Abstract
The transcription termination factor Rho is a global regulator of RNA polymerase (RNAP). Although individual Rho-dependent terminators have been studied extensively, less is known about the sites of RNAP regulation by Rho on a genome-wide scale. Using chromatin immunoprecipitation and microarrays (ChIP-chip), we examined changes in the distribution of Escherichia coli RNAP in response to the Rho-specific inhibitor bicyclomycin (BCM). We found ≈200 Rho-terminated loci that were divided evenly into 2 classes: intergenic (at the ends of genes) and intragenic (within genes). The intergenic class contained noncoding RNAs such as small RNAs (sRNAs) and transfer RNAs (tRNAs), establishing a previously unappreciated role of Rho in termination of stable RNA synthesis. The intragenic class of terminators included a previously uncharacterized set of short antisense transcripts, as judged by a shift in the distribution of RNAP in BCM-treated cells that was opposite to the direction of the corresponding gene. These Rho-terminated antisense transcripts point to a role of noncoding transcription in E. coli gene regulation that may resemble the ubiquitous noncoding transcription recently found to play myriad roles in eukaryotic gene regulation.
Keywords: chromatin immunoprecipitation, RNA polymerase
Transcription termination is critical for maintaining control over gene expression. Bacteria employ 2 distinct types of termination: (i) intrinsic termination, for which a GC-rich RNA hairpin followed by a U-tract dissociates RNA polymerase (RNAP) without the need for accessory proteins, and (ii) factor-dependent termination caused by the Rho protein. Rho was originally identified as a factor that increased the “accuracy” of in vitro transcription by terminating RNAP at specific positions on a bacteriophage λ DNA template (1). Later, Rho was found to be the cause of polarity, whereby the uncoupling of transcription and translation by premature stop codons decreases gene expression of downstream genes in an operon (2). Rho is a homohexameric protein with RNA-dependent ATPase activity (3). Rho binds to the nascent RNA and translocates 5′ to 3′ along RNA using energy derived from ATP hydrolysis (4). At certain sites, Rho contacts RNAP, and terminates the elongation complex (EC) by an unknown mechanism (5).
Bicyclomycin (BCM) is a specific inhibitor of Rho (6). BCM blocks Rho-dependent termination in vivo (7) and in vitro (8) through noncompetitive inhibition of the RNA-dependent ATPase activity of Rho (9). Biochemical and structural analyses show that BCM binds adjacent to the ATPase of Rho (8) and prevents ATP hydrolysis by interfering with a key glutamic acid residue that is involved in catalysis (10). Treatment of wild-type E. coli K-12 with high concentrations of BCM is lethal (6), because rho is an essential gene (11). However, sublethal doses of BCM are sufficient to perturb Rho termination in vivo (7).
Genome-wide studies have documented the role of Rho as a global regulator of RNAP. Chromatin immunoprecipitation assays using tiling microarrays (ChIP-chip) revealed remarkably similar global distributions of RNAP and Rho on DNA (12). These similar distributions suggest that Rho contacts ECs soon after initiation, interacts with ECs throughout elongation, and interacts with ECs on nearly all transcription units (TUs), rather than having specificity for a small set of genes. Cardinale et al. (13) used expression array analysis to gauge the effect of BCM treatment on mRNA levels. Their findings showed changes in abundance of a subset of transcripts, particularly for genes integrated into the genome by horizontal transfer. Thus, Rho termination occurs preferentially on a subset of genes, even though its physical distribution is widespread. However, the specific locations of BCM-inhibited Rho-dependent terminators have not yet been determined.
We used ChIP-chip to examine changes in the distribution of RNAP in response to Rho inhibition by BCM. We found ≈200 Rho-terminated loci where BCM shifted the distribution of RNAP downstream of the apparent termination site. Half of the Rho-dependent terminators were located at the 3′ ends of genes (intergenic), including small RNAs (sRNAs) and transfer RNAs (tRNAs). The other half were found within the coding sequence of annotated genes (intragenic). For one set of intragenic terminators, the readthrough event was in the opposite direction of the gene, indicating antisense transcription.
Results
BCM Alters the Distribution of RNAP.
To determine the contribution of Rho to the genome-wide distribution of RNAP, ChIP was performed on cells grown in the presence or absence of BCM at 20 μg/mL. This concentration of BCM was chosen because it did not alter the growth rate of cells under the conditions used in these experiments (14), and thus limited the potential indirect effects that could result from inhibiting Rho. DNAs from ChIP experiments targeting the β or β′ subunit of RNAP and “input” genomic DNA were differentially labeled with Cy3 and Cy5 dyes, then hybridized to a tiling microarray (see Materials and Methods), revealing the RNAP distribution in BCM-treated and untreated conditions. Independent biological replicates showed good agreement (Pearson's r = 0.9).
