Abstract
Human induced pluripotent stem (iPS) cells hold great promise for cardiovascular research and therapeutic applications, but the ability of human iPS cells to differentiate into functional cardiomyocytes has not yet been demonstrated. The aim of this study was to characterize the cardiac differentiation potential of human iPS cells generated using OCT4, SOX2, NANOG, and LIN28 transgenes compared to human embryonic stem (ES) cells. The iPS and ES cells were differentiated using the embryoid body (EB) method. The time course of developing contracting EBs was comparable for the iPS and ES cell lines, although the absolute percentages of contracting EBs differed. RT-PCR analyses of iPS and ES cell-derived cardiomyocytes demonstrated similar cardiac gene expression patterns. The pluripotency genes OCT4 and NANOG were downregulated with cardiac differentiation, but the downregulation was blunted in the iPS cell lines due to residual transgene expression. Proliferation of iPS and ES cell derived-cardiomyocytes based on BrdU labeling was similar, and immunocytochemistry of isolated cardiomyocytes revealed indistinguishable sarcomeric organizations. Electrophysiology studies indicated that iPS cells have a capacity like ES cells for differentiation into nodal-, atrial-, and ventricular-like phenotypes based on action potential characteristics. Both iPS and ES cell-derived cardiomyocytes exhibited responsiveness to β-adrenergic stimulation manifest by an increase in spontaneous rate and a decrease in action potential duration. We conclude that human iPS cells can differentiate into functional cardiomyocytes, and thus iPS cells are a viable option as an autologous cell source for cardiac repair and a powerful tool for cardiovascular research.
Keywords: induced pluripotent stem cells, embryonic stem cells, cardiomyocyte, action potential, differentiation
Introduction
The generation of induced pluripotent stem (iPS) cells from mouse somatic cells using a combination of four retrovirally transduced transcription factors (Oct3/4, Sox2, Klf4, and c-Myc) has opened remarkable new avenues for basic research and regenerative medicine applications.1 The promise of applying this technology to human cells was rapidly realized with the successful generation of human iPS cells from human fibroblasts using either the same combination of transcription factors or an independently determined combination of lentivirally transduced genes (OCT4, SOX2, NANOG and LIN28).2, 3 This approach has also recently been applied to generate disease-specific iPS cell lines from patients with a variety of diseases.4, 5
Like mouse ES cells, mouse iPS cells have most rigorously been proven to be pluripotent by generation of chimeric animals following blastocyst injection with subsequent germline transmission.6, 7 In comparison, ethical constraints preclude human embryo experiments for human iPS cells, and so formation of teratomas following transplantation of human iPS cells to immmunocompromised mice has provided evidence for pluripotency based on finding derivatives of the three embryonic germ layers.2, 3 In addition, in vitro differentiation studies are critically important for demonstrating pluripotency of human iPS cells and for characterizing the properties of the committed cell types that form. In vitro differentiation studies of various human iPS cell lines have identified derivatives of the three primary germ layers,2, 3, 8 but detailed characterization of the ability of human iPS cells to form specific cell lineages with functional characterization of the resulting cells are generally lacking. Critical issues such as viral integration, the combination of reprogramming genes, and residual transgene expression could fundamentally impact the differentiation potential of each iPS cell line.
Cardiac differentiation of human ES cells has been well-described using embryoid body (EB) formation or more recently using directed differentiation approaches.9-13 Detailed molecular and functional characterization of the resulting ES-cell derived cardiomyocytes revealed multiple cell types including nodal, atrial and ventricular cardiomyocytes typically found in the human heart.12, 14, 15 Given the promise of human iPS cells to supply large quantities of patient-specific cells for cardiac repair without the risk of immune rejection, it is essential to evaluate of the ability of human iPS cells to undergo cardiogenesis. Furthermore, use of iPS cell-derived cardiomyocytes as in vitro models for cardiac disease or other research applications will require careful characterization of the properties of the cardiomyocytes. The purpose of this study was to provide a detailed evaluation of the cardiac differentiation potential of recently described human iPS cell lines induced by OCT4, SOX2, NANOG, and LIN28 in comparison to well-studied human ES cell lines, H1 and H9.3, 16
Materials and Methods
Human iPS and ES Cell Culture
Human iPS cell lines reprogrammed by the lentiviral-mediated transduction of four transcription factors (OCT4, SOX2, NANOG and LIN28) were previously described.3 In the present study, we utilized a subset of those iPS clones of fetal origin including IMR90 clone 1 (IMR90 C1), IMR90 clone 4 (IMR90 C4), and of newborn origin including Foreskin clone 1 (Foreskin C1) and Foreskin clone 2 (Foreskin C2). In addition, we used two human ES cell lines, H1 and H9.16 The iPS cells and ES cells were maintained on irradiated mouse embryonic fibroblasts (MEFs) at a density of 19,500 cells/cm2 in 6-well culture plates (Nunc) as previously described.3, 12 In brief, both iPS and ES cells were maintained in DMEM/F12 culture medium supplemented with 20% KnockOut serum replacer, 0.1 mmol/L non-essential amino acids, 1 mmol/L L-glutamine (all from Invitrogen), and 0.1 mmol/L β-mercaptoethanol (Sigma). In addition, the medium was supplemented with 100 ng/ml zebrafish basic fibroblast growth factor (zbFGF) (purified from a bacterial expression system by us) for iPS cells, and with 4 ng/ml human recombinant bFGF (Invitrogen) for hES cells.
EB Formation and Cardiac Differentiation
Prior to EB formation, the iPS cells and ES cells were passaged onto a lower density of MEFs (∼13,000 cells/cm2) and expanded for 3 – 4 days. Colonies were detached from 6-well culture plates by incubating with 1mg/ml dispase (Gibco) solution at 37°C for 8-15 minutes and placed in ultra-low attachment plates (Corning, Cat. No. 3471) in suspension culture for 4 days. Differentiation medium - EB20, consisting of 80% DMEM/F12, 0.1 mmol/L non-essential amino acids, 1 mmol/L L-glutamine, 0.1 mmol/L β-mercaptoethanol, and 20% fetal bovine serum (FBS) that was pre-tested for cardiac differentiation (Gibco, Cat. No. 16000-044, Lot No. 291526), was used to initiate cardiac differentiation. During suspension culture, the medium was changed at day 1 followed by culture for another 3 days without medium change. EBs were then plated on 0.1% gelatin-coated 6-well culture plates (Nunc) at the density of 50-100 EBs /well, and cultured in EB20 medium (changed daily) for a total of 10 days from EB formation. After 10 days of differentiation, the FBS concentration was reduced to 2% (EB2 medium). The number of contracting EBs and contraction rates were measured at day 10, 20, 30 and 60 from EB formation using a microscope with a heated stage (37°C).
RT-PCR and Quantitative RT-PCR
Total RNA was isolated using Trizol Reagent (Invitrogen) from one well of a 6-well plate of undifferentiated iPS cells and ES cells and from 30-40 contracting areas microdissected from day 60 iPS or ES cell EBs. Total RNA from human heart tissue (obtained from donor hearts rejected for transplant due to technical reasons, following a protocol approved by the University of Institutional Review Board) was isolated from 1 g of left ventricular tissue using RNAzol B solution (Tel-Test, Friendswood, TX).17 Possible genomic DNA contamination was removed by DNase I (Invitrogen) treatment for 15 minutes at room temperature. 500 ng of total RNA was used for Oligo(dT)20 – primed reverse transcription using SuperScript™ III First-Strand Synthesis System (Invitrogen). RT-PCR was carried out using Platinum™ Taq DNA Polymerase (Invitrogen). Genes of interest in RT-PCR and Q-PCR are listed in Table 1. PCR conditions included denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 1 minute, for 35 cycles, with 72°C extension for 7 minutes at the end. ACTB (β-actin) was used as an endogenous control in RT-PCR. Quantitative RT-PCR was performed using Power SYBR® Green PCR Master Mix (Applied Biosystems) in triplicate for each sample and each gene. One μl of 1:5 dilution of cDNA from RT reaction was added as template for each RT-PCR or Q-PCR reaction. For Q-PCR, the cDNA from undifferentiated H1 ES cells was used as a relative standard for the measurement of total and endogenous expression of OCT4 and NANOG. The expression of genes of interest was normalized to that of GAPDH in Q-PCR.
