Abstract
Vibrio cholerae, the causative agent of cholera, is a bacterium autochthonous to the aquatic environment, and a serious public health threat. V. cholerae serogroup O1 is responsible for the previous two cholera pandemics, in which classical and El Tor biotypes were dominant in the sixth and the current seventh pandemics, respectively. Cholera researchers continually face newly emerging and reemerging pathogenic clones carrying diverse combinations of phenotypic and genotypic properties, which significantly hampered control of the disease. To elucidate evolutionary mechanisms governing genetic diversity of pandemic V. cholerae, we compared the genome sequences of 23 V. cholerae strains isolated from a variety of sources over the past 98 years. The genome-based phylogeny revealed 12 distinct V. cholerae lineages, of which one comprises both O1 classical and El Tor biotypes. All seventh pandemic clones share nearly identical gene content. Using analogy to influenza virology, we define the transition from sixth to seventh pandemic strains as a “shift” between pathogenic clones belonging to the same O1 serogroup, but from significantly different phyletic lineages. In contrast, transition among clones during the present pandemic period is characterized as a “drift” between clones, differentiated mainly by varying composition of laterally transferred genomic islands, resulting in emergence of variants, exemplified by V. cholerae O139 and V. cholerae O1 El Tor hybrid clones. Based on the comparative genomics it is concluded that V. cholerae undergoes extensive genetic recombination via lateral gene transfer, and, therefore, genome assortment, not serogroup, should be used to define pathogenic V. cholerae clones.
Keywords: genomic islands, cholera toxin prophage, lateral gene transfer
Vibrio cholerae, a bacterium autochthonous to the aquatic environment, is the causative agent of cholera, a severe, watery, life-threatening diarrheal disease. Historically, cholera bacteria have been serogrouped based on their somatic O antigens, with >200 serogroups identified to date (1). Although strains from many of the serogroups of V. cholerae have caused either individual cases of mild gastroenteritis or local outbreaks of gastroenteritis, only the toxigenic strains of serogroups O1 and O139 have been identified as agents of cholera epidemics. Genes coding for cholera toxin, ctxAB, and other virulence factors have been shown to reside in bacteriophages and various mobile genetic elements. In addition, V. cholerae serogroup O1 is differentiated into two biotypes, classical and El Tor, by a combination of biochemical traits and sensitivity to specific bacteriophages (2).
Throughout human history cholera pandemics have been recorded with seven such pandemics characterized over the past hundred or more years. Today the disease remains endemic only in developing countries, even though V. cholerae is native to estuaries and river systems throughout the world (3). Isolates of the sixth pandemic were almost exclusively of the O1 classical biotype, whereas the current (seventh) pandemic is dominated by V. cholerae O1 El Tor biotype, a transition occurring between 1905 and 1961. The six pandemics previous to the current pandemic are considered to have originated in the Indian subcontinent, whereas the seventh pandemic strain was first isolated in the Indonesian island of Sulawesi in 1961, and subsequently in Asia, Africa, and Latin America.
Over the last 20 years, several new epidemic lineages of V. cholerae O1 El Tor have emerged or reemerged. In 1992, a new serogroup of V. cholerae, O139, was identified as the cause of epidemic cholera in India and Bangladesh (4). That is, both V. cholerae O1 El Tor and O139 consistently have been isolated where the major cholera epidemics have occurred since 1992, although V. cholerae O139 appears still to be restricted to Asia. Additionally, V. cholerae “hybrid” O1 El Tor variants that carry the classical type CTX prophage, or produce classical type cholera toxin subunit B have been isolated repeatedly in Bangladesh (5, 6) and Mozambique (7). These new variants have replaced the prototype seventh pandemic V. cholerae O1 El Tor strains in Asia and Africa, with respect to frequency of isolation from clinical cases of cholera.
It is clear that the dynamics of V. cholerae, like other enteric pathogens, involve extensive lateral gene transfer via transduction, conjugation, and transformation (2, 8, 9). However, the evolutionary history of this bacterium remains to be documented. Here, we compare the genome sequences of 23 V. cholerae strains (Table 1), representing diverse serogroups isolated at various times over the past 98 years from a variety of sources and geographical locations. We conclude that the current pandemic is caused by strains belonging to a single phyletic line, diversified mainly by lateral gene transfer occurring in the natural environment.
Table 1.