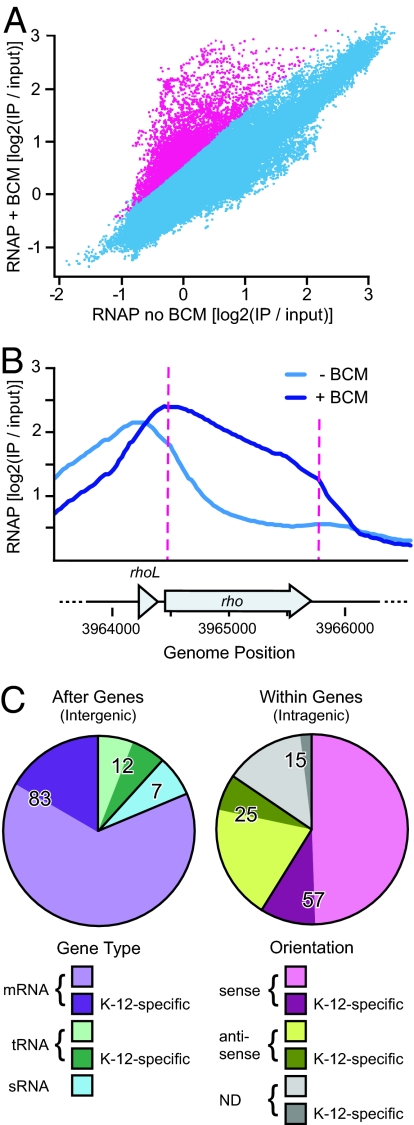
Changes in the distribution of RNAP upon BCM treatment were readily apparent by visual inspection of the data, and were quantified statistically. A moving average method implemented in the program CMARRT (15) was used to identify regions where at least 3 consecutive probes exhibited increased ChIP-chip signal in BCM-treated cells versus untreated cells (see Materials and Methods; no BCM-induced reductions in RNAP occupancy were detected). This analysis revealed a total of 199 BCM significant regions (BSRs) dispersed throughout the E. coli K-12 chromosome. Most of the probes with increased ChIP-chip signal in BCM-treated cells were within BSRs, but they represented only a small percentage of the total probes (≈3%; Fig. 1A). This suggests that the effects of BCM were mostly direct consequences of Rho inhibition rather than a large-scale redistribution of RNAP in response to cellular stresses or other pleiotropic effects.
Fig. 1.
Global effects of Rho inhibition on the distribution of RNAP. (A) Scatterplot of ChIP-chip data from untreated cells versus cells grown in BCM. Probes within BSRs are colored magenta. (B) BCM effect on the distribution of RNAP at the rho locus. RNAP ChIP-chip data from untreated (blue) and BCM-treated conditions (navy) were smoothed using 2 rounds of sliding-window averaging over 500 bp. Magenta dashed lines represent left and right boundaries of the rhoL BSR. Genes are shown as labeled arrows. (C) BSR classifications. Intergenic and intragenic BSRs are represented as separate pie charts, with corresponding keys below each chart.
The BSR dataset was compared to previously characterized Rho-dependent terminators, which confirmed that BCM effectively inhibited Rho in our experiment. For instance, the rho gene is autoregulated by a Rho-dependent terminator immediately upstream of its coding sequence (16). As expected, a BSR was found at the rho locus just after the rhoL gene (Fig. 1B). In untreated cells, ChIP-chip signal for RNAP was highest at the rho promoter, situated just upstream of rhoL. After the rhoL gene, the signal for RNAP decreased, indicative of Rho-dependent termination. When BCM was used to inhibit Rho function, however, the RNAP signal remained high throughout the Rho-dependent terminator region and gradually decreased across the rho gene, indicating readthrough of the rhoL terminator.
Our findings also are broadly consistent with effects of BCM on global mRNA expression reported by Cardinale et al. (13), but provide high-resolution positional information that could not be accessed through mRNA expression analysis alone. Based on genomic position, approximately half of all BSRs were located within 300 bp of an expression array probeset that was upregulated at least 2-fold in mRNA expression (supporting information (SI) Fig. S1). However, the mRNA expression analysis did not detect a large fraction of the BSRs identified in our dataset (49%). The lower resolution of the expression array data relative to the tiling array-based ChIP-chip data likely explains this discordance, although differences in experimental growth conditions could also contribute. Importantly, the ChIP-chip-derived BSR data define the locations at which Rho-dependent termination normally occurs.
To understand the roles of these Rho-dependent terminators, we next sought to associate each BSR with a specific gene. Although ChIP-chip experiments do not provide strand information per se, the “directionality” of terminator readthough was used to assess the orientation of RNAP on DNA. An example of directionality can be found at the rho locus (Fig. 1B). The distribution of RNAP shifts to the right downstream of rhoL in ChIP-chip data from BCM-treated cells compared with untreated cells. Therefore, the terminator must be on the “plus” strand at the 3′ end of rhoL. This logic was extended to assign each BSR to a particular gene (Table S1). When directionality could not be determined (as was the case in 15 BSRs), the BSR was assigned to the gene that contained the majority of significant probes for that BSR. Quantitative PCR of ChIP DNA was used to confirm the array results at 3 of the BSR-associated loci (rho, valVW, and rygD; Fig. S2).
Our analysis revealed a diverse set of Rho targets in the E. coli genome (Figs. 1C and 2, and Table 1). Half of the targets (n = 102) were after genes (intergenic targets), where Rho would be expected to terminate transcription. Most of these followed protein-coding genes (83 mRNAs), but 12 followed tRNA genes and 7 followed sRNA genes. However, the other half (n = 97) were within coding regions (intergenic targets), including 25 that could be assigned to antisense transcripts. This distribution suggests that Rho plays important roles in E. coli transcription in addition to termination at the ends of operons or mediation of polarity.
Fig. 2.
Locations of BSRs and BSR-associated genes across the E. coli chromosome. Genome features are represented to scale as colored bars.
Table 1.