Table 1.
Primers for RT-PCR and Q-PCR
| Genes | Sequences (5′ - 3′) | Size (bp) | ||
|---|---|---|---|---|
| Primers for RT-PCR | ||||
| OCT43 | F: CAGTGCCCGAAACCCACAC | 161 | ||
| R: GGAGACCCAGCAGCCTCAAA | ||||
| NANOG3 | F: CAGAAGGCCTCAGCACCTAC | 111 | ||
| R: ATTGTTCCAGGTCTGGTTGC | ||||
| NKX2-59 | F: GCGATTATGCAGCGTGCAATGAGT | 220 | ||
| R: AACATAAATACGGGTGGGTGCGTG | ||||
| TNNT29 | F: TTCACCAAAGATCTGCTCCTCGCT | 165 | ||
| R: TTATTACTGGTGTGGAGTGGGTGTGG | ||||
| MYH615 | F: GGGGACAGTGGTAAAAGCAA | 542 | ||
| R: TCCCTGCGTTCCACTATCTT | ||||
| ACTN215 | F: GGCGTGCAGTACAACTACGTG | 580 | ||
| R: AGTCAATGAGGTCAGGCCGGT | ||||
| MYL715 | F: GAGGAGAATGGCCAGCAGGAA | 449 | ||
| R: GCGAACATCTGCTCCACCTCA | ||||
| MYL29 | F: ACATCATCACCCACGGAGAAGAGA | 164 | ||
| R: ATTGGAACATGGCCTCTGGATGGA | ||||
| HPPA15 | F: GAACCAGAGGGGAGAGACAGAG | 406 | ||
| R: CCCTCAGCTTGCTTTTTAGGAG | ||||
| PLN15 | F: ACAGCTGCCAAGGCTACCTA | 191 | ||
| R: GCTTTTGACGTGCTTGTTGA | ||||
| ACTB15 | F: CCTGAACCCTAAGGCCAACCG | 400 | ||
| R: GCTCATAGCTCTTCTCCAGGG | ||||
| Primers for quantitative RT-PCR | ||||
| GAPDH | F: GTGGACCTGACCTGCCGTCT | 152 | ||
| R: GGAGGAGTGGGTGTCGCTGT | ||||
| OCT4 (total) | F: CAGTGCCCGAAACCCACAC | 161 | ||
| R: GGAGACCCAGCAGCCTCAAA | ||||
| OCT4 (endogenous) | F: AGTTTGTGCCAGGGTTTTTG | 113 | ||
| R: ACTTCACCTTCCCTCCAACC | ||||
| NANOG (total) | F: CAGAAGGCCTCAGCACCTAC | 111 | ||
| R: ATTGTTCCAGGTCTGGTTGC | ||||
| NANOG (endogenous) | F: TTTGGAAGCTGCTGGGGAAG | 194 | ||
| R: GATGGGAGGAGGGGAGAGGA | ||||
Immunolabeling
Undifferentiated iPS cells were plated on coverslips with MEF feeders as described for iPS cell culture. Single cardiomyocytes were isolated from day 60 microdissected contracting areas using 0.25% trypsin-EDTA (Invitrogen) plus 2% chick serum (Sigma) for 5-10 minutes at 37°C. The cells were washed and plated on coverslips coated with 0.1% gelatin solution in EB20 medium for 3 days to allow attachment. Cells were fixed in 4% paraformaldehyde for 15 minutes at room temperature, rinsed twice in PBS, and permeabilized in 0.2% Triton X-100 (Sigma) for 1 hour at room temperature. Samples were blocked with 5% non-fat dry milk (Bio-Rad) in 0.2% Triton X-100 solution and incubated for 2 hours at room temperature on a rotator followed by two washes with PBS. Primary antibodies, including monoclonal anti-Oct4 (IgG2b, Santa Cruz Biotechnology, 1:100 dilution), polyclonal anti-Nanog (IgG, Cosmo Bio Co., Ltd, 1:100 dilution), monoclonal anti-α-actinin (IgG1, Sigma, 1:500 dilution), monoclonal anti-cTnT (IgG1, Thermo Scientific, 1:200 dilution), monoclonal anti-MLC2a (IgG2b, Synaptic Systems, Germany, 1:400 dilution) and polyclonal anti-MLC2v (IgG, ProteinTech Group, 1:200 dilution) were added in 0.1% Triton X-100, 1% BSA in PBS solution and incubated overnight at 4°C. The samples were washed with 0.2% Tween 20 in PBS twice and 1× PBS twice. The isotype specific secondary antibodies, Alexa Fluor 488 goat anti-mouse IgG1 (Invitrogen) and Alexa Fluor 594 goat anti-mouse IgG2b (Invitrogen) were used for IgG1 and IgG2b isotype primary antibodies. Alexa Fluor 488 or 594 goat anti-rabbit IgG (H+L) (Invitrogen) was used for polycolonal antibodies. The secondary antibodies were diluted in the same solution as the primary antibodies and incubated at room temperature for 1.5 hours in the dark on a rotator. The samples were washed with 0.2% Tween 20 in PBS twice and 1× PBS twice. Nuclei were stained with Hoechst or DAPI (Invitrogen, 1:1000 dilution) for 5 minutes at room temperature followed by three washes with PBS. One drop of antifade reagent (Invitrogen) was placed on each slide, and coverslips were applied with cell surface down. The slides were examined with an epifluorescence microscope (Leica DM IRB) and imaged using QImaging® Retiga 4000R camera.
Analysis of Cardiomyocyte Proliferation
Single cardiomyocytes were isolated from early (day 10-20) and late (day 60) contracting EBs followed by plating on glass coverslips as described above for 2 days. Cells were pulsed for 17 hours with 10 μmol/L BrdU (Invitrogen) in EB2 medium. Cells were fixed in 4% paraformaldehyde and washed twice with PBS solution. DNA was denatured in 2 mol/L HCL, 0.1% Triton X-100 in PBS solution for 15 minutes at room temperature and washed for three times with PBS solution. Primary antibodies for sarcomeric myosin, MF20 (IgG2b, Developmental Studies Hybridoma Bank, Iowa City, IA, 1:20 dilution), and BrdU (IgG1, Invitrogen, 1:200 dilution) were used to co-label the cells as described above. Alexa Fluor 594 goat anti-mouse IgG2b (Invitrogen) was used to detect MF20+ cells, and Alexa Fluor 488 goat anti-mouse IgG1 (Invitrogen) was used to detect BrdU+ cells. Nuclei were stained with Hoechst as decribed above. The BrdU+ nuclei were counted relative to the total number of nuclei (Hoechst) in MF20+ cells. At least three coverslips of at least 100 isolated cells were analyzed for each condition.
Electrophysiology
Single spontaneously contracting EB outgrowths were microdissected and plated on glass coverslips. Microdissected outgrowths were maintained in EB2 media for 1 – 10 days prior to recording. Cardiomyocyte activity was assessed from 56 – 70 days post-EB formation using sharp microelectrodes (50 – 100 MΩ; 3 M KCl) in a 37°C bath continuously perfused with Tyrode's solution (mmol/L): 140 NaCl, 5.4 KCl, 1.8 CaCl2, 1 MgCl2, 10 Hepes, 10 glucose, pH 7.4 NaOH). Junction potentials and capacitance were nulled and data were acquired at 10 kHz using an AxoClamp2A amplifier and pClamp 9.2 software (Molecular Devices, Sunnyvale CA). Electrical field stimulation was performed using two platinum electrodes coupled to a Grass SD-9 stimulator (Quincy, MA). For analysis, data were filtered off-line using a low pass Gaussian filter with a cut-off frequency of 2 kHz.