Characteristics of Vibrio cholerae strains analyzed in this study
| Strain | Genome code | Serogroup | Biotype | Geographical Origin | Source of isolation | Year of isolation | Sequencing status* | No. of contigs | Accession |
|---|---|---|---|---|---|---|---|---|---|
| N16961 | VCN16961 | O1 Inaba | El Tor | Bangladesh | Clinical | 1975 | Complete | 2 | AE003852/AE003853 |
| RC9 | VCRC9 | O1 Ogawa | El Tor | Kenya | Clinical | 1985 | S/4/E | 11 | ACHX00000000 |
| MJ-1236 | VCMJ1236 | O1 Inaba | El Tor | Matlab, Bangladesh | Clinical | 1994 | Complete | 2 | CP001485/CP001486 |
| B33 | VCB33 | O1 Ogawa | El Tor | Beira, Mozambique | Clinical | 2004 | S/4/E | 17 | ACHZ00000000 |
| CIRS 101 | VCCIRS101 | O1 Inaba | El Tor | Dhaka, Bangladesh | Clinical | 2002 | S/4/E | 18 | ACVW00000000 |
| MO10 | VCMO10 | O139 | Madras, India | Clinical | 1992 | S/4 | 84 | AAKF03000000 | |
| 2740–80 | VC274080 | O1 Inaba | El Tor | US Gulf Coast | Water | 1980 | Sanger | 257 | AAUT01000000 |
| BX330286 | VCBX330286 | O1 Inaba | El Tor | Australia | Water | 1986 | Complete | 8 | ACIA00000000 |
| MAK757 | VCMAK757 | O1 Ogawa | El Tor | Celebes Islands | Clinical | 1937 | Sanger | 206 | AAUS00000000 |
| NCTC 8457 | VC8457 | O1 Inaba | El Tor | Saudi Arabia | Clinical | 1910 | Sanger | 390 | AAWD01000000 |
| O395 | VCO395 | O1 Ogawa | Classical | India | Clinical | 1965 | Complete | 2 | CP000626/CP000627 |
| V52 | VCV52 | O37 | Sudan | Clinical | 1968 | Sanger | 268 | AAKJ02000000 | |
| 12129(1) | VC12129 | O1 Inaba | El Tor | Australia | Water | 1985 | S/4/E | 12 | ACFQ00000000 |
| TM 11079-80 | VCTM11079 | O1 Ogawa | El Tor | Brazil | Sewage | 1980 | S/4/E | 35 | ACHW00000000 |
| VL426 | VCVL426 | non-O1/O139 | Albensis | Maidstone, Kent, UK | Water | Unknown | Complete | 5 | ACHV00000000 |
| TMA21 | VCTMA21 | non-O1/O139 | Brazil | Seawater | 1982 | S/4/E | 20 | ACHY00000000 | |
| 1587 | VC1587 | O12 | Lima, Peru | Clinical | 1994 | Sanger | 254 | AAUR01000000 | |
| RC385 | VCRC385 | O135 | Chesapeake Bay | Plankton | 1998 | Sanger | 550 | AAKH02000000 | |
| MZO-2 | VCMZO2 | O14 | Bangladesh | Clinical | 2001 | Sanger | 162 | AAWF01000000 | |
| V51 | VCV51 | O141 | USA | Clinical | 1987 | Sanger | 360 | AAKI02000000 | |
| MZO-3 | VCMZO3 | O37 | Bangladesh | Clinical | 2001 | Sanger | 292 | AAUU01000000 | |
| AM-19226 | VCAM19226 | O39 | Bangladesh | Clinical | 2001 | Sanger | 154 | AATY01000000 | |
| 623–39 | VC62339 | non-O1/O139 | Bangladesh | Water | 2002 | Sanger | 314 | AAWG00000000 |
*Sanger, Draft assemblies by Sanger sequencing; S/4, Sanger sequencing and 454 pyrosequencing were combined; S/4/E, S/4 followed by quality improvement by standard genome sequencing procedures.
Results and Discussion
Phylogeny and Gene Content of Vibrio cholerae.
Phylogenetic analysis, accomplished using ≈1.4 million bp of orthologous protein-coding regions for 23 V. cholerae strains, revealed 12 distinct phyletic lineages. Strains belonging to non-O1/non-O139 serogroups from various sources showed significant genomic diversity (Fig. 1A). In fact, each unique phyletic line adds 206 new genes to the pan-genome of V. cholerae, on average (See SI Text and Fig. S1). In contrast, all V. cholerae serogroup O1 strains, except for two, comprised a monophyletic clade, designated V. cholerae phylocore genome (PG) clade. Strains of both the sixth and seventh pandemics are concluded to have evolved from a common ancestor of this PG clade.
Fig. 1.
Neighbor-joining trees showing phylogenetic relationships of 23 V. cholerae strains representing diverse serogroups. (A) All V. cholerae strains based on 1,676 genes (1,370,469 bp). (B) Phylocore genome (PG) clade based on 2,663 genes (2,567,393 bp). (C) Seventh pandemic (7P) clade based on 3,364 genes (3,291,577 bp). Bootstrap supports, as percentage, are indicated at the branching points. Bars represent the numbers of substitution per site, respectively. Only orthologous genes showing >95% nucleotide sequence similarity to those of V. cholerae N16961 were selected. The tree was rooted using Vibrio vulnificus YJ016 and Vibrio parahaemolyticus RIMD 2210633.