BSR Annotation summary
| BSR type | tRNAa | sRNAb | mRNAc | Total | K-12 specificd | Prophaged | |
|---|---|---|---|---|---|---|---|
| Intergenic | 12 | 7 | 83 | 102 | 21 | 15 | |
| Intragenic | |||||||
| Sense | 0 | 0 | 57 | 57 | 10 | 4 | |
| Antisense | 0 | 0 | 25 | 25 | 6 | 3 | |
| NDe | 0 | 0 | 15 | 15 | 2 | 1 | |
| Total | 12 | 7 | 180 | 199 | 39 | 23 |
aNumber of BSRs associated with annotated tRNA genes.
bNumber of BSRs associated with annotated sRNA genes.
cNumber of BSRs associated with annotated mRNA genes.
dNumber of BSRs associated with E. coli K-12-specific genes or prophageDNA (ASAP Database, http://www.genome.wisc.edu/tools/asap.htm).
eDirectionality was not determined.
Rho Termination at tRNAs.
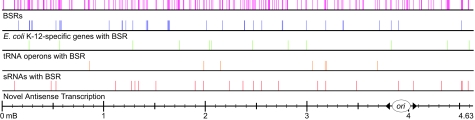
Many tRNA operons appeared to be terminated by Rho. Of the 36 tRNA-containing TUs located outside of rrn (and thus subject to termination), 12 had a BSR immediately downstream of the mature 3′ end of the last tRNA in the TU (Table S2). Rho termination had been previously demonstrated in vivo and in vitro at one of these tRNA TUs (tyrTV) (17). Two tRNA loci that show the effects of BCM treatment on the distribution of RNAP are valVW and thrW (Fig. 3A and Fig. S3). Although the RNAP ChIP-chip signal is restricted to the tRNA operon itself in untreated cells, BCM treatment caused the distribution of RNAP to extend downstream past the presumed Rho-dependent termination point. The ChIP-chip signals on tRNA operons without significant BCM effects, such as lysT-valT-lysW-valZ-lysYZQ, were qualitatively and quantitatively distinct from tRNA operons affected by BCM (cf. Fig. 3A and Fig. S3 to Fig. 3B).
Fig. 3.
Rho termination at tRNAs and sRNAs. BCM affects the distribution of RNAP at (A) the valVW tRNA operon but not (B) the lysT-valT-lysW-valZ-lysYZQ tRNA operon. The distribution of RNAP at (C) the sroG sRNA is affected by BCM. Colors, labels, and data smoothing are as described in Fig. 1B, except that noncoding RNA genes are colored yellow.
To determine the distinctions between tRNA operons that were affected by BCM and those that were not, we analyzed the sequence within and surrounding the BSR. The number of tRNAs in an operon, and the direction of transcription in genes downstream of the operon, had no relationship with Rho termination (Table S2, Fig. 3A, and Fig. S3). Additionally, no obvious “termination sequence” could be ascribed to tRNA BSRs using motif-finding algorithms (e.g., MEME; http://meme.sdsc.edu/meme4/). However, the first 50 nucleotides after the mature 3′ end of the tRNA differed significantly in GC content for tRNAs affected by BCM (Table S3). Although these sequences were only 25% C on average, they were significantly more enriched for C than their non-Rho terminated counterparts (Student's t test, P = 0.01) and significantly depleted in G. The average G content was only 12% within the first 50 nucleotides after these tRNAs, which was highly significant compared with tRNAs without corresponding BSRs (Student's t test, P = 0.001). These patterns are consistent with previous studies that noted a bias toward C and away from G in cases of Rho-dependent polarity after premature stop codons (18).
Unsurprisingly, the feature that most distinguished tRNA operons affected by BCM from those that were not was the presence or absence of a putative intrinsic terminator hairpin RNA structure. Of the 24 tRNA operons that lacked associated BSRs, 22 (92%) encoded putative intrinsic terminator hairpin structures and corresponding U-tracts within 150 bp of the 3′ end of the tRNA. Potential hairpins were identified by examining the RNA secondary structure in silico using the mfold algorithm (19) (Table S2). The 2 exceptions lacking both a BSR and putative intrinsic terminator were ileY and the thrU-tyrU-glyT-thrT operon. The ileY tRNA gene produced very little RNAP ChIP-chip signal in both BCM-treated and untreated conditions, and likely fell below the limits of detection. The thrU-tyrU-glyT-thrT operon is known to be cotranscribed with the downstream tufB gene (20). Although a small drop in RNAP ChIP-chip signal occurred between thrT and tufB, apparently, the majority of ECs were not terminated. Eleven of the 12 (92%) tRNA operons with an associated BSR lacked putative intrinsic terminator hairpin structures. The exception, asnU, contained a putative RNA structure that resembled an intrinsic terminator hairpin despite being affected by BCM treatment (Table S2). However, the purported terminator contained an unpaired A residue between the hairpin stem and U-tract. Systematic substitutions of U-tract residues with A in the canonical pyrBI intrinsic terminator revealed that mutations closer to the hairpin stem caused progressively greater termination defects (although the first U of the U-tract was not tested) (21). Also, weakening the base of the hairpin stem reduces termination markedly (22). Therefore, this deviation from a canonical intrinsic terminator would likely disrupt the function of the terminator hairpin. This finding raises the intriguing possibility that Rho-dependent termination is a “default” termination pathway in E. coli, taking over when intrinsic terminator hairpins are disrupted by mutation or removed by horizontal transfer events.
Rho Termination of sRNA Synthesis.
