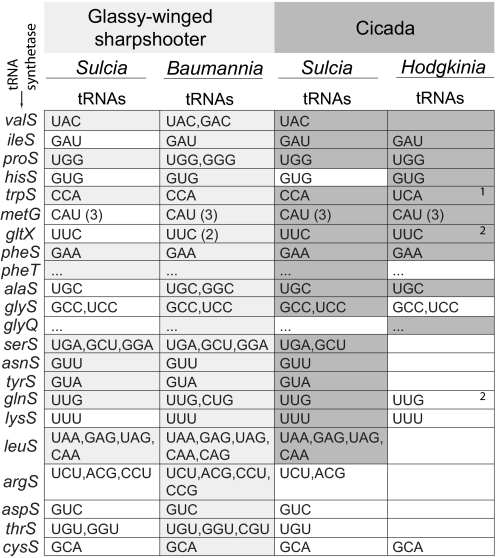
Because Sulcia's cosymbionts in GWSS and cicada differ drastically in genome size (Baumannia, 686 kb; Hodgkinia, 144 kb) and gene content (Fig. 1), one might expect the patterns of loss and retention in the two Sulcia genomes to reflect the genome complexity of the cosymbiont—that is, Sulcia from cicada would be larger and contain genes that compensate for the genes missing in Hodgkinia compared to Baumannia. Comparisons of genome sizes suggested this might be true, as the Sulcia genome is indeed bigger in cicada (277 kb vs. 246 kb) and contains more protein-coding genes (242 vs. 227), but further analysis did not produce significant evidence for any compensatory patterns. Many of the genes retained in cicada Sulcia while lost from GWSS Sulcia are involved in replication, transcription, and translation (17 of 24 identifiable genes, or 71%); in particular, six are aminoacyl tRNA synthetases (Fig. 3). This retention of aminoacyl tRNA synthetase genes suggested the hypothesis that, in cicada, Sulcia might be compensating for the tiny genome of its companion symbiont Hodgkinia by providing a source of aminoacylated tRNAs. Comparisons of the aminoacyl tRNA synthetases and tRNA populations of the two bacteria did not give strong evidence for this, as four of the six synthetases retained in cicada Sulcia are also present in Hodgkinia (Fig. 3). Furthermore, the arginyl-, aspartyl-, threonyl-, and cysteinyl-tRNA synthetases cannot be identified by sequence comparisons in either Sulcia or Hodgkinia, and only 15 tRNAs can be identified computationally in Hodgkinia, raising questions as to how translation occurs in these organisms. Other systems with non-canonical translation apparatuses suggest some possible solutions to these seemingly intractable problems. For example, some Archaea have split tRNA genes, some of which are not identifiable by standard computational searches (31). Additionally, many methanogenic archaea form cysteninyl-tRNA in the absence of cysteinyl-tRNA synthetase by an unusual biochemical route (32).
Fig. 3.
The aminoacyl tRNA synthetases and tRNAs found in cicada and GWSS symbiont genomes. Presence of an aminoacyl tRNA synthetase is indicated by shading in the column for each organism, light gray for GWSS symbionts and darker gray for cicada symbionts. The tRNAs that could be identified by computational methods are listed by anticodon sequences, and a parenthetical number indicates that many with an identical anticodon. Ellipses are shown for the second gene in a two component aminoacyl tRNA synthetase; refer to the box directly above for the relevant anticodon(s). 1The tRNA-tryptophan in Hodgkinia is predicted to read both UGA and GGA codons because of the UGA Stop→Trp recoding in that genome (4). 2Hodgkinia has homologs of the gatA and gatB genes involved in generating tRNA-Gln from tRNA-Glu (44), potentially eliminating the need for glnS in this genome.