Twelve strains of the PG clade were further divided into two subgroups, as shown in the phylogenetic tree constructed using ≈2.6 million bp alignment (Fig. 1 A and B). The PG-1 subclade is comprised of most of the V. cholerae O1 El Tor strains and one V. cholerae O139 strain, whereas the PG-2 subclade contains strains of V. cholerae O1 classical and O37 serogroups. Interestingly, all clinical isolates associated with the current seventh cholera pandemic formed a very tight, monophyletic clade within the PG-1 subclade, which we have designated the seventh pandemic (7P) clade (Fig. 1C). V. cholerae O1 El Tor and O139 strains isolated from the Indian subcontinent and Africa epidemics during 1975 to 2004 are located in the 7P clade. We use the terms shift and drift in a manner similar in some respects to their use in studies of the influenza virus. In particular, we use shift to refer to long-term accumulation of numerous base pair mutations whereas drift to refer to short-term changes resulting from horizontal acquisition of genomic islands.
O Serogrouping in the Context of Genome Evolution.
The lipopolysaccharide (LPS) of V. cholerae consists of three major regions: lipid A, core oligosaccharide (OS), and O antigen. V. cholerae synthesizes core OS and O antigen using wav and wb* gene clusters, respectively (10, 11). Molecular phylogeny and genetic organization of the wav and wb* gene clusters are summarized in SI Text, and Figs. S2 and S3.
In contrast to the limited diversity observed in the wav gene cluster (5 major types), 11 different types of wb* gene clusters were observed among the 23 strains. Phylogeny and genetic organization, based on the whole genome (Fig. 1), core OS, and O antigen gene clusters (Fig. S2A), clearly indicate both core OS and O antigen gene clusters have been horizontally transferred. The relatively stable gene order (synteny) of the core OS gene cluster suggests that it transfers as an entity. In contrast, the region coding for the O antigen is comprised of combinations of several smaller gene sets of different origin, leading to a remarkable diversity of the various O antigens seen in nature (Fig. S2B). This finding is in good agreement with the study in ref. 12 showing that the gene cluster coding for the O139 antigen is similar to that of V. cholerae serogroup O22, where substitution of a part of the cluster occurred, but not a deletion.
Genome phylogeny (Fig. 1A) revealed that strains of O1 serogroup are found in three different phyletic lineages, namely the PG clade, and the V. cholerae O1 El Tor 12129 (1) and TM11079-80 strains, in which the coding region for the O1 antigen is nearly identical. It is concluded that the O1 antigen phenotype arose by lateral gene exchange at least three times in the evolution of V. cholerae presented here. Furthermore, we hypothesize that the ancestor of the PG clade possessed a combination of the type 1 core OS and the O1 antigen gene clusters, giving rise to the present 12 V. cholerae PG strains, including the two V. cholerae non-O1 strains (V52 and MO10). The latter two became different serogroups by gene replacement, via lateral gene transfer, with strain V52 receiving both type 1 core OS and O37 antigen gene clusters from a V. cholerae O37 strain and V. cholerae MO10 receiving only the V. cholerae O139 antigen gene cluster from an unknown source, most likely a variant of the V. cholerae O22 serogroup (12).
The V. cholerae O1 strains not belonging to the PG group, V. cholerae 12129 (1) and TM11079-80, are environmental isolates from Australia isolated in 1985 (13), and from Brazil, isolated in 1980 (14). They showed the typical El Tor phenotype, but unlike other V. cholerae O1 El Tor strains in the PG-1 subclade, lack the two major virulence-related genomic islands, i.e., CTX prophage containing ctxAB and Vibrio pathogenicity island-1 (VPI-1) containing genes for biosynthesis of the toxin coregulated pilin (TCP). By comparing genome phylogenies based on the whole genome (Fig. 1) and gene clusters coding for the core OS (Fig. S2A) and O1 antigen (Fig. S2C), it is clear that genesis of these nontoxigenic V. cholerae O1 El Tor strains can be attributed to independent lateral gene transfer events, most probably transfer of only the O1 antigen gene cluster, but not the core OS region.