Genes in a second class uncovered in the BSR analysis encoded known sRNAs. Seven annotated sRNA genes were found to have BSRs associated with the 3′ end of the gene (Fig. 1C). Two types of Rho-dependent terminators were found at sRNAs. The first type was primarily involved in sRNA 3′ end formation. The rygD gene (also known as sibD) produces a noncoding stable RNA product that regulates the toxicity of the short, hydrophobic IbsD protein (23). An extension in the distribution of RNAP at rygD is seen in the presence of BCM, indicating that this sRNA is terminated by Rho (Fig. S4).
The second type of Rho-dependent terminator found at sRNAs appeared to play a role in the regulation of downstream genes. The sroG sRNA is situated in between the promoter and protein coding sequence of the ribB gene, which is involved in riboflavin synthesis (24). Although the exact function of the SroG RNA has not been demonstrated experimentally, sequence alignments suggest that it contains a flavin mononucleotide (FMN) binding riboswitch known as an RFN (riboflavin) element (25). Based on the absence of an intrinsic terminator hairpin, and complementarity between the Shine-Dalgarno (SD) of ribB and upstream sequences in the RNA, the riboswitch contained in sroG was proposed to operate by blocking translation of ribB in conditions of high FMN concentration (25). Interestingly, a BSR occurred at the 3′ end of sroG, implicating Rho-dependent termination as a mechanism for tightening the regulation of this riboswitch (Fig. 3C). The ribB transcript, when left untranslated, is logically a good substrate for Rho action, and termination by Rho would prevent synthesis of the full-length ribB mRNA. This would ensure that RibB protein could not be produced, even if SD pairing is lost by FMN release from the riboswitch. This system is similar to the Bacillus subtilis trp operon, where Rho termination occurs after translation initiation is blocked by a hairpin that occludes the SD of trpE (26). Our findings indicate that Rho termination at sRNAs can be involved both in 3′-end formation and in the mechanism by which sRNAs regulate their target genes. Just 7 of the ≈80 known sRNAs are terminated by Rho. Previous studies have identified sRNAs by searching for promoter-intrinsic terminator pairs in intergenic regions (see ref. 27), suggesting that only a fraction of Rho-terminated sRNAs have been discovered. Therefore, identifying Rho-dependent terminators with associated promoters could function as an additional method for finding novel sRNAs.
Rho Inhibition Reveals Antisense Transcription.
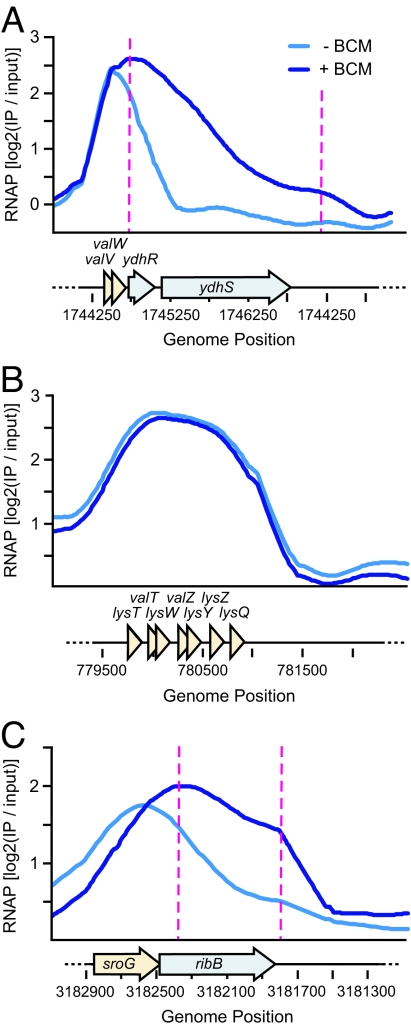
Although half of the BSRs were found at the 3′ ends of genes, as would be predicted if Rho functions to terminate RNAP at the ends of TUs (intergenic), the other half were located within genes (intragenic). In many cases, we found that the directionality of intragenic terminator readthrough was opposite to the direction of the annotated gene (Fig. 4 A and B). These observations were indicative of antisense transcription by RNAP. In total, we found 25 instances of antisense transcription in the BSR dataset, 24 of which were previously uncharacterized transcripts (Table S4). A majority (17/25) of the antisense transcripts had an associated σ70 peak in ChIP-chip data from Mooney et al. (12) that indicated a putative promoter for the transcript. We estimated the approximate lengths of the antisense transcripts by finding the distance between the start of the BSR and the midpoint of its associated σ70 peak. The average antisense transcript length was 456 nt. This number likely overstates the transcript length, because the same analysis applied to tRNAs overestimated their lengths by 50–150 nucleotides. Thus, the average length of antisense transcripts found in this study falls within the range of 50 to 400 nt typically assigned to sRNAs (28).
Fig. 4.
Rho inhibition reveals antisense transcription. (A) An antisense transcript within the eutB gene was detected based on the direction of terminator readthrough by RNAP in BCM-treated cells. σ70 ChIP-chip data (orange) from Mooney et al. (12) suggests a promoter location for the antisense transcript. (B) Unique antisense transcription within the bglF gene. Colors, labels, and data smoothing are as described in Fig. 1B, except that putative antisense transcripts are represented by red arrows.
Reppas et al. (29) had previously identified an antisense transcript on the opposite strand of the eutB gene. This transcript is also apparent in the BSR dataset due to the directionality of terminator readthrough (Fig. 4A), and a corresponding peak in σ70 ChIP-chip data suggests a promoter location for the transcript. An example of a previously uncharacterized antisense transcript found in the BSR dataset lies within the cryptic bgl operon on the opposite strand of the bglF gene (Fig. 4B). The ambiguous directionality of the σ70 and RNAP peaks in bglF was made clear by readthrough of an antisense, Rho-dependent terminator in BCM-treated cells, establishing the existence of antisense transcription in bglF.