Statistics
Data are presented as mean ± standard error of the mean (SEM). Statistical significance was determined by Student's t-test (two-tail) for two groups or one-way ANOVA for multiple groups with post hoc testing using Tukey method using Microcal Origin, v7.5. P < 0.05 was considered statistically significant.
Results
Cardiac Differentiation of Human iPS Cells in Embryoid Bodies
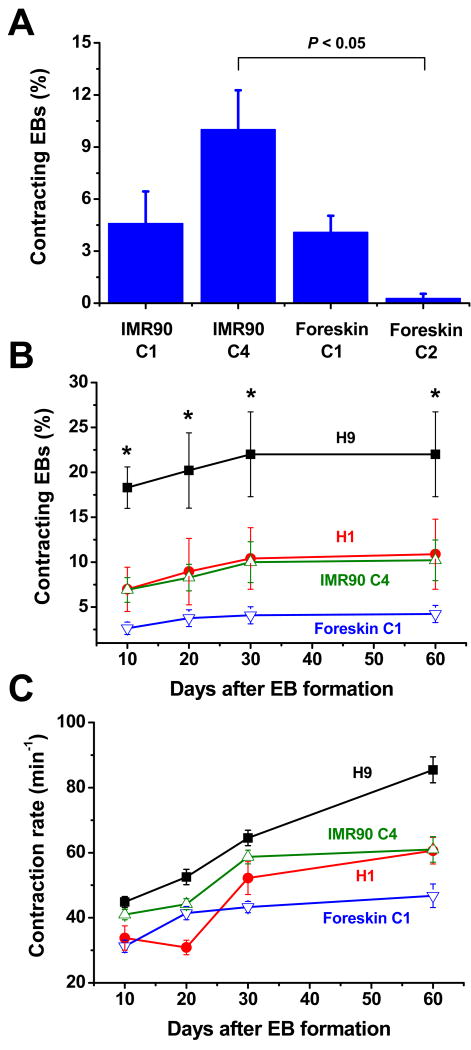
As an initial test of the ability of human iPS cells to undergo cardiac differentiation, we generated embryoid bodies (EBs) from four previously described iPS cell clones (IMR90 C1, IMR90 C4, Foreskin C1, Foreskin C2).3 A fraction of the EBs generated from each of the tested iPS cell lines formed contracting outgrowths of cells suggesting cardiac differentiation. The efficiency of forming contracting EBs after 30 days in culture varied significantly among iPS cell clones from less than 1% to 10% of the EBs having contracting regions (Fig. 1A). Based on these results, we chose two iPS clones of different origins (IMR90 C4 and Foreskin C1) which readily underwent cardiogenesis for comparison to two well-characterized human ES cell lines, H1 and H9.
Figure 1.
Development of contracting EBs from human iPS and ES cells. (A) Percentage of contracting EBs formed from four different iPS clones (IMR90 C1, n=11; IMR90 C4, n=23; Foreskin C1, n=13; Foreskin C2, n=8) after 30 days of differentiation. (B) Time course of forming contracting EBs from iPS and ES cell lines (IMR90 C4, n=23; Foreskin C1, n=13; H1, n=3; H9, n=3). (C) Comparison of the contraction rate of iPS and ES cell contracting EBs over time (IMR90 C4, n=72-139; Foreskin C1, n=97-157; H1, n=14-38; H9 n=106-118). Error bars represent SEM. Data were compared using one-way ANOVA and Tukey post tests with * indicating H9 is significantly different from Foreskin C1, P < 0.05.
We first compared the in vitro differentiation of the iPS and ES cell lines by determining the time courses for formation of spontaneously contracting EBs and their associated rates of contraction. Contractions were first observed 8-9 days after EB formation for all of the iPS and hES cell lines (supplemental movies 1-4). As shown in Fig. 1B, contracting EBs developed over a similar time course during 60 days of observation, although the overall efficiency varied from line to line. H9 ES cells showed the greatest efficiency, reaching a maximum of 22%, in contrast to the least efficient, Foreskin C1, which reached 4.2%. The human iPS cell line IMR90 C4 and ES cell line H1 displayed nearly identical efficiencies (∼10%) for the formation of contracting outgrowths. Comparison of the rates of spontaneous contractions showed an increase in rate over the period of observation with some differences observed between the lines (Fig. 1C). H9 EBs exhibited the highest contraction rates while Foreskin C1 was slowest at most time points. Again, IMR90 C4 and H1 EBs performed comparably. In summary, there was no observable difference in the time course for the development of contraction for iPS- and ES- derived EBs, although the efficiency of forming contracting EBs varied for both iPS and ES cell lines.
Cardiac and Pluripotency Gene Expression in Cardiomyocytes Derived from iPS Cells
To provide a more detailed comparison of the cardiomyocytes derived from iPS cells and ES cells, we examined the gene expression patterns present in microdissected contracting regions from EBs using RT-PCR. Pluripotency and cardiac muscle gene expression was analyzed in day 60 EBs when cardiogenesis reached a stable plateau. The gene expression patterns of stem cell-derived cardiomyocytes were compared to those of undifferentiated iPS and ES cells as well as adult human left ventricular myocardium.
We examined a variety of cardiac genes: the transcription factor Nkx2.5 (NKX2-5); several myofilament protein genes including cardiac troponin T (TNNT2), α-myosin heavy chain (MYH6), α-actinin (ACTN2), myosin light chain 2 atrial isoform (MYL7) and myosin light chain 2 ventricular isoform (MYL2); atrial natriuretic factor (HPPA); and phospholamban (PLN). The cardiac genes showed no detectable expression in undifferentiated iPS or ES cells with the exception of low levels of transcript detected for TNNT2 and MYL7. In contrast, by day 60, there was robust expression of the full range of cardiac genes in cardiomyocytes derived from iPS and ES cell lines, comparable to that observed in adult human ventricular myocardium (Fig. 2A). Overall, the cardiac gene expression pattern was quite similar in iPS and ES cell-derived cardiomyocytes, with a strong increase in expression following cardiac differentiation.
Figure 2.
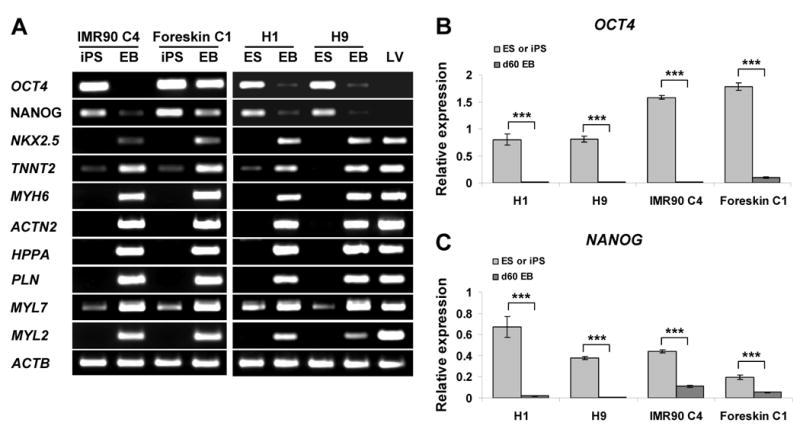
Cardiac and Pluripotency Gene Expression in Cardiomyocytes Derived from iPS and ES Cells. (A) RT-PCR analyses of pluripotency genes, OCT4 and NANOG, and cardiac genes in undifferentiated iPS and ES cells, day 60 EBs, and adult left ventricular myocardium (LV). (B and C) Quantitative RT-PCR analyses of total OCT4 (B) and NANOG (C) expression in undifferentiated iPS and ES cells compared to differentiated contracting areas from day 60 EBs. Error bars represent SEM (n=3), *** indicate P < 0.001 comparing gene expression in undifferentiated cells and d60 EBs using t-test.