Four O serogroup conversions, from non-O1 to O1 (twice), O1 to O139, and O1 to O37, were detected among the 23 V. cholerae. Several previous studies suggested such conversions take place in nature (11, 14, 15), and chitin-induced natural transformation has been proposed as the mechanism in the natural environment (16). V. cholerae O1 to O139 serogroup conversion by a single-step exchange of large fragments of DNA was demonstrated in a microcosm experiment (9), and is supported by the conclusion of this study that O serogroup conversion occurs frequently in nature. Mobility of the O phenotype in V. cholerae was first proposed by Colwell et al. (17), and the cumulative results of both in vivo and in vitro experiments are compelling. Given the inconsistency between O serogroup typing and genome-based phylogeny, we conclude that, at the very least, the term “O1 El Tor” is both misleading and inaccurate for describing a set of phylogenetically coherent V. cholerae strains, in light of the frequency of serogroup conversion. Therefore, we propose a new terminology based on genome sequence; namely the phylocore genome (PG) clade, PG-1 and PG-2 subclades, and seventh pandemic (7P) clade, to describe homologous intraspecific groups of V. cholerae (Fig. 1).
Virulence-Associated Prophage and Genomic Islands Within the Context of the Genome.
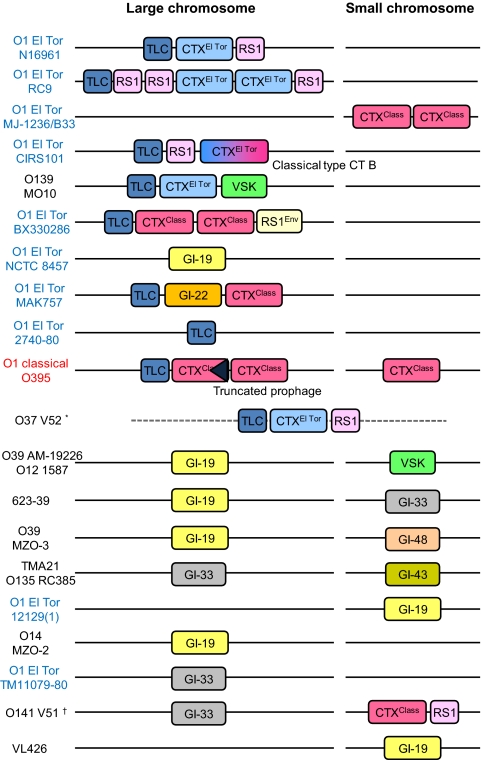
V. cholerae possesses several known virulence factors, of which the cholera toxin (CT) and TCP are considered the most significant. Genes coding for CT (ctxAB) are part of a temperate filamentous bacteriophage CTXφ (8) that can be incorporated into both chromosomes of V. cholerae at specific positions. The CTXφ genes were found to be present in members of the PG clade, except for V. cholerae NCTC 8457 and 2740–80. Among non-PG strains, only V. cholerae serogroup O141 (V51) contains this prophage.
The CTXφ found in classical and El Tor biotypes differs in the sequence of their repressor gene, rstR, and are classified as CTXφClass and CTXφEl Tor, according to the biotype of the original hosts in which they were described (18). From the genome sequences, we found that CTXφClass is not restricted to the classical biotype, but is also widely distributed in V. cholerae O1 El Tor and O141 strains (Fig. 2). Given that V. cholerae O1 El Tor MAK 757, a clinical strain isolated in 1937, has this type of prophage, correlation of host biotype and prophage type is not considered significant. A recent study(19) showing the infection of CTXφClass to V. cholerae non-O1 supports this finding.
Fig. 2.
Schematic representation of various prophages and genetic elements present in the target regions of CTXφ insetion. *, TLC, El Tor type CTXφ, RS1 element are found, but no positional information can be obtained from genome assemblies. †, classical type CTXφ and RS1 are present, but no positional information can be obtained.
Chromosomal attachment sites for CTXφ are known to harbor other genetic elements, including toxin-linked cryptic (TLC), RS1 elements, and VSK(=pre-CTX) prophages (20, 21). We have discovered five genomic islands (GI-19, GI-22, GI-33, GI-43, GI-48; for details see Table S1) in the region of the CTXφ attachment sites on both chromosomes. In total, nine distinct genetic elements were found in these regions, where they appear in different combinations (Fig. 2). Seven strains possess GI-19 in either chromosome, which is similar but not identical to KSF-1φ, previously discovered in an environmental V. cholerae strain (22). It is evident that more bacteriophages/genetic elements are located in the CTXφ attachment regions of PG strains than non-PG strains. The ability to harbor more, especially toxigenic, bacteriophage-like elements in these regions of the PG strains might explain why only PG strains have been the agents of the pandemics. We found no two toxigenic (CTXφ-harboring) strains with identical GI organization and combination, with the exception of two hybrid strains (the only 7P members harboring CTXφClass). It is evident from Fig. 2 that the two CTXφ attachment sites serve as an engine of genetic diversity for the V. cholerae PG clade.