Our finding of ≈100 intragenic Rho-dependent terminators shows that transcription in E. coli is much more complex than previously envisioned, with many transcripts terminated within coding sequences and a greater amount of antisense transcription. Intragenic terminators are associated with both sense and antisense transcription. Intragenic antisense transcripts terminated by Rho represent a mostly uncharacterized group of RNAs with unknown functions. Intragenic sense Rho-dependent terminators may be associated with transcriptional attenuation (30), premature termination due to failed translation, or synthesis of sRNAs that lie within larger genes.
Discussion
Our findings lead to 3 insights into the role of Rho in global gene regulation. First, Rho terminates synthesis of small noncoding RNAs, including tRNAs to a much greater extent than previously realized. This is significant because the extensive structure of such RNAs is thought to inhibit Rho binding. Second, Rho terminates synthesis of intragenic transcripts, including antisense transcripts, of unknown function. Many of these likely represent previously uncharacterized, noncoding transcripts in E. coli. Finally, the strong effect of Rho on horizontally transferred genes may reflect the propensity of such genes to insert at tRNA-encoding loci, rather than Rho-targeting of foreign DNA per se.
The BSR dataset is likely to reveal only a subset of Rho-dependent terminators in E. coli. Detection of Rho terminators by ChIP-chip requires sufficient occupancy of RNAP before the terminator to see the readthrough event. For instance, the well-characterized Rho-dependent trp t′ terminator (31) was barely discernable, and failed to meet the statistical cutoff due to low RNAP signal at the 3′ end of the trp operon. Many condition-specific Rho terminators also were likely missed (e.g., the tnaC terminator in the catabolite-repressed tna operon) (32). Finally, cryptic Rho terminators that occur only when transcription and translation are uncoupled would not be found because translation should be efficient under our assay conditions.
To estimate the total extent to which Rho terminates mRNA synthesis, we examined 109 high-quality TUs for which the RNAP ChIP-chip signal was significantly above background and could readily be distinguished from adjacent TUs (12). Of these 109 TUs, 18 were associated with intergenic BSRs, indicating that 17% of these TUs are terminated at their 3′ ends by Rho. We extrapolated this percentage out to the total predicted number of TUs in E. coli (2,271) (33), which gave 386 as the estimated number of intergenic Rho-dependent terminators. Based on this estimate, Rho-dependent termination is likely to account for ≈20% of the total mRNA 3′-end formation in E. coli, rather than the 50% estimate that is often cited (34). We note that the 50% estimate does not appear to be based on a genome-scale analysis.
Rho-Dependent Termination and Stable RNA Synthesis.
Stable RNA transcripts are surprising substrates for Rho action because they are typically highly structured, whereas Rho is thought to bind unstructured RNA. However, ChIP-chip analysis reveals Rho occupancy across most TUs, including sRNAs, tRNAs, and rRNAs (12). Thus, Rho appears capable of association with structured transcripts, consistent with our finding that Rho terminates these transcripts.
Rho-dependent termination of tRNA and sRNA transcripts is also unexpected because Rho generates heterogeneous transcript 3′ ends that would seemingly be problematic for the function of these RNAs. Extraneous 3′ nucleotides may interfere with folding or enzymatic modifications of stable RNAs, many of which require specific secondary structures for biological activity. However, extra 3′ nucleotides can be removed by multiple 3′ → 5′ exonucleases that exist in E. coli. For instance, a 3′ tail on the Rho-terminated valVW tRNA transcript becomes detectable only in a pnp rnb double mutant, implying that 3′ ends generated by Rho termination are rapidly degraded by redundant 3′ → 5′ exonucleases encoded by pnp and rnb (35). Thus, heterogeneous 3′ tails generated by Rho can be readily removed to avoid interfering with RNA function.
Rho inhibition had no effect on RNAP occupancy of rRNA TUs, whereas the recent proposal that Rho removes paused RNAPs predicts a 3′-proximal decrease (36). However, the lower level of rrn transcription expected for minimal medium could preclude detection of this effect.
Rho Terminates a Set of Antisense Transcripts of Unknown Function in E. coli.
We find that Rho is involved in termination of a set of antisense transcripts with unknown function. These antisense transcripts are likely to be noncoding, because the protein-coding sequence on the opposite strand greatly constrains the sequence of the antisense RNA. The 25 antisense transcripts we detected likely represent only a small fraction of a larger set of similar antisense transcripts in E. coli. As noted previously for intergenic transcripts, our method will miss a significant number on which the effect of BCM fails to generate a BSR. Additionally, to be detectable, antisense TUs within genes must also generate signals significantly above the level of the corresponding genic TU. Thus, E. coli likely possesses a large set of intragenic, antisense TUs of which the 25 we detected are only a limited, highly transcribed subset.
Some antisense transcripts may encode small RNAs with specific regulatory functions. For instance, antisense RNAs are known to block translation by pairing to a sense transcript (e.g., RyhB and IS10 in bacteria), to block formation of persistent RNA-DNA hybrids (e.g., RNAI in ColE1-type plasmids), or to interfere with sense transcription during their synthesis (37, 38). Some of these transcripts could conceivably produce sRNAs with functions unrelated to the genes within which they are embedded.