The Sulcia genome is among the smallest of any cellular organism, and it has been suggested that other symbiotic bacteria with tiny genomes, such as Carsonella (160 kb) and B. aphidicola Cc (422 kb), have crossed an evolutionary threshold that will ultimately result in their extinction and possible replacement by another symbiont (7, 33). Were that the case in Sulcia, one would expect a relaxation of purifying selection (negative selection against deleterious mutations) on its protein-coding sequences. The completion of a second Sulcia genome allows us to gain some insight into genome degradation by calculating the ratios of synonymous changes to nonsynonymous changes (dN/dS) for pairs of protein-coding genes shared between the two genomes. This analysis shows that Sulcia genes are subject to strong purifying selection (average dN/dS = 0.066, ±0.045); however this value should be interpreted cautiously because the dS value is at or near saturation (5.942 ± 17.292). Nonetheless, the low average dN value (0.142 ± 0.082) indicates efficient purifying selection despite the remarkably small genome size. Thus, Sulcia shows no signs of having crossed any threshold that would result in its elimination. Indeed, the two Sulcia genomes are remarkably conserved considering that they have been evolving separately for at least 200 million years (18). In particular, they show complete co-retention of genes involved in essential amino acid biosynthesis. Additionally, some of the most abundant proteins in the Sulcia proteome are involved in essential amino acid biosynthesis (18/28, or 64% of identifiable proteins), exactly what would be expected for a symbiont whose central function in the symbiosis is to make essential amino acids (Table 1).
Table 1.
Protein abundance in the Sulcia proteome ranked by emPAI value (42)
| Gene | Pathway | General function | emPAI | Number of peptides |
|---|---|---|---|---|
| AroG/PheA | Phe and Trp synthesis | Amino acids | 4.89 | 17 |
| GroL | Chaperonin Hsp60 | Protein folding | 3.21 | 19 |
| IlvC | Branched-chain aa synthesis | Amino acids | 2.76 | 10 |
| DnaK | Chaperonin Hsp70 | Protein folding | 2.48 | 18 |
| GapA | Glycolysis | General metabolism | 2.00 | 8 |
| Asd | Branched-chain aa synthesis | Amino acids | 1.72 | 9 |
| AroB | Phe and Trp synthesis | Amino acids | 0.79 | 6 |
| LeuA | Leu synthesis | Amino acids | 0.62 | 5 |
| LeuC | Leu synthesis | Amino acids | 0.55 | 2 |
| TktA | Pentose phosphate synthesis | General metabolism | 0.54 | 2 |
| ArgC | Arg synthesis | Amino acids | 0.45 | 4 |
| TrpB/TrpF | Trp synthesis | Amino acids | 0.43 | 6 |
| IlvD | Branched-chain aa synthesis | Amino acids | 0.41 | 6 |
| Fba | Glycolysis | General metabolism | 0.41 | 4 |
| TrpA | Trp synthesis | Amino acids | 0.27 | 2 |
| DapA | Lys synthesis | Amino acids | 0.26 | 2 |
| TrpC | Trp synthesis | Amino acids | 0.25 | 2 |
| CarB | Arg synthesis | Amino acids | 0.22 | 7 |
| HtpG | Heat shock protein 90 | Protein folding | 0.21 | 4 |
| TktB | Pentose phosphate synthesis | General metabolism | 0.21 | 2 |
| ArgF | Arg synthesis | Amino acids | 0.21 | 2 |
| IlvE | Branched-chain aa synthesis | Amino acids | 0.19 | 2 |
| SucB | TCA cycle | General metabolism | 0.18 | 2 |
| AroC | Phe and Trp synthesis | Amino acids | 0.18 | 2 |
| ThrC | Branched-chain aa synthesis | Amino acids | 0.15 | 2 |
| Lpd | TCA cycle | General metabolism | 0.14 | 2 |
| ArgG/ArgA | Arg synthesis | Amino acids | 0.11 | 2 |
| SucA | TCA cycle | General metabolism | 0.07 | 2 |
Protein abundance in the Sulcia proteome ranked by emPAI value (42). Only proteins with at least 2 high-quality peptides are listed. This should not be considered a complete list of expressed proteins, because the complexity of the sample does not allow an exhaustive search for all peptides. Sulcia is not currently culturable, and therefore the sample is derived from insect tissue and is a mixture of host proteins together with Sulcia and Hodgkinia proteins.