Genes coding for TCP are part of a genomic island, VPI-1 present in all PG strains. Among the non-PG strains, only V. cholerae O141 V51 contained VPI-1 but with less sequence similarity. Because TCP serves as a receptor for CTXφ (8), it explains why only this strain, of all non-PG strains, possesses CTXφ. Results of phylogenetic analysis using the 24 genes of VPI-1 indicate that the original GI of V. cholerae NCTC 8457 was replaced by VPI-1 of a non-PG strain (Fig. S4). Interestingly, GI-47, but not VPI-1, was found in strains MZO-3, 1587, MZO-2, and VL426 in the same genomic region. This cassette-like property of GI mobility was also observed for the other known pathogenicity islands, including VPI-2, VSP-1, and VSP-2 (Table S2).
Extensive Lateral Gene Transfer in V. cholerae.
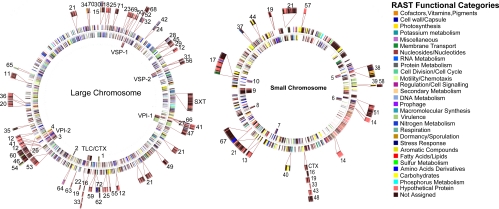
Because it is generally accepted that lateral gene transfer plays an important role in the evolution of many pathogenic bacteria, V. cholerae serves as a useful paradigm. For purposes of this study, a GI is defined as a genomic region containing five or more ORFs, where transfer, but not deletion, is obvious from comparison of genome phylogeny and its presence/absence among test strains. A total of 73 GIs were identified (Table S1) and their chromosomal locations are shown in Fig. 3. As discussed above, with respect to GIs associated with O antigen biosynthesis, CTXφ, VPI-1,2 and VSP-2, a total of 13 genomic regions (eight in the large and five in the small chromosome) were found to have a cassette-like property, whereby different GIs occupy the same or a similar region (Table S2). Most GIs were singletons in a given genome, although two (GI-12, GI-21) were present as four and two copies, respectively. Thus, we conclude that genetic diversity of V. cholerae derives most significantly from lateral gene transfer, of which several transfers are cassettes.
Fig. 3.
Genomic representation of genomic islands of both V. cholerae chromosomes. The two circles in the middle represent the genes in V. cholerae O1 El Tor N16961. The inner circle indicates genomic islands found in strain N16961, whereas the outer circles are those absent in strain N16961.
Genomic Definition of the V. cholerae Phylocore Genome (PG) Clade and Pandemic Strains.
The V. cholerae PG clade, with both sixth and seventh pandemic strains, is defined by gene content. Twenty-seven genes are present exclusively in the genomes of the PG strains, but only five genes are unique to the PG-1 subclade. Four of these (VCA0198–VCA0201) comprise a genomic island (GI-5) on the small chromosome, including genes coding for cytosine-specific DNA methyltransferase (23) and hypothetical proteins, adjacently located to the IS1004 transposase gene. The 7P strains are differentiated in harboring two unique GIs, the Vibrio seventh pandemic island-1 (VSP-1) and VSP-2, first discovered by microarray analysis (24). In addition to the 7P strains, a variant of VSP-1 was found in V. cholerae biovar albensis VL426 (Fig. S5). Similarly, VSP-2 like GIs were detected in three non-PG strains (TMA21, O39 MZO-3, O135 RC385). Interestingly, a similar GI was also detected in Vibrio vulnificus YJ016 and Vibrio splendidus 12B01, suggesting that VSP-2 may be widespread among vibrios (Fig. S6). It should be noted that the stability of these well known pathogenicity islands among 7P members is questionable, because most of VPI-2 and VSP-2 were deleted in MO10 and CIRS 101, respectively.
V. cholerae contains a superintegron, a large integron island (gene capture system), in the small chromosome (≈120 Kbp), comprising predominantly hypothetical genes and proposed as a source of genetic variation (25). All V. cholerae strains examined in this study have this integron, a source of much of the variation in gene content (Fig. S7). Interestingly, if this region is excluded, all six members of the 7P clade have an identical gene content, with the exception of a few genomic islands, including those found in the CTXφ attachment region. An SXT element belonging to a family of conjugative transposon-like mobile genetic elements encodes multiple antibiotic resistance genes and is present only in V. cholerae MO10, CIRS 101, MJ-1236, and B33, but not in the other V. cholerae strains. V. cholerae O139 MO10 differs from other members of the 7P clade in having an O139 antigen specific genomic island, a finding strongly supporting the conclusion of several previous studies that V. cholerae O139 derives from a seventh pandemic V. cholerae O1 El Tor strain (26). No other V. cholerae O139-specific genes were found in V. cholerae MO10.