For pluripotency genes, we focused on the most extensively studied genes, OCT4 and NANOG. Expression of OCT4 and NANOG was high in undifferentiated iPS and ES cells, and the expression greatly decreased with differentiation, with the exception of the Foreskin C1 iPS line. In the Foreskin C1 line, expression of OCT4 and NANOG persisted to some extent (Fig. 2A). To provide a more quantitative assessment of OCT4 and NANOG expression during differentiation, we performed quantitative RT-PCR. OCT4 gene expression was significantly downregulated in EBs from all lines compared to undifferentiated iPS or ES cells (Figure 2B, P < 0.001), and the degree of downregulation was similar for H1 (114-fold), H9 (105-fold) and IMR90 C4 (105-fold), but less for Foreskin C1 (14-fold). There was also significant downregulation of NANOG expression in all lines during differentiation (Fig. 2C, P < 0.001), although the decrease in NANOG in differentiated cardiomyocytes was relatively less than that of OCT4 (H1: 34-fold; H9: 45-fold; IMR90 C4: 4-fold; Foreskin C1: 4-fold). These results confirmed a reduction in pluripotency gene expression for both iPS and ES cells during cardiogenesis in EBs, but the reduction may be variably blunted in the iPS cell lines.
To investigate if persistent expression of lentiviral transgenes contributes to the blunted downregulation of OCT4 and NANOG during cardiogenesis of iPS cells, we measured the total OCT4 or NANOG gene expression using primers located in the coding region and compared it to the expression of endogenous OCT4 or NANOG using primers located in 3′ UTR. The difference between total and endogenous gene expression is due to expression of the transgene. In undifferentiated ES cells, the total and endogenous OCT4 and NANOG expression was not different as predicted given the lack of transgenes (Fig. 3A and B). The iPS cells exhibited significantly greater total than endogenous OCT4 expression indicating residual transgene expression most prominently in the case of the Foreskin iPS cell line (Fig. 3A). For NANOG, the range of total expression varied in the undifferentiated cells, but there was no significant difference between total and endogenous expression of NANOG in either of the iPS cell lines (Fig. 3B).
Figure 3.
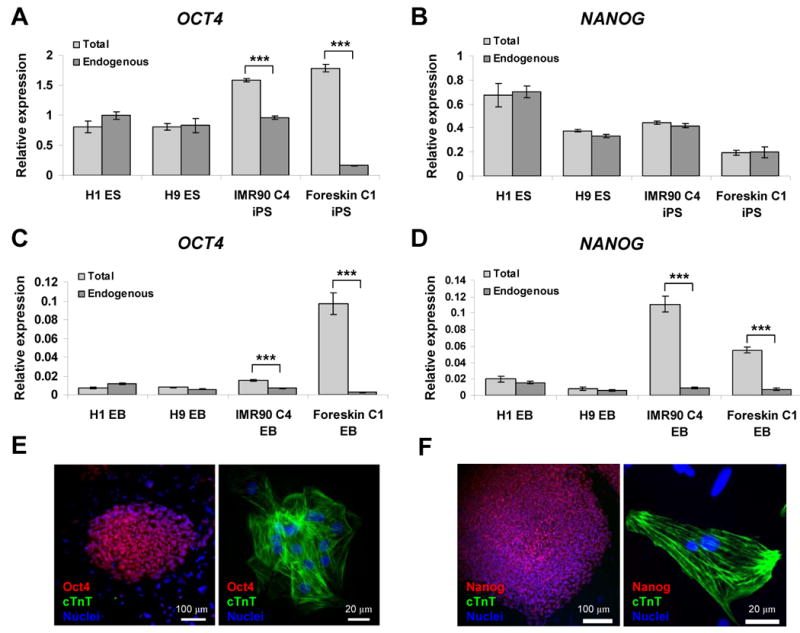
Transgene expression of OCT4 and NANOG in undifferentiated and differentiated iPS cells. Quantitative RT-PCR analyses of total and endogenous OCT4 and NANOG expression in undifferentiated iPS and ES cells (A and B), and in day 60 EB contracting areas (C and D). Error bars represent SEM (n=3), and *** indicate P < 0.001 comparing total and endogenous gene expression using t-test. (E) Double immunolabeling for Oct4 and cTnT in undifferentiated Foreskin C1 iPS cells (left panel) and Foreskin C1 iPS cell-derived cardiomyocytes from day 60 EBs (right panel). (F) Double immunolabeling for Nanog and cTnT in undifferentiated IMR90 C4 iPS cells (left panel) and IMR90 C4 iPS cell-derived cardiomyocytes from day 60 EBs (right panel).
To determine whether transgene expression accounted for the difference in iPS and ES cell expression of pluripotency genes following differentiation, we compared total and endogenous expression of OCT4 and NANOG in contracting outgrowths isolated from each of the four cell lines. Total expression of OCT4 was comparable in differentiated H1, H9 and IMR90 C4 EBs, but the level of OCT4 transcript in Foreskin C1 EBs was significantly greater (P < 0.001) which could largely be accounted for by persistent expression of the transgene (Fig. 3C). In the case of NANOG, the relative total expression in EBs also varied from line to line (Fig. 3D), and both IMR90 C4 and Foreskin C1 EBs exhibited a relatively higher total expression of NANOG than observed for EBs from H1 and H9 (P < 0.001), an effect which, again, was largely attributable to transgene expression. Together, these results demonstrate that there is some persistent expression of the OCT4 and NANOG transgenes in the two iPS cell lines studied following differentiation, and this is especially evident for the OCT4 transgene in the Foreskin C1 iPS cell line. Nevertheless, a strong downregulation in total OCT4 and NANOG gene expression occurs during cardiogenesis of iPS cells. Although we did not evaluate LIN28 and SOX2 transgene expression, it is possible that some level of expression for these transgenes persists as well.
Given the findings of persistent OCT4 and NANOG transgene expression in differentiated iPS cell-derived cardiomyocytes, we examined whether Oct4 and Nanog protein expression could be detected using standard immunolabeling approaches. Immunolabeling for Oct4 and Nanog was examined in undifferentiated iPS cells and differentiated cells from day 60 EBs which were co-labeled for cardiac troponin T (cTnT) to detect cardiomyocytes. In experiments using Foreskin C1 cells (Fig. 3E), we detected nuclear localized Oct4 immunolabeling in the undifferentiated iPS cells but not in the surrounding mouse fibroblast feeder layer cells. There was no detectable cTnT immunolabeling in the undifferentiated cells. In contrast, cells isolated from contracting outgrowths from Foreskin C1 EBs subjected to the identical immunolabeling protocol revealed that the majority of cells were cTnT positive and showed DAPI labeled nuclei without detectable Oct4 labeling. Nor was Oct4 labeling detected in any surrounding cTnT negative cells. Similar results were observed for Nanog and cTnT immunolabeling in the IMR90 C4 line (Fig. 3 F). Nanog was detected in the undifferentiated iPS cells but not detectable in IMR90 iPS cell-derived cardiomyocytes. These results show that Oct4 and Nanog protein expression is strongly downregulated during differentiation of Foreskin C1 and IMR90 C4 iPS cells despite some persistent mRNA expression. This apparent discrepancy between mRNA and protein expression could be due to a variety of regulatory effects such as microRNA regulation, which may be particularly important for pluripotency genes.18
Proliferation of Cardiomyocytes Derived from iPS and ES Cells
Because the proliferative activity of cells may be impacted by overexpression of genes associated with pluripotency, we next compared cellular proliferation in iPS cell- and ES cell-derived cardiomyocytes. In normal embryonic cardiac development, the proliferative activity of cardiomyocytes declines steeply at later stages, and a similar phenomena has also been observed during in vitro differentiation of human ES cell-derived cardiomyocytes.11, 19, 20 In this experiment, cardiomyocytes were isolated from early (day 10-20) and late (day 60) contracting EBs. Immunofluorescence analysis showed that a fraction of the MF20 positive cells contained BrdU positive nuclei indicating proliferating cardiomyocytes for both iPS and ES cell-derived populations (Fig. 4A). In early EBs, there was no difference in the percentage of BrdU positive cardiomyocyte (MF20 positive) nuclei for H9 and IMR90 C4 (∼15%), but Foreskin C1 showed a significantly lower percentage of dividing cardiomyocytes (∼11%, P < 0.05) compared to the other two cell lines (Fig. 4B). Cells from late EBs revealed significantly less proliferation compared to early EBs in all the ES and iPS cells tested (P < 0.01). H9 late EBs exhibited greater proliferation of cardiomyocytes (4%) compared to late EBs from IMR90 C4 and Foreskin C1 (∼1%, P < 0.05). These results demonstrate that iPS cell-derived cardiomyocytes like ES-cell derived cardiomyocytes show a marked reduction in proliferation during 60 days in culture, and the proliferative activity of the iPS cardiomyocytes from the cell lines studied tended to be slightly less than for cardiomyocytes formed from H9 ES cells.