Cobalamin biosynthesis genes have never before been found in an insect symbiont genome. The likely reason these genes occur in Hodgkinia is its use of MetH, the cobalamin-dependent version of methonine synthase, instead of MetE for the last step in methionine synthesis, as in other known insect symbionts. No other cobalamin-requiring enzymes can be found in the Hodgkinia genome. Additionally, insects are not thought to require cobalamin, as none of the 19 complete insect genomes contain any cobalamin-dependent homologs (34). Most γ-Proteobacteria with large genomes have copies of both metE and metH in their genomes, while the ability to make cobalamin de novo from uroporphyrinogen-III shows a patchy phylogenetic distribution (e.g., E. coli can only carry out the last few steps of cobalamin biosynthesis (35), and H. influenzae cannot do any, but both have both metE and metH in their genome). Previously studied γ-Proteobacterial insect symbionts with methionine synthase capabilities have lost metH and retained metE in the process of genome reduction (Table 2), allowing the last step in methionine biosynthesis to be carried out without the metabolic burden of cobalamin biosynthesis or import.
Table 2.
Distribution of methionine synthase genes in bacterial genomes
| Bacterial class | Organism | Genome size (Mb) | metE | metH | No. of Cobalamin biosynthesis genes |
|---|---|---|---|---|---|
| γ-Proteobacteria | Pseudomonas | 6.26 | + | + | 22 |
| Salmonella | 4.86 | + | + | 19 | |
| Escherichia | 4.64 | + | + | 5 | |
| Haemophilus | 1.83 | + | + | 0 | |
| Blochmannia | 0.71 | + | – | 0 | |
| Baumannia | 0.69 | + | – | 0 | |
| Buchnera | 0.64 | + | – | 0 | |
| α-Proteobacteria | Bradyrhizobium | 9.11 | + | + | 21 |
| Mesorhizobium | 7.04 | + | + | 21 | |
| Agrobacterium | 4.92 | + | + | 20 | |
| Sinorhizobium | 3.65 | – | + | 20 | |
| Brucella | 3.29 | – | + | 21 | |
| Rickettsia | 1.11 | – | – | 0 | |
| Hodgkinia | 0.14 | – | + | 13 |
Distribution of methionine synthase genes in bacterial genomes. The bacterial genomes are listed in order of decreasing genome size within each class. Cobalamin-independent methionine synthase, metE; cobalamin-dependent methionine synthase, metH.
The patterns of retention of metE, metH, and cobalamin biosynthesis genes are less clear in the Rhizobiales, the order of α-Proteobacteria which encompasses Hodgkinia and its closest free-living relatives (4). Some members of this group have large genomes and retain both metE and metH, while others retain only metH, and most have complete cobalamin biosynthetic capabilities (Table 2). Given this patchy distribution of metE and metH, two evolutionary scenarios can be constructed to explain the presence of metH and cobalamin biosynthesis genes in Hodgkinia. The simplest explanation is that the ancestor of Hodgkinia had a gene complement similar to extant Sinorhizobium meliloti, which has genes for the biosynthesis of cobalamin and metH, but no metE (Table 2). Alternatively, the Hodgkinia ancestor could have been similar in gene content to modern Bradyrhizobium japonicum, which has metH, metE, and cobalamin synthesis genes, but then lost metE during the random events of genome reduction. In either case, a requirement for methionine by all three organisms in the symbiosis would impose a strong selective pressure to maintain metH along with the complex cobalamin biosynthetic pathway. A parallel case of metE loss followed by cobalamin auxotrophy was recently demonstrated in algae (36), giving some plausibility to the second more complicated metE loss scenario. During algal diversification, various unrelated lineages have lost metE but retained metH, thereby imposing a requirement for an exogenous source of cobalamin, which has been shown to come from an extracellular symbiosis with bacteria (36).