The hybrid strains, possessing an El Tor biotype phenotype, but classical biotype CTXφ, were isolated during current cholera epidemics in Asia and Africa (6, 7). Two hybrid strains (B33 and MJ-1236) share a virtually identical genome backbone. Among 3,587,239 bp of orthologous protein-coding regions, only 106 nucleotide positions are different and the only significant difference is the presence of a V. cholerae MJ-1236 specific 19,729 bp genomic island (GI-12). This GI occurs four times as an almost identical sequence in the large chromosome, with 14 genes including those coding for the putative phage integrase and type I restriction-modification system, probably a recently introduced temperate bacteriophage. It is not clear why the hybrid strains outcompete V. cholerae O1 El Tor/O139 in the clinical setting, but a key to this puzzle surely lies in differences among closely related strains, i.e., tandem copies of CTXφClass, GI-14 and single nucleotide polymorphisms. In addition to these hybrid clones, V. cholerae O1 El Tor strains producing the classical type of cholera toxin B repeatedly have been isolated from patients in Asia and Africa (6). The genome sequence of a representative of this newly emerged group, i.e., V. cholerae CIRS 101, reveals that these strains also have a typical 7P gene content, but with CTXφEl Tor, not CTXφClass, albeit expressing the classical type subunit B protein (Fig. 2).
The comparative genomics of phylogenetically diverse strains has permitted analysis of the mechanism by which current seventh pandemic clones may have arisen. An highly conserved gene content, synteny, and significant similarity among the six strains of the 7P clade indicate that these V. cholerae strains share an almost identical genome “backbone,” having evolved very recently from a common ancestral strain. An hypothetical evolutionary pathway proposed for V. cholerae (Fig. 4), with GI migration matched to a genome-based phylogenetic tree, allows the conclusion that the ancestor for the 7P clade was a V. cholerae O1 El Tor strain containing several GIs (VPI-1,2, GI-1 to GI-10), receiving VSP-1, VSP-2 and GI-11 by lateral gene transfer, and finally giving rise to the contemporary V. cholerae O1 El Tor and O139 strains. Interestingly, such an hypothetical ancestral strain shows a gene content similar to V. cholerae O1 El Tor BX330286, isolated from a water sample collected in Australia in 1986, a geographic location near Indonesia where the first seventh pandemic V. cholerae O1 El Tor was reported in 1961.
Fig. 4.
Proposed hypothetical evolutionary pathway of the V. cholerae species. Probable insertions and deletions of genomic islands (Table S1) found in 23 V. cholerae strains are indicated by black and red arrows, respectively, along the phylogenetic tree based on genome sequence data. Hypothetical ancestral strains are indicated by open circles.
Mechanism of V. cholerae Evolution.
There are only a few human pathogens for which the complete sequences of many isolates are available (27, 28, 29). Because V. cholerae is both highly pathogenic for humans and an autochthonous inhabitant of estuaries worldwide, it provides a unique opportunity to elucidate evolutionary mechanisms. Furthermore, it is the natural inhabitant of the estuarine environment of both cholera epidemic and nonepidemic countries (3).
Unlike Salmonella enterica serovar Typhi and Bacillus anthracis, bacterial species showing clonal properties, V. cholerae, with Streptococcus agalactiae and Escherichia coli, offers a prime example of the important role of lateral gene transfer in the evolution of a bacterial species. The transition from sixth to seventh cholera pandemic genome type is concluded to result from a change from V. cholerae O1 classical to O1 El Tor biotype. We propose the term shift for the event occurring between two distinct phyletic lineages (Fig. 1B). It should be noted that only one genome of O1 classical biotype was included in this study, therefore more isolates of this biotype should be examined to determine its population structure.
In contrast, the present cholera global pandemic is ascribed to a change among 7P strains, e.g., emergence of V. cholerae O139, V. cholerae O1 El Tor hybrid, and V. cholerae O1 El Tor with altered cholera toxin subunit B. These represent transitions among genetically nearly identical clones, with a few different GIs, for which we propose the term drift. Much as in the case of influenza viruses, cholera bacteria undergo a shift/drift cycle over time, although the drift in V. cholerae is derived mainly from lateral gene transfer, most likely occurring in the natural environment in association with its plankton hosts (3, 30).
The present cholera global pandemic is concluded to have been initiated by multiple descendants of a V. cholerae O1 El Tor ancestor, diversified and continuously rapidly evolving, mainly via lateral gene transfer and most likely driven by environmental factors. Most importantly, the common genome backbone and variable genomic islands of the 7P clade of V. cholerae require that a reevaluation be done of the epidemiological practice that employs serogroups as the primary marker for V. cholerae. The so-called pandemic clones, identified by serogroup, instead, should be defined by gene content, the description of which offers significantly greater potential for development of reliable and useful diagnostics, vaccines, and therapeutics for cholera. Without doubt, more variants of the 7P clade, as a result of drift, will be encountered in the future, yielding new serogroups (other than O1 and O139) and phenotypic combinations. Public health workers will be unprepared if the evolution of this species remains unappreciated as an ongoing process in the natural environment, where V. cholerae is autochthonous and plays an important role in the nutrient cycles of the natural aquatic ecosystem.