Figure 4.
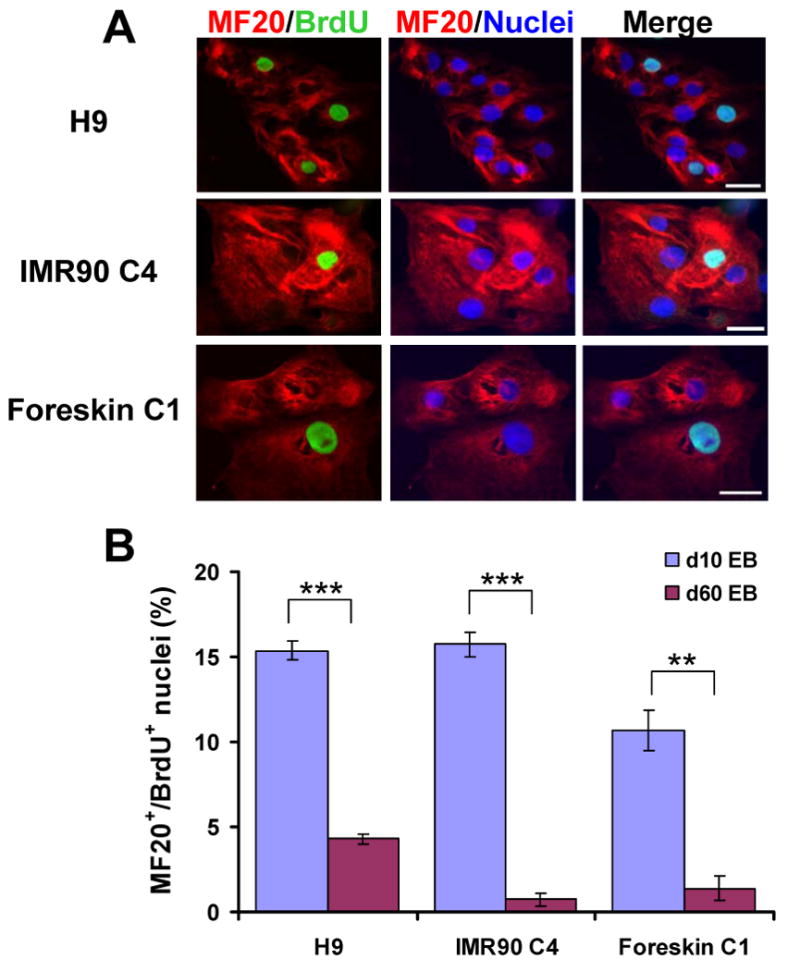
Proliferation of cardiomyocytes differentiated from iPS and ES cells. (A) Co-labeling for sarcomeric myosin with the MF20 antibody (red) and BrdU (green) in cardiomyocytes isolated from contracting areas of early EBs (10-20 days) from H9 and IMR90 C4 following a 17-hour pulse of BrdU to identify dividing cells. Nuclei were stained with Hoechst (blue). Scale bars are 50 μm. (B) Average percentage of BrdU positive nuclei in cardiomyocytes (MF20 positive) differentiated from H9 ES cells, IMR90 C4 and Foreskin C1 iPS cells from early and late (day 60) EBs. Error bars represent SEM (n=3), ** for P < 0.01 and *** indicate P < 0.001 in t-test comparisons of early and late EBs.
iPS Cell-derived Cardiomyocytes Exhibit Sarcomeric Organization
To evaluate the expression of myofilament proteins and the sarcomeric organization in iPS cell-derived cardiomyocytes, we performed immunolabeling with antibodies for specific myofilament proteins using enzymatically isolated cells from day 60 EBs. Cells were co-labeled with the antibodies for α-actinin, which is present at the Z-line of the sarcomere, and myosin light chain 2 atrial isoform (MLC2a) which is typically present at the A-band of the sarcomere. Immunofluorescence analysis revealed a clear striated pattern for α-actinin labeling in cardiomyocytes from IMR90 C4 and Foreskin C1 comparable to that observed for H1 and H9 cardiomyocytes (Fig. 5). Striated MLC2a labeling was also observed in cardiomyocytes from all cell lines. Overlap of α-actinin and MLC2a labeling demonstrated an alternating pattern in the sarcomeres in agreement with the known localization of MLC2a to the A-band of the sarcomere, which lies between the Z-lines highlighted by the α-actinin labeling.
Figure 5.
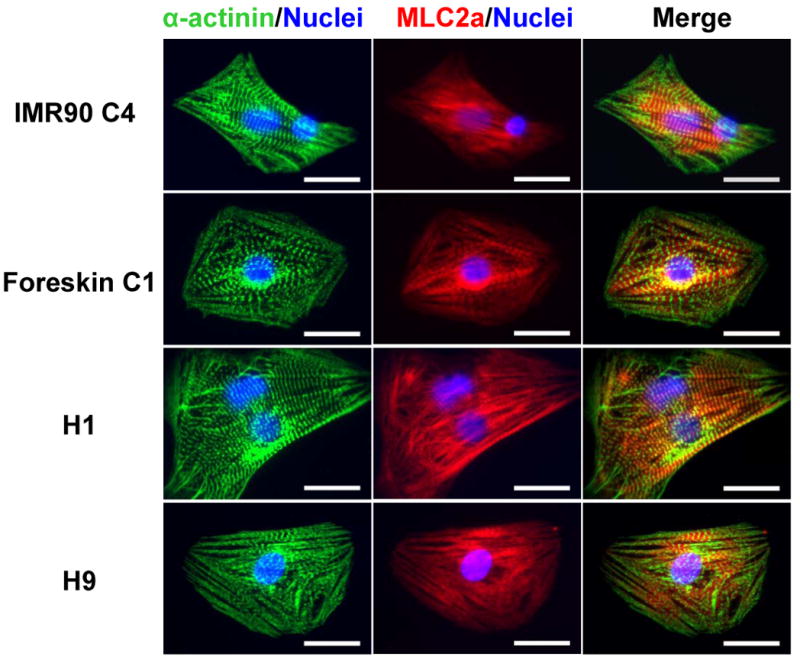
Sarcomeric organization in cardiomyocytes derived from iPS and ES cells. Cardiomyocytes isolated from contracting areas of day 60 EBs from IMR90 C4, Foreskin C1, H1 and H9 were co-labeled for α-actinin (green) and MLC2a (red). Nuclei were stained with Hoechst (blue). Overlap of α-actinin and MLC2a labeling demonstrated an alternating pattern of sarcomeric labeling consistent with MLC2a present in the A band of the sarcomere between the Z-lines demonstrated by the α-actinin labeling. Scale bars are 20 μm.
We also performed immunolabeling for cTnT, which is a highly cardiac-specific myofilament protein. We observed comparable sarcomeric labeling of iPS and hES cell cardiomyocytes (Fig. 6). Likewise, immunolabeling with an antibody to the ventricular specific protein, myosin light chain 2 ventricular isoform (MLC2v), detected presumed ventricular cardiomyocytes derived from iPS cells and ES cells (Fig. 6). In summary, immunolabeling of multiple myofilament proteins indicates that a well-organized sarcomeric structure can similarly develop in iPS and ES cell-derived cardiomyocytes.