GWSS and cicadas are thought to feed exclusively on xylem sap (11, 14, 37, 38). It is therefore reasonable to expect that their symbiont pairs would collectively make the same nutritional contributions. Analysis of inferred metabolic capabilities of the four symbiont genomes indicates that this is true for essential amino acids, but not for vitamins, as Hodgkinia is missing most vitamin and cofactor biosynthetic pathways (Fig. 1), implying that the cicada and its symbionts have access to external sources of these required micronutrients. Both sharpshooters and cicadas typically have broad host plant ranges, and this is true for both GWSS (11) and Diceroprocta (13, 37) in particular. The fundamental difference in feeding habits between cicadas and sharpshooters is that cicadas spend most of their lives underground and feed primarily on plant roots (14, 16). The plant root-soil interface, or rhizosphere, is a complex environment. Roots form symbioses with microbes, extract water and nutrients from the soil, and exude large amounts of metabolites, including amino acids and simple amides, carbohydrates, fatty acids, and vitamins into the surrounding soil (21, 22). While cicadas are only known to feed on xylem sap (14), it could be the concentration of metabolites in plant root xylem is different from that in other parts of the plant, arising from compounds from the root exudate or soil, and that cicadas are somehow able to assimilate the necessary vitamins in this way. Another hypothesis is that an additional symbiotic microorganism is present, but this possibility was excluded by a computational screen of our sequence data. The only genomes present are those of Hodgkinia, Sulcia, and host, suggesting that all nutritional needs must be satisfied based solely on the diet and the metabolic capabilities of these three organisms.
Hodgkinia is present in distantly related cicadas (genus Magicicada) (4), suggesting that the original infection is ancient and that the extremely tiny Hodgkinia genome reflects a long period of genome reduction. Some of Hodgkinia's closest relatives are rhizosphere-associated or root nodule-forming bacteria (4), and it is tempting to speculate that these could have been the initial source of bacteria for the establishment of the cicada-Hodgkinia symbiosis. Regardless of Hodgkinia's origin, a comparison of its amino acid biosynthetic capabilities with Baumannia's reveals a remarkable case of convergent evolution, especially considering the vast differences between the two genomic architectures.
Materials and Methods
Genome Sequencing and Annotation.
Female cicadas were collected in and around Tucson, Arizona, and their bacteriomes were dissected in 95% ethanol. Genomic DNA was purified using Qiagen DNeasy Blood & Tissue Kits and prepared for Roche 454 GS FLX pyrosequencing as directed by the manufacturer. The sequencing run generated 523,979 reads totaling 116,176,938 bases that assembled into 1,029 contigs greater than 500 bases using the GS De novo Assembler (version 1.1.03.24). Contigs expected to belong to the Sulcia genome were identified by BLASTX searches against the GenBank non-redundant database, and reads associated with these contigs were extracted and reassembled to generate the Sulcia genome. Reassembly produced 41 contigs representing 269,151 bases with an average depth of 29× and a GC content of 22.9%. The order and orientation of some of the 41 contigs were predicted using the “.fm” and “.to” information appended to read names encoded in the 454Contigs.ace file. All contig joins were confirmed using PCR amplification followed by Sanger sequencing.
Errors in homopolymeric run lengths were corrected by mapping 12,965,640 Illumina/Solexa reads of 39 bases to the genome using either MUMMER (nucmer -b 10 -c 30 -g 2 -l 12; show-snps -rT -x 30) or BLASTN (-G 2 -E 1 -F F -e 1e-8 -W 7 -b 1 -v 1). Average coverage in Illumina reads on the Sulcia genome was 164×. Any remaining uncertainties in homopolymer lengths, in particular those that shifted a seemingly conserved coding sequence out of frame and were not well covered by Illumina reads, were checked by PCR followed by Sanger sequencing.
Post Genome Analysis.
Whole genome alignments were generated with promer from the MUMMER package (39). The genome was annotated as described previously (6).