Materials and Methods
Genome Sequencing.
Draft sequences were obtained from a blend of Sanger and 454 sequences and involved paired end Sanger sequencing on 8-kb plasmid libraries to 5× coverage, 20× coverage of 454 data, and optional paired end Sanger sequencing on 35-kb fosmid libraries to 1–2× coverage (depending on repeat complexity). To finish the genomes, a collection of custom software and targeted reaction types were used. In addition to targeted sequencing strategies, Solexa/Illumina data in an untargeted strategy were used to improve low quality regions and to assist gap closure. Repeat resolution was performed using in-house custom software. Targeted finishing reactions included transposon bombs (31), primer walks on clones, primer walks on PCR products, and adapter PCR reactions. Gene-finding and annotation were achieved using the RAST server (32) and details are given in Table S3.
Comparative Genomics.
Genome to genome comparison was performed using three approaches, because completeness and quality of nucleotide sequences varied from strain to strain (Table 1). First, ORFs of a given pair of genomes were reciprocally compared each other, using the BLASTN, BLASTP and TBLASTX programs (ORF-dependent comparison). Second, a bioinformatic pipeline was constructed to identify homologous regions of a given query ORF. Initially, a segment on target contig, which is homologous to a query ORF, was identified using the BLASTN program. This potentially homologous region was expanded in both directions by 2,000 bp. Nucleotide sequences of the query ORF and selected target homologous region were then aligned using a pairwise global alignment algorithm (33), and the resultant matched region in the subject contig was extracted and saved as a homolog (ORF-independent comparison). Orthologs and paralogs were differentiated by reciprocal comparison. In most cases, both ORF-dependent and -independent comparisons yielded the same orthologs, although ORF-independent method performed better for draft sequences of low quality, in which sequencing errors, albeit rare, hampered identification of correct ORFs.
Identification and Annotation of Genomic Islands.
In this study, we defined genomic islands (GIs) as a continuous array of five or more ORFs that were found to be discontinuously distributed among genomes of test strains. Correct transfer or insertion of GIs was readily differentiated from deletion event by comparing genome-based phylogenetic tree and full matrices showing pairwise detection of orthologous genes between test strains. Identified GIs were designated, and annotated using the BLASTP search of its member ORFs against GenBank NR database.
Phylogenetic Analyses Based on Genome Sequences.
A set of orthologues for each ORF of V. cholerae N16961 was obtained for different sets of strains, and then aligned using the CLUSTALW2 (34) program. The resultant multiple alignments were concatenated to generate genome scale alignments, which were subsequently used to reconstruct the neighbor-joining phylogenetic tree (35). The evolutionary model of Kimura (36) was used to generate the distance matrix. The program MEGA (37) was used for phylogenetic analysis.
Supplementary Material
Acknowledgments.
This work was supported in part by Korea Science and Engineering Foundation National Research Laboratory Program Grant R0A-2005-000-10110-0 (to J.C.); National Institutes of Health Grant 1RO1A139129-01 (to R.R.C.); National Oceanic and Atmospheric Administration, Oceans and Human Health Initiative Grant S0660009 (to R.R.C.); Department of Homeland Security Grant NBCH2070002 (to R.R.C.); Intelligence Community Post-Doctoral Fellowship Program (to C.J.G.); and the Korean and Swedish governments (to I.V.I.). Funding for genome sequencing was provided by the Office of the Chief Scientist and National Institute of Allergy and Infectious Diseases Microbial Sequencing Centers Grants N01-AI-30001 and N01-AI-40001.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907787106/DCSupplemental.