Figure 6.
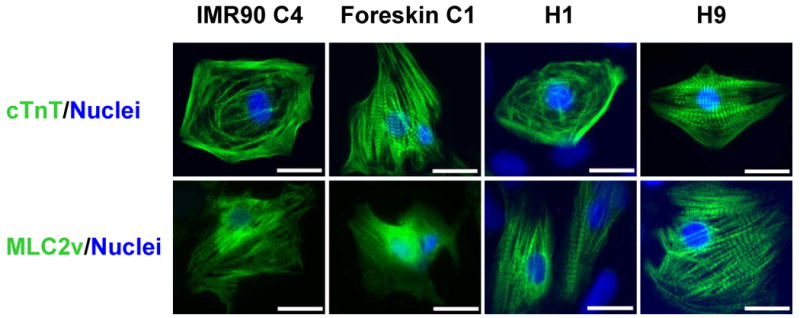
Immunolabeling of cTnT and MLC2v in iPS and ES cell-derived cardiomyocytes. Single cardiomyocytes isolated from contracting areas of day 60 EBs from IMR90 C4, Foreskin C1, H1 and H9 were immunolabeled for cTnT, a cardiac-specific myofilament protein, and MLC2v, a ventricular-specific protein. Nuclei were stained with Hoechst (blue). Scale bars are 20 μm.
Action Potentials Reveal Multiple Types of Cardiomyocytes Derived from iPS Cells
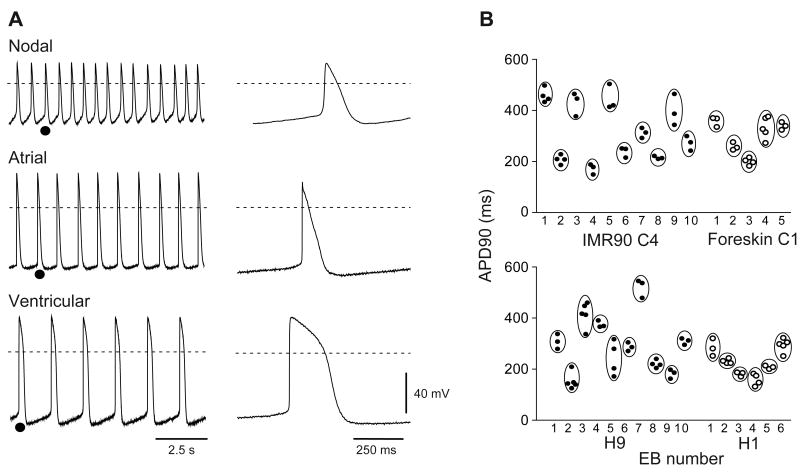
To provide an initial assessment of the functional competence of iPS cell-derived cardiomyocytes, we performed sharp microelectrode recordings from spontaneously contracting EB outgrowths at 56 – 70 days post-EB formation. A total of 54 and 47 stable recordings from 23 and 20 IMR90 C4 and Foreskin C1 derived EBs, respectively, were obtained. Three major types of action potential were observed: ventricular, atrial, and nodal. Cells with ventricular-like action potentials (Fig. 7A, bottom) were the most frequently encountered in both IMR90 C4 and Foreskin C1 derived EBs, and typically displayed a more negative maximum diastolic potential (MDP), a rapid action potential upstroke, and a distinct plateau phase. Atrial-like cells were distinguished from ventricular-like cells by the absence of a distinct plateau during repolarization and typically also exhibited spontaneous activity that was higher in frequency than that observed for ventricular cells (Fig. 7A, middle). Finally, nodal-like cells were distinguished by MDPs that were less negative than those of ventricular- and atrial- like cells, smaller amplitude action potentials, a slower action potential upstroke, and a pronounced phase 4 depolarization preceding the action potential upstroke (Fig. 7A, top).
Figure 7.
Electrophysiological characterization of iPS cell-derived cardiomyocytes. (A) Representative recordings from 3 iPS cell-derived EB outgrowths demonstrating that each of the 3 major action potentials types were observed. Right, action potentials shown at an expanded timescale taken from the region indicated (•) on the left. Dotted lines indicate 0 mV. (B) Cardiomyocytes within a given EB outgrowth display similar electrophysiological properties: Action potential durations at 90% repolarization (APD90s) are shown for consecutively studied iPS (top) or ES (bottom) cell-derived EBs from which 3 or more cells were examined.
Comparison of recordings from cardiomyocytes within the same EB revealed that a given action potential phenotype was predominant in each EB, as has previously been shown for human ES cell-derived EBs.12 Figure 7B plots action potential durations measured at 90% repolarization from the action potential peak (APD90) for EBs from which three or more recordings were obtained. APD90 values obtained from myocytes within the same EB clustered together much more closely than would be predicted if EBs contained myocytes of each class. The relative proportions of EBs exhibiting the nodal, atrial, and ventricular phenotypes are shown in Table 2 and compared to the proportions observed for EB outgrowths derived from the H1 and H9 cell lines. Although sample sizes are limited due to the difficulty inherent in recording, iPS and ES cells appear comparably efficient in generating cardiac myocytes of each major class.
Table 2.
Comparison of cardiomyocyte types in H9, H1, IMR90 C4, and Foreskin C1 derived cardiomyocytes.
| Total | Nodal-like | Atrial-like | Ventricular-like | ||||
|---|---|---|---|---|---|---|---|
| N, EBs | N, EBs | % of total | N, EBs | % of total | N, EBs | % of total | |
| iPS cell-derived | |||||||
| IMR90 C4 | 23 | 3 | 13 | 3 | 13 | 17 | 74 |
| Foreskin C1 | 20 | 4 | 20 | 2 | 10 | 14 | 70 |
| ES cell-derived | |||||||
| H9 | 27 | 3 | 11 | 8 | 30 | 16 | 59 |
| H1 | 15 | 3 | 20 | 3 | 20 | 9 | 60 |
Table indicates the number of EBs examined for each cell line and the proportion of EBs exhibiting nodal, atrial, and ventricular-like properties.
Table 3 presents a comparison of the properties of iPS and ES cell-derived action potentials for ventricular-like cells, which were the predominant class of cardiomyocytes encountered in EBs formed from each line. Specifically, we compared spontaneous beating rate (in beats per minute, BPM), action potential durations (APD), the maximum rates of rise during the action potential upstroke (dV/dtmax), action potential amplitudes (APA), and maximum diastolic potentials (MDP). As observed for H9 and H1 cardiomyocytes, the action potentials of IMR90 C4 and Foreskin C1 cardiomyocytes exhibited properties that were clearly more comparable to those of human embryonic, than neonatal or adult, cardiac muscle. Measurements from IMR90 C4- and Foreskin C1-derived cells fell within range of the measurements obtained for cardiomyocytes from H9 and H1 ES cell lines and, although there were modest differences in means, these were no larger than the differences between H9 and H1 cardiomyocytes.
Table 3.
Comparison of action potential properties of ventricular-like cardiomyocytes derived from H9, H1, IMR90 C4, and Foreskin C1 cell lines.
| N (cells) |
Rate (bpm) |
APD50 (ms) |
APD90 (ms) |
dV/dtmax (V/s) |
APA (mV) |
MDP (mV) |
|
|---|---|---|---|---|---|---|---|
| iPS cell-derived | |||||||
| IMR90 C4 | 37 | 43.8 ± 2.7 | 240.7 ± 15.0 | 320.1 ± 17.0 | 40.5 ± 4.6 | 87.7 ± 2.6 | -63.5 ± 1.7 |
| Foreskin C1 | 39 | 44.2 ± 3.5 | 238.9 ± 10.0 | 312.5 ± 11.2 | 27.2 ± 3.7 | 87.9 ± 2.4 | -63.3 ± 1.5 |
| ES cell-derived | |||||||
| H9 | 45 | 44.6 ± 2.8 | 223.9 ± 13.4 | 312.6 ± 15.9 | 33.5 ± 3.7 | 83.5 ± 2.4 | -64.3 ± 3.2 |
| H1 | 27 | 33.3 ± 2.3 | 233.9 ± 11.4 | 298.7 ± 13.4 | 17.6 ± 2.6 | 82.7 ± 3.0 | -60.8 ± 2.8 |
Data are mean ± SEM. APD50 and APD90, action potential durations at 50% and 90% repolarization. dV/dtmax, maximum rate of rise of action potential upstroke. APA, action potential amplitude. MDP, average maximum diastolic potential for action potentials during the time period examined.