However, the possibility that some intragenic transcripts result from “transcriptional noise” must be considered. The involvement of Rho is itself compellingly analogous to some types of noncoding transcription in eukaryotes. Bacterial RNAP and eukaryotic RNAPII are both terminated by at least 2 distinct pathways. In many bacteria, intrinsic termination appears to be the dominant mechanism for termination of mRNA synthesis; indeed, our findings suggest Rho terminates only a minority of full-length E. coli mRNAs. RNAPII termination is coupled to transcript cleavage and polyadenlyation for most mRNAs (39), but can instead occur by the Nrd1/Nab3/Sen1-dependent pathway for small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and some short mRNAs (40). Sen1 contains an ATP-dependent, 5′ → 3′ RNA/DNA helicase activity and may function similarly to Rho (41). Thus, Rho-dependent termination in bacteria appears to be analogous to Sen1-dependent termination in eukaryotes. The Nrd1/Nab3/Sen1 pathway is implicated in the termination of cryptic unstable transcripts (CUTs) that become detectable in S. cerevisiae mutants defective for nuclear RNA degradation (42). Similar to pervasive noncoding transcription in other eukaryotes, the biological function of CUTs is unknown; however, CUTs may simply reflect transcriptional noise that is an unavoidable consequence of robust gene expression, and the Nrd1/Nab3/Sen1 pathway may play a role in “genome surveillance” by suppressing them. Given the similarities of Rho-dependent and Sen1-dependent termination, one possibility is that at least some antisense transcription terminated by Rho in bacteria may also reflect transcriptional noise.
Rho-Dependent Termination and Horizontal Transfer.
Our findings are consistent with a connection between Rho and suppressed expression of horizontally transferred, “foreign” genes (13), but suggest an indirect mechanism underlies this relationship. The connection is evident from the significant association of BSRs with E. coli K-12 genes lacking homologs in E. coli 0157:H7 EDL 933 (Mann-Whitney U test; P < 0.001). However, specific targeting of Rho to AU-rich RNA in horizontally transferred genes (13) can be ruled out, as the global distribution of Rho lacks bias toward any particular set of TUs (12).
Three non-mutually exclusive ideas may explain why Rho termination is associated with “foreign” DNA. First, foreign genes acquired from distantly related organisms may not be adapted to the E. coli translation apparatus, allowing Rho to act on poorly translated RNAs. Second, some foreign DNA may contain specific Rho-dependent terminators. For instance, the rac prophage contains the Rho-dependent timm terminator upstream of the lethal kilR gene (13); BCM causes readthrough of timm and the appearance of a BSR (Table S1).
Third, foreign DNA may preferentially insert into active TUs, and thereby produce readthrough transcription into foreign DNA that is terminated by Rho. Of the 63 E. coli K-12-specific genes associated with a BSR, 24 are inserted into active TUs at which Rho terminated transcription into the horizontally transferred DNA. This phenomenon is apparent at tRNA operons terminated by Rho (Fig. S5). Half the tRNAs terminated by Rho have associated BSRs that read into E. coli K-12-specific genes or prophage elements (Fig. 1C and Table S1). Indeed, the majority of prophages and other horizontally transferred elements in Gammaproteobacteria encode integrases that specifically target tRNAs genes as attachment sites (43). Thus, horizontally transferred elements may integrate into the chromosome by disruption of tRNA genes, causing loss of their intrinsic terminators. In such cases, Rho can supply an alternate termination mechanism to prevent transcription of potentially toxic foreign genes from the tRNA gene promoter. Williams (43) categorized horizontally transferred elements that use tRNA as insertion points across several species of proteobacteria and Gram-positive bacteria. Of the 54 characterized horizontally transferred elements that insert into tRNA, 22 (41%) lacked an intrinsic terminator within 400 bases of the mature 3′ end of the tRNA (43). These data suggest that Rho termination provides a general mechanism for guarding the borders of tRNA transcription against the deleterious consequences of foreign gene expression in a diverse set of bacteria.
Conclusion.
Rho-dependent termination plays many roles in bacterial transcription, including generation of full-length mRNA 3′ ends (1), establishment of polarity (2), resolution of extended RNA-DNA hybrids (44), and protection of cells from harmful expression of foreign genes (13). Our findings suggest Rho plays additional, and possibly more significant, roles by halting RNA chain elongation in a previously uncharacterized antisense transcriptome and by terminating synthesis of stable RNAs, including tRNAs and sRNAs. Rho-dependent termination is especially well suited for halting antisense transcription. The stringent sequence requirements of intrinsic terminators would be incompatible with a protein-coding gene on the opposite strand. In contrast, Rho-dependent terminators exhibit modest sequence specificity (C enriched and G depleted), which would place few limitations on codon usage in a protein-coding gene. Taken together, these data suggest Rho may play a principal role in halting transcription at locations where intrinsic terminators could not readily evolve (e.g., horizontally transferred DNA and antisense transcripts). Thus, further study of the targets of Rho may help elucidate the scope of the noncoding transcriptome of E. coli.
Materials and Methods
Growth Conditions.
E. coli K-12 MG1655 was grown in MOPS minimal medium containing 0.2% glucose at 37 °C with vigorous agitation in the presence or absence of 20 μg/mL BCM (12). BCM was obtained from Fujisawa Pharmaceutical Co.
ChIP-Chip.