The genomes used in the generation of Table 2 were: Pseudomonas aeruginosa PAO1 (NC_002516), Salmonella enterica LT2 (NC_003197), Escherichia coli K-12 substr. MG1655 (NC_000913), Haemophilus influenzae Rd KW20 (NC_000907), Blochmannia floridanus (NC_005061), Baumannia cicadellinicola Hc (NC_007984), Buchnera aphidicola APS (NC_002528), Bradyrhizobium japonicum USDA 110 (NC_004463), Mesorhizobium loti MAFF303099 (NC_002678), Agrobacter tumefaciens C58 (NC_003062, NC_003063), Sinorhizobium meliloti 1021 (NC_003047), Brucella melitensis 16M (NC_003317, NC_003318), and Rickettsia prowazekii str. Madrid E (NC_000963). The values for the number of genes involved in cobalamin biosynthesis were from (26) and (40).
The dN/dS values for 203 pairs of protein-coding sequences from Sulcia of GWSS and cicada were calculated using the CODEML package from PAML (41) after aligning protein sequences using the linsi module of MAFFT (42) and making nucleotide-based alignments from these protein alignments using PAL2NAL (43).
Proteomics was performed as described (4). The exponentially modified protein abundance index (emPAI) is a rough measure of relative protein amounts in complex mixtures, derived from the number of sequenced peptides and normalized by the expected number per protein. Only those proteins with two high-quality peptide hits [as described in (4)] were included in Table 1.
To screen for other bacteriome-dwelling symbionts, the 454AllContigs.fna file from the 454 assembly (containing all contigs composed of at least two reads; 31,890 sequences comprising 7,732,561 bps) was searched against small ribosomal subunit RNA sequences (SSUs) from the Ribosomal Database Project release 8.1 (34,530 SSU sequences comprising 29,402,220 bps from bacterial, archaeal, and eukaryal sources) using blastn (-b 10 -v 10 -e 1e-10). The only hits were to sequences from insect 18S rDNA, insect mitochondrial 16S rDNA, Bacteroidetes 16S rDNA, and α-Proteobacterial 16S rDNA. Any other symbiont present in the bacteriome at an appreciable level should have been well sampled owing to the massive depth of coverage generated by 454 sequencing.
Acknowledgments.
We thank F. Chen, K. Barry, and colleagues at the DOE Joint Genome Institute for 454 and Solexa sequencing runs and Q. Lin at the State University of New York at Albany Proteomics facility for performing the proteomic analysis. This work was supported by National Science Foundation Microbial Genome Sequencing award 0626716 (to N.A.M.) and the University of Arizona's Center for Insect Science through National Institutes of Health Training Grant 1K 12 GM00708 (to J.P.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. CP001605).
References
- 1.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 2.Williams BA, Hirt RP, Lucocq JM, Embley TM. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- 3.Adams KL, Palmer JD. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- 4.McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakabachi A, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 6.McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Brocal V, et al. A small microbial genome: The end of a long symbiotic relationship? Science. 2006;314:312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- 8.Buchner P. Endosymbiosis of Animals with Plant Microorganisms. New York, NY: Interscience; 1965. [Google Scholar]
- 9.Douglas AE. Mycetocyte symbiosis in insects. Biol Rev Camb Philos Soc. 1989;64:409–434. doi: 10.1111/j.1469-185x.1989.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 10.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 11.Redak RA, et al. The biology of xylem fluid-feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology. Annu Rev Entomol. 2004;49:243–270. doi: 10.1146/annurev.ento.49.061802.123403. [DOI] [PubMed] [Google Scholar]
- 12.Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: An ancient symbiont of sap-feeding insects from the Bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005;71:8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu D, et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 2006;4:e188. doi: 10.1371/journal.pbio.0040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White J, Strehl CE. Xylem feeding by periodical cicada nymphs on tree roots. Ecol Entomol. 1978;3:323–327. [Google Scholar]