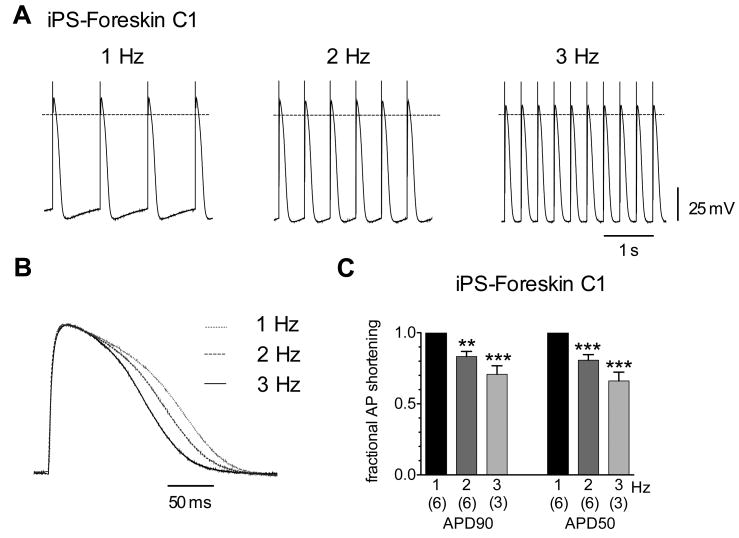
Cardiomyocytes typically respond to increases in heart rate with a compensatory decrease in action potential duration, and this property is likewise present in ES cell-derived cardiomyocytes.12 Therefore, we tested whether iPS cell-derived cardiomyocytes exhibit this typical rate adaptation in response to changing rates of electrical stimulation. As shown in Figure 8 (A and B), field stimulation of microdissected Foreskin C1-derived contracting outgrowths at 1, 2, and 3 Hz resulted in incremental shortening of the observed action potential durations. Increasing the stimulus frequency from 1 to 2 and 3 Hz decreased the average APD90 and APD50 by approximately 20 and 30 percent (Fig. 8C), respectively, with little or no change in other action potential features.
Figure 8.
Action potentials of iPS cell-derived cardiomyocytes exhibit rate adaptation. (A) Electrical field stimulation of a ventricular-like cardiomyocyte derived from the Foreskin C1 line at three frequencies as indicated. Dashed line represents 0 mV. (B) Overlay of single action potentials from the cell in (A) obtained at 1, 2, and 3 Hz stimulation rates. Action potentials were normalized to correct for a slight differences in amplitude (for this cell the average amplitudes were 79.2, 83.1, and 84.7 mV at 1, 2, and 3 Hz, respectively). Electrical artifacts corresponding to the stimulus were manually removed for normalization. (C) Average (± SEM) fractional changes in APD90 and APD50 during 2 and 3 Hz stimulation. Durations were normalized to the respective values at 1 Hz stimulation. The number of cells is given in parentheses below each bar. Data were compared using a one-way ANOVA and Tukey post tests to the durations at 1 Hz with ** P < 0.01 and *** P < 0.005.
β-adrenergic Regulation of iPS Cell-derived Cardiomyocytes
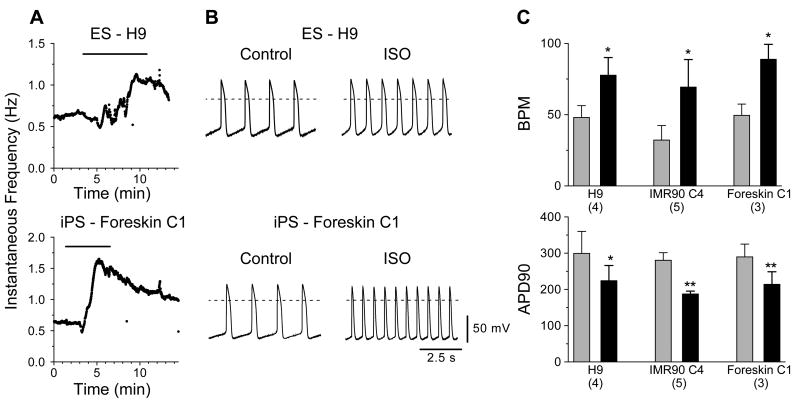
The above experiments suggest that iPS and ES cell-derived cardiomyocytes exhibit similar functional potential in regard to their electrical activity. We next sought to determine whether β-adrenergic signaling, a canonical cardiomyocyte signaling pathway, is operational in iPS cell-derived myocytes by examining the responsiveness to isoproterenol (ISO), a nonselective β-adrenergic receptor agonist. In spontaneously active myocytes such as those derived from embryonic or neonatal hearts as well as nodal cells in adult hearts, β-adrenergic receptor stimulation, via protein kinase A-mediated regulation of several different ion channels, results in a positive chronotropic effect which is accompanied by a shortening of APD.21 Figure 9 panels A and B show individual responses to isoproterenol for ventricular-like myocytes derived from H9 (top) and Foreskin C1 (bottom) lines. Both cells displayed initial contraction frequencies slightly above 0.6 Hz during perfusion of control Tyrode's solution and responded to perfusion of ISO (1 μmol/L) with a 2-fold or greater increase in rate. As shown in Figure 9B, the increases in rate were also accompanied by decreases in action potential duration as observed in native cardiomyocytes. Summary data are presented in Figure 9C; perfusion with ISO resulted in statistically significant increases in rate and decreases in action potential duration for IMR90 C4- and Foreskin C1-, as well as H9-, derived cardiomyocytes. These results suggest that β-adrenergic receptors and their associated intracellular signaling partners are present and functional in cardiomyocytes derived from iPS cells.
Figure 9.
Effect of Isoproterenol on spontaneous electrical activity of iPS cell-derived cardiomyocytes. (A) Time course of responses for an H9- (top) and a Foreskin C1- derived (bottom) cardiomyocyte before, during, and after perfusion with Tyrode's solution containing 1 μmol/L ISO as indicated by measurement of the instantaneous frequencies. Bar at the top of each graph shows the duration of ISO application. (B) Action potentials recorded from the cells in (A) before and during perfusion of ISO. (C) Bar graphs of the average rate and APD90 data for H9, IMR90 C4, and Foreskin C1 derived cardiomyocytes before (light gray bars) and during (black bars) application of ISO. Number of cells examined for each cell type is indicated in parentheses below each graph. * P < 0.05, ** P < 0.01 in paired t-test comparisons.
Discussion
The purpose of this study was to evaluate the cardiac differentiation potential of the recently described human iPS cell lines in comparison to the potential observed for the well-studied human ES cell lines, H1 and H9.3, 16 We compared iPS and ES cell lines in terms of the relative efficiency in forming contracting EBs, the contraction rates of developing EBs, the upregulation of cardiac genes and concomitant downregulation of pluripotency genes, proliferation, sarcomeric organization, electrical activity, and chronotropic regulation by the beta-adrenergic signaling pathway. Our findings suggest that iPS cells are a viable alternative to ES cells as an autologous cell source for cardiac repair and for a variety of research applications. Although limited by the number of cell lines examined, our results generally indicate that the differences between iPS cells and ES cells are no greater than the differences already noted between ES cell lines. In addition, the contractile properties of the iPS and ES cell-derived cardiomyocytes appeared similar, but we did not perform specific measurements of contraction or Ca2+ transients in these cells. Our findings are discussed in more detail below, along with the single notable exception, namely that of continued transgene expression in the iPS cell lines.