ChIP-chip assays were performed as previously described (12). Briefly, cells were grown to an apparent OD600 of ≈0.4 and cross-linked by the addition of formaldehyde at 1% final concentration with continued shaking at 37 °C for 5 min before quenching with glycine (100 mM final). Cells were then lysed and DNAs were sheared by sonication followed by treatment with micrococcal nuclease and RNase A. RNAP crosslinked to DNA was immunoprecipitated using antibodies against either the β or β′ subunit (antibodies 8RB13 and NT73, respectively; Neoclone) using Sepharose protein A and G beads. Enriched ChIP DNA and input DNA were amplified by linker-mediated PCR (45) and processed by NimbleGen, Inc. to incorporate Cy3 or Cy5 dyes, hybridized to a tiling array, and quantified by fluorescence scanning. Two biological replicates were obtained for both BCM-treated and untreated conditions.
Array Designs.
We used 2 distinct isothermal tiling arrays that cover the entire E. coli K-12 MG1655 genome. The first array contained 187,204 oligonucleotide probes based on the sequence of the plus strand that were synthesized on the array in duplicate with ≈24-bp spacing (12), whereas the second contained 374,408 probes that alternated strands with ≈12-bp spacing.
Data Analysis.
We performed locally weighted linear regression (LOWESS) normalization (46) on raw Cy3 and Cy5 signals to correct for intensity-dependent dye effects within each array using the “normalizeWithinArrays” function (47) in the limma package (48) for the statistical program R (49). Normalized log2 ratios were then averaged over probe positions found in the 187,204 probe array to make the 2 array formats directly comparable. Next, biological replicates for BCM-treated or untreated conditions were quantile normalized between arrays using the “normalize.quantiles” function in the R package affy (50). For each of the BCM-treated (Trt) and untreated (UnTrt) conditions, we computed the average of the 2 biological replicates for each probe position. The analysis to identify regions enriched in Trt relative to UnTrt was performed using CMARRT (15) on the difference between the average of treated and untreated conditions (AveTrt − AveUnTrt) at the FDR level of 0.05.
Quantitative PCR.
Quantitative PCR was performed on ChIP DNA using SYBR Green JumpStart Taq ReadyMix for Real-Time PCR (Sigma-Aldrich) in an ABI 7500 Real-Time PCR System thermal cycler (Applied Biosystems). Two primers pairs were designed for each BSR locus tested. The first primer pair annealed before the BSR, and the second annealed within the BSR. Primer sequences are available upon request. Cycle threshold values obtained from quantitative PCR were converted to a relative quantity of DNA based on a standard curve created for each primer pair. The relative DNA quantity within the BSR was then normalized to the quantity before the BSR.
Supplementary Material
Acknowledgments.
We thank Yann Dufour for array design, and Nicole Perna for assistance in defining E. coli K-12-specific genes. We also thank Richard Gourse, David Brow, Charles Turnbough, Jr., and members of the Landick Lab for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM38660 to R.L.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nim.nih.gov/geo (accession no. GSE16562).
This article contains supporting information online at www.pnas.org/cgi/content/full/0903846106/DCSupplemental.
References
- 1.Roberts JW. Termination factor for RNA synthesis. Nature. 1969;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- 2.Richardson JP, Grimley C, Lowery C. Transcription termination factor Rho activity is altered in Escherichia coli with suA gene mutations. Proc Natl Acad Sci USA. 1975;72(5):1725–1728. doi: 10.1073/pnas.72.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galluppi GR, Richardson JP. ATP-induced changes in the binding of RNA synthesis termination protein Rho to RNA. J Mol Biol. 1980;138(3):513–539. doi: 10.1016/s0022-2836(80)80016-7. [DOI] [PubMed] [Google Scholar]
- 4.Richardson JP. How Rho exerts its muscle on RNA. Mol Cell. 2006;22(6):711–712. doi: 10.1016/j.molcel.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee S, Chalissery J, Bandey I, Sen R. Rho-dependent transcription termination: More questions than answers. J Microbiol. 2006;44(1):11–22. [PMC free article] [PubMed] [Google Scholar]
- 6.Zwiefka A, Kohn H, Widger WR. Transcription termination factor Rho: The site of bicyclomycin inhibition in Escherichia coli. Biochemistry. 1993;32(14):3564–3570. doi: 10.1021/bi00065a007. [DOI] [PubMed] [Google Scholar]
- 7.Yanofsky C, Horn V. Bicyclomycin sensitivity and resistance affect Rho factor-mediated transcription termination in the tna operon of Escherichia coli. J Bacteriol. 1995;177(15):4451–4456. doi: 10.1128/jb.177.15.4451-4456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magyar A, Zhang X, Kohn H, Widger WR. The antibiotic bicyclomycin affects the secondary RNA binding site of Escherichia coli transcription termination factor Rho. J Biol Chem. 1996;271(41):25369–25374. doi: 10.1074/jbc.271.41.25369. [DOI] [PubMed] [Google Scholar]
- 9.Park HG, et al. Bicyclomycin and dihydrobicyclomycin inhibition kinetics of Escherichia coli Rho-dependent transcription termination factor ATPase activity. Arch Biochem Biophys. 1995;323(2):447–454. doi: 10.1006/abbi.1995.0066. [DOI] [PubMed] [Google Scholar]
- 10.Skordalakes E, Brogan AP, Park BS, Kohn H, Berger JM. Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic bicyclomycin. Structure. 2005;13(1):99–109. doi: 10.1016/j.str.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Bubunenko M, Baker T, Court DL. Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J Bacteriol. 2007;189(7):2844–2853. doi: 10.1128/JB.01713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mooney RA, et al. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33(1):97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardinale CJ, et al. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320(5878):935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ederth J, Mooney RA, Isaksson LA, Landick R. Functional interplay between the jaw domain of bacterial RNA polymerase and allele-specific residues in the product RNA-binding pocket. J Mol Biol. 2006;356(5):1163–1179. doi: 10.1016/j.jmb.2005.11.080. [DOI] [PubMed] [Google Scholar]