- 15.Glinski RL, Ohmart RD. Factors of reproduction and population densities in the Apache cicada (Diceroprocta apache) Southwest Nat. 1984;29:73–79. [Google Scholar]
- 16.Williams KS, Simon C. The ecology, behavior, and evolution of periodical cicadas. Annu Rev Entomol. 1995;40:269–295. [Google Scholar]
- 17.Davis WT. Cicadas belonging to the genus Diceroprocta with descriptions of new species. J NY Entomol Soc. 1928;36:439–460. [Google Scholar]
- 18.Shcherbakov DE, Popov YA. Superorder Cimicidea Laicharting, 1781; Order Hemiptera Linne, 1758. The bugs, cicadas, plantlice, scale insects, etc. In: Rasnitsyn AP, Quicke DLJ, editors. History of Insects. Dordrecht: Kluwer; 2002. pp. 143–157. [Google Scholar]
- 19.Tamas I, et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 20.Degnan PH, Lazarus AB, Wernegreen JJ. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 2005;15:1023–1033. doi: 10.1101/gr.3771305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertin C, Yang X, Weston LA. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil. 2003;256:67–83. [Google Scholar]
- 22.Uren NC. Types, amounts and possible functions of compounds released into the rhizosphere by soil grown plants. In: Pinton R, Varani Z, Nanniperi P, editors. The Rhizosphere: Biochemistry and Organic Substances at the Soil Interface. New York: Marcel Dekker Inc; 2000. pp. 19–40. [Google Scholar]
- 23.Roth JR, Lawrence JG, Bobik TA. Cobalamin (coenzyme B12): Synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 24.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. The biosynthesis of adenosylcobalamin (vitamin B12) Nat Prod Rep. 2002;19:390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- 25.Raux E, Schubert HL, Warren MJ. Biosynthesis of cobalamin (vitamin B12): A bacterial conundrum. Cell Mol Life Sci. 2000;57:1880–1893. doi: 10.1007/PL00000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- 27.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 28.van Ham RC, et al. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci USA. 2003;100:581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gil R, et al. The genome sequence of Blochmannia floridanus: Comparative analysis of reduced genomes. Proc Natl Acad Sci USA. 2003;100:9388–9393. doi: 10.1073/pnas.1533499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva FJ, Latorre A, Moya A. Why are the genomes of endosymbiotic bacteria so stable? Trends Genet. 2003;19:176–180. doi: 10.1016/S0168-9525(03)00041-6. [DOI] [PubMed] [Google Scholar]
- 31.Randau L, Munch R, Hohn MJ, Jahn D, Soll D. Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature. 2005;433:537–541. doi: 10.1038/nature03233. [DOI] [PubMed] [Google Scholar]
- 32.Sauerwald A, et al. RNA-dependent cysteine biosynthesis in Archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 33.Tamames J, et al. The frontier between cell and organelle: Genome analysis of Candidatus Carsonella ruddii. BMC Evol Biol. 2007;7:181. doi: 10.1186/1471-2148-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Rodionov DA, Gelfand MS, Gladyshev VN. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics. 2009;10:78. doi: 10.1186/1471-2164-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrence JG, Roth JR. The cobalamin (coenzyme B12) biosynthetic genes of Escherichia coli. J Bacteriol. 1995;177:6371–6380. doi: 10.1128/jb.177.22.6371-6380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 37.Leopold RA, Freeman TP, Buckner JS, Nelson DR. Mouthpart morphology and stylet penetration of host plants by the glassy-winged sharpshooter, Homalodisca coagulata, (Homoptera: Cicadellidae) Arthropod Struct Dev. 2003;32:189–199. doi: 10.1016/S1467-8039(03)00047-1. [DOI] [PubMed] [Google Scholar]
- 38.Cheung WWK, Marshall AT. Water and ion regulation in cicadas in relation to xylem feeding. J Insect Physiol. 1973;19:1801–1816. [Google Scholar]
- 39.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleischmann RD, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 41.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 42.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suyama M, Torrents D, Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Curnow AW, et al. Glu-tRNAGln amidotransferase: A novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]