The efficiency of cardiogenesis, based on the percentage of contracting EBs, varied among the different tested iPS and hES cell lines. Significant variability in the efficiency of cardiogenesis among human ES cell lines has previously been described.22-24 The cause of this variability is not understood and may relate to subtle differences in basal gene expression or epigenetic state. The iPS cell clones studied here tended to have a lower efficiency of forming contracting EBs than typically observed for human ES cells, but there was significant overlap in the behavior of different ES and iPS cell lines. Thus at least some human ES cell lines and iPS cell lines show comparable abilities to form cardiomyocytes in EBs. The overall efficiency of the EB protocol to generate human iPS cell or ES cell-derived CMs is low with a minority of EBs developing CMs and only a small region of the EBs containing contracting cardiomyocytes. Although EB-mediated cardiac differentiation is the classical in vitro approach, directed differentiation approaches have more recently been applied with promising results for cardiogenesis from human ES cells that likely will also be useful for iPS cardiac differentiation.9, 13, 25
The time course of in vitro cardiac differentiation was not different comparing the human iPS and ES cell lines based on a similar time required to form contracting EBs. In contrast, one of the first studies to examine the time course of differentiation of mouse iPS cells to cardiomyocytes noted a substantial delay in cardiogenesis compared to mouse ES cells,26 but another study using different mouse iPS cell lines found no difference between the mouse iPS and mouse ES cell lines in cardiac differentiation.27 Given that first generation iPS cells are all subject to viral integration effects and that integration sites are highly variable,28 from a probabilistic perspective, it seems likely that some iPS cell lines will have altered cardiogenesis. However, our results and results with mouse iPS cells suggest that iPS cell lines can be readily obtained that undergo cardiogenesis in a highly similar fashion to ES cells.
Although functional cardiomyocytes have been obtained from human ES cells and iPS cells, significant challenges remain in optimizing these cell preparations for experimental and potentially clinical applications. The heterogeneity of cells produced in differentiation protocols can be great even if one succeeds in isolating cardiomyocytes. For example, using a mixed population of cardiomyocytes including nodal, atrial and ventricular cells in attempts at left ventricular repair raises concerns for pro-arrhythmia effects. Likewise, a preparation including undifferentiated cells could lead to tumorigenesis. Thus, approaches to produce homogenous or well-characterized mixed cell preparations remain a great need. Techniques to isolate cardiovascular progenitor cell populations from human ES cells have recently been described which help address this concern.9 Another limitation observed with the in vitro differentiation of human ES and iPS cells is that the myocytes even after 2 months of standard 2-dimensional tissue culture conditions remain embryonic in phenotype based on their size, organization and electrical properties. Techniques to promote the maturation of these cells such as three-dimension culture methodology as well application of electrical and/or mechanical stimulation will likely help address this need. The ability of human ES or iPS cell-derived cardiomyocytes to mature following transplantation has not been demonstrated, but this seems a reasonable expectation.
The technology to generate iPS cells is rapidly evolving and overcoming some of the challenges associated with the first generation iPS cells used in this study. Mouse iPS cells have now been generated using adenovirus or plasmid-mediated transfections which avoid the potential problems associated with viral integration of transgenes.29, 30 Small molecules have been used to increase the efficiency of generating iPS cell lines and allowed the use of only two transcription factors (OCT4/SOX2 or OCT4/KLF4) to generate iPS cell lines.31, 32 Given the pace of progress, it seems likely that techniques to generate human iPS cells will continue to rapidly improve; nevertheless, the current study provides evidence that with existing OCT4/SOX2/NANOG/LIN28 human iPS cells, functional cardiomyocytes can be generated which exhibit properties highly similar to human ES cell derived cardiomyocytes. Thus iPS cell-derived cardiomyocytes hold significant promise for research applications studying cardiac disease models, drug development and as an autologous source of cells for myocardial repair.
Supplementary Material
Acknowledgments
The authors acknowledge Jason Foell for providing the total RNA of human heart tissue, and the assistance of Thankful Sanftleben and Deborah Faupel in manuscript preparation.
Sources of Funding: Supported by NIH RO1 HL0846150-01 (TJK), NIH RO1 EB007534 (SPP), NSF EFRI-0735903 (SPP), Wisconsin Institutes for Discovery Seed Grant (TJK, SPP)
Footnotes
Disclosures: T.J.K. and J.A.T. are founding shareholders in Cellular Dynamics International, Inc.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 6.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 7.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 8.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 9.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 10.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, Livne E, Binah O, Itskovitz-Eldor J, Gepstein L. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91:501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- 12.He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res. 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- 13.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O'Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 14.Cao F, Wagner RA, Wilson KD, Xie X, Fu JD, Drukker M, Lee A, Li RA, Gambhir SS, Weissman IL, Robbins RC, Wu JC. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS ONE. 2008;3:e3474. doi: 10.1371/journal.pone.0003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, van den BS, Hassink R, van der HM, Opthof T, Pera M, de la Riviere AB, Passier R, Tertoolen L. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 16.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 17.Foell JD, Balijepalli RC, Delisle BP, Yunker AM, Robia SL, Walker JW, McEnery MW, January CT, Kamp TJ. Molecular heterogeneity of calcium channel beta-subunits in canine and human heart: evidence for differential subcellular localization. Physiol Genomics. 2004;17:183–200. doi: 10.1152/physiolgenomics.00207.2003. [DOI] [PubMed] [Google Scholar]
- 18.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 19.McDevitt TC, Laflamme MA, Murry CE. Proliferation of cardiomyocytes derived from human embryonic stem cells is mediated via the IGF/PI 3-kinase/Akt signaling pathway. J Mol Cell Cardiol. 2005;39:865–873. doi: 10.1016/j.yjmcc.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snir M, Kehat I, Gepstein A, Coleman R, Itskovitz-Eldor J, Livne E, Gepstein L. Assessment of the ultrastructural and proliferative properties of human embryonic stem cell-derived cardiomyocytes. Am J Physiol Heart Circ Physiol. 2003;285:H2355–H2363. doi: 10.1152/ajpheart.00020.2003. [DOI] [PubMed] [Google Scholar]
- 21.Kamp TJ, Hell JW. Regulation of cardiac L-type calcium channels by protein kinase A and protein kinase C. Circulation Research. 2000;87:1095–1102. doi: 10.1161/01.res.87.12.1095. [DOI] [PubMed] [Google Scholar]
- 22.Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 23.Moore JC, Fu J, Chan YC, Lin D, Tran H, Tse HF, Li RA. Distinct cardiogenic preferences of two human embryonic stem cell (hESC) lines are imprinted in their proteomes in the pluripotent state. Biochem Biophys Res Commun. 2008;372:553–558. doi: 10.1016/j.bbrc.2008.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 25.Yao S, Chen S, Clark J, Hao E, Beattie GM, Hayek A, Ding S. Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc Natl Acad Sci USA. 2006;103:6907–6912. doi: 10.1073/pnas.0602280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauritz C, Schwanke K, Reppel M, Neef S, Katsirntaki K, Maier LS, Nguemo F, Menke S, Haustein M, Hescheler J, Hasenfuss G, Martin U. Generation of functional murine cardiac myocytes from induced pluripotent stem cells. Circulation. 2008;118:507–517. doi: 10.1161/CIRCULATIONAHA.108.778795. [DOI] [PubMed] [Google Scholar]
- 27.Narazaki G, Uosaki H, Teranishi M, Okita K, Kim B, Matsuoka S, Yamanaka S, Yamashita JK. Directed and systematic differentiation of cardiovascular cells from mouse induced pluripotent stem cells. Circulation. 2008;118:498–506. doi: 10.1161/CIRCULATIONAHA.108.769562. [DOI] [PubMed] [Google Scholar]
- 28.Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 29.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 31.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 32.Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.