- 15.Kuan PF, Chun H, Keles S. CMARRT: A tool for the analysis of ChIP-Chip data from tiling arrays by incorporating the correlation structure. Pacific Symposium on Biocomputing. 2008;13:515–526. [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto Y, Shigesada K, Hirano M, Imai M. Autogenous regulation of the gene for transcription termination factor Rho in Escherichia coli: Localization and function of its attenuators. J Bacteriol. 1986;166(3):945–958. doi: 10.1128/jb.166.3.945-958.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kupper H, Sekiya T, Rosenberg M, Egan J, Landy A. A Rho-dependent termination site in the gene coding for tyrosine tRNA su3 of Escherichia coli. Nature. 1978;272(5652):423–428. doi: 10.1038/272423a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alifano P, Rivellini F, Limauro D, Bruni CB, Carlomagno MS. A consensus motif common to all Rho-dependent prokaryotic transcription terminators. Cell. 1991;64(3):553–563. doi: 10.1016/0092-8674(91)90239-u. [DOI] [PubMed] [Google Scholar]
- 19.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JS, An G, Friesen JD, Fill NP. Location of the tufB promoter of E. coli: Cotranscription of tufB with four transfer RNA genes. Cell. 1981;25(1):251–258. doi: 10.1016/0092-8674(81)90250-6. [DOI] [PubMed] [Google Scholar]
- 21.Sipos K, Szigeti R, Dong X, Turnbough CL., Jr Systematic mutagenesis of the thymidine tract of the pyrBI attenuator and its effects on intrinsic transcription termination in Escherichia coli. Mol Microbiol. 2007;66(1):127–138. doi: 10.1111/j.1365-2958.2007.05902.x. [DOI] [PubMed] [Google Scholar]
- 22.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132(6):971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fozo EM, et al. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol Microbiol. 2008;70(5):1076–1093. doi: 10.1111/j.1365-2958.2008.06394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel J, et al. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31(22):6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002;30(14):3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yakhnin H, Babiarz JE, Yakhnin AV, Babitzke P. Expression of the Bacillus subtilis trpEDCFBA operon is influenced by translational coupling and Rho termination factor. J Bacteriol. 2001;183(20):5918–5926. doi: 10.1128/JB.183.20.5918-5926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livny J, Waldor MK. Identification of small RNAs in diverse bacterial species. Curr Opin Microbiol. 2007;10(2):96–101. doi: 10.1016/j.mib.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Vogel J, Sharma CM. How to find small non-coding RNAs in bacteria. Biol Chem. 2005;386(12):1219–1238. doi: 10.1515/BC.2005.140. [DOI] [PubMed] [Google Scholar]
- 29.Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell. 2006;24(5):747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 30.Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: How RNA provides instructions for transcription termination/antitermination decisions. Bioessays. 2002;24(8):700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 31.Wu AM, Christie GE, Platt T. Tandem termination sites in the tryptophan operon of Escherichia coli. Proc Natl Acad Sci USA. 1981;78(5):2913–2917. doi: 10.1073/pnas.78.5.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart V, Landick R, Yanofsky C. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J Bacteriol. 1986;166(1):217–223. doi: 10.1128/jb.166.1.217-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salgado H, et al. RegulonDB (version 3.0): Transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 2000;28(1):65–67. doi: 10.1093/nar/28.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu AQ, von Hippel PH. Rho-dependent termination within the trp t′ terminator. I. Effects of Rho loading and template sequence. Biochemistry. 1998;37(32):11202–11214. doi: 10.1021/bi9729110. [DOI] [PubMed] [Google Scholar]
- 35.Mohanty BK, Kushner SR. Ribonuclease P processes polycistronic tRNA transcripts in Escherichia coli independent of ribonuclease E. Nucleic Acids Res. 2007;35(22):7614–7625. doi: 10.1093/nar/gkm917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klumpp S, Hwa T. Stochasticity and traffic jams in the transcription of ribosomal RNA: Intriguing role of termination and antitermination. Proc Natl Acad Sci USA. 2008;105(47):18159–18164. doi: 10.1073/pnas.0806084105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136(4):615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward DF, Murray NE. Convergent transcription in bacteriophage lambda: Interference with gene expression. J Mol Biol. 1979;133(2):249–266. doi: 10.1016/0022-2836(79)90533-3. [DOI] [PubMed] [Google Scholar]
- 39.Lykke-Andersen S, Jensen TH. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie. 2007;89(10):1177–1182. doi: 10.1016/j.biochi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Steinmetz EJ, et al. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24(5):735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 41.Steinmetz EJ, Brow DA. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol. 1996;16(12):6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006;23(6):841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 43.Williams KP. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: Sublocation preference of integrase subfamilies. Nucleic Acids Res. 2002;30(4):866–875. doi: 10.1093/nar/30.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harinarayanan R, Gowrishankar J. Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J Mol Biol. 2003;332(1):31–46. doi: 10.1016/s0022-2836(03)00753-8. [DOI] [PubMed] [Google Scholar]
- 45.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11(3):709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 46.Yang YH, et al. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30(4):e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31(4):265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 48.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21(9):2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 49.Team RDC. R: A language and environment for statistical computing. 2008 Available at R Foundation for Statistical Computing, http://www.R-project.org/ [Google Scholar]
- 50.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.