References
- 1.Chatterjee SN, Chaudhuri K. Lipopolysaccharides of Vibrio cholerae. I. Physical and chemical characterization. Biochim Biophys Acta. 2003;1639:65–79. doi: 10.1016/j.bbadis.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Kaper JB, Morris JG, Jr, Levine MM. Cholera Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colwell RR. Global climate and infectious disease: The cholera paradigm. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- 4.Ramamurthy T, et al. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 5.Nair GB, et al. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J Clin Microbiol. 2002;40:3296–3299. doi: 10.1128/JCM.40.9.3296-3299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nair GB, et al. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol. 2006;44:4211–4213. doi: 10.1128/JCM.01304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ansaruzzaman M, et al. Cholera in Mozambique, variant of Vibrio cholerae. Emerg Infect Dis. 2004;10:2057–2059. doi: 10.3201/eid1011.040682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 9.Blokesch M, Schoolnik GK. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 2007;3:e81. doi: 10.1371/journal.ppat.0030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nesper J, et al. Comparative and genetic analyses of the putative Vibrio cholerae lipopolysaccharide core oligosaccharide biosynthesis (wav) gene cluster. Infect Immun. 2002;70:2419–2433. doi: 10.1128/IAI.70.5.2419-2433.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Shimada T, Morris JG, Jr, Sulakvelidze A, Sozhamannan S. Evidence for the emergence of non-O1 and non-O139 Vibrio cholerae strains with pathogenic potential by exchange of O-antigen biosynthesis regions. Infect Immun. 2002;70:2441–2453. doi: 10.1128/IAI.70.5.2441-2453.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamasaki S, et al. The genes responsible for O-antigen synthesis of Vibrio cholerae O139 are closely related to those of Vibrio cholerae O22. Gene. 1999;237:321–332. doi: 10.1016/s0378-1119(99)00344-3. [DOI] [PubMed] [Google Scholar]
- 13.Safa A, et al. Multilocus genetic analysis reveals that the Australian strains of Vibrio cholerae O1 are similar to the pre-seventh pandemic strains of the El Tor biotype. J Med Microbiol. 2009;58:105–111. doi: 10.1099/jmm.0.004333-0. [DOI] [PubMed] [Google Scholar]
- 14.Farfan M, Minana D, Fuste MC, Loren JG. Genetic relationships between clinical and environmental Vibrio cholerae isolates based on multilocus enzyme electrophoresis. Microbiology. 2000;146(Pt 10):2613–2626. doi: 10.1099/00221287-146-10-2613. [DOI] [PubMed] [Google Scholar]
- 15.Bik EM, Gouw RD, Mooi FR. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: A tool to identify epidemic strains. J Clin Microbiol. 1996;34:1453–1461. doi: 10.1128/jcm.34.6.1453-1461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 17.Colwell RR, Huq A, Chowdhury MA, Brayton PR, Xu B. Serogroup conversion of Vibrio cholerae. Can J Microbiol. 1995;41:946–950. doi: 10.1139/m95-131. [DOI] [PubMed] [Google Scholar]
- 18.Davis BM, Moyer KE, Boyd EF, Waldor MK. CTX prophages in classical biotype Vibrio cholerae: Functional phage genes but dysfunctional phage genomes. J Bacteriol. 2000;182:6992–6998. doi: 10.1128/jb.182.24.6992-6998.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Udden SM, et al. Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc Natl Acad Sci USA. 2008;105:11951–11956. doi: 10.1073/pnas.0805560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubin EJ, Lin W, Mekalanos JJ, Waldor MK. Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol Microbiol. 1998;28:1247–1254. doi: 10.1046/j.1365-2958.1998.00889.x. [DOI] [PubMed] [Google Scholar]
- 21.Faruque SM, et al. Genomic analysis of the Mozambique strain of Vibrio cholerae O1 reveals the origin of El Tor strains carrying classical CTX prophage. Proc Natl Acad Sci USA. 2007;104:5151–5156. doi: 10.1073/pnas.0700365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faruque SM, et al. CTXphi-independent production of the RS1 satellite phage by Vibrio cholerae. Proc Natl Acad Sci USA. 2003;100:1280–1285. doi: 10.1073/pnas.0237385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banerjee S, Chowdhury R. An orphan DNA (cytosine-5-)-methyltransferase in Vibrio cholerae. Microbiology. 2006;152(Pt 4):1055–1062. doi: 10.1099/mic.0.28624-0. [DOI] [PubMed] [Google Scholar]
- 24.Dziejman M, et al. Comparative genomic analysis of Vibrio cholerae: Genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci USA. 2002;99:1556–1961. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karaolis DK, Lan R, Reeves PR. Molecular evolution of the seventh-pandemic clone of Vibrio cholerae and its relationship to other pandemic and epidemic V. cholerae isolates. J Bacteriol. 1994;176:6199–6206. doi: 10.1128/jb.176.20.6199-6206.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasko DA, et al. The pangenome structure of Escherichia coli: Comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol. 2008;190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome.”. Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holt KE, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Constantin de Magny G, et al. Environmental signatures associated with cholera epidemics. Proc Natl Acad Sci USA. 2008;105:19676–19681. doi: 10.1073/pnas.0809654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goryshin IY, Reznikoff WS. Tn5 in vitro transposition. J Biol Chem. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 32.Aziz RK, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers EW, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 34.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S, Nei M, Dudley J, Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.