SUMMARY
The plasma membrane calcium extrusion mechanism, PMCA (plasma membrane calcium ATPase) isoform 2 is richly expressed in the brain and particularly the cerebellum. Whilst PMCA2 is known to interact with a variety of proteins to participate in important signalling events (Strehler et al, 2007), its molecular interactions in brain synapse tissue are not well understood. An initial proteomics screen and a biochemical fractionation approach identified PMCA2 and potential partners at both pre- and post-synaptic sites in synapse-enriched brain tissue from rat. Reciprocal immunoprecipitation and GST pull down approaches confirmed that PMCA2 interacts with the post-synaptic proteins PSD95 and the NMDA glutamate receptor subunits NR1 and NR2a, via its C-terminal PDZ (PSD95/Dlg/ZO-1) binding domain. Since PSD95 is a well known partner for the NMDA receptor this raises the exciting possibility that all three interactions occur within the same post-synaptic signalling complex. At the pre-synapse, where PMCA2 was present in the pre-synapse web, reciprocal immunoprecipitation and GST pull down approaches identified the pre-synaptic membrane protein syntaxin-1A, a member of the SNARE complex, as a potential partner for PMCA2. Both PSD95-PMCA2 and syntaxin-1A-PMCA2 interactions were also detected in the molecular and granule cell layers of rat cerebellar sagittal slices by immunohistochemistry. These specific molecular interactions at cerebellar synapses may allow PMCA2 to closely control local calcium dynamics as part of pre-and post-synaptic signalling complexes.
Keywords: synapse, NMDA receptor, PSD95, PDZ domain, syntaxin-1A
INTRODUCTION
Plasma membrane calcium ATPases (PMCAs) are a family of membrane-spanning proteins that play an important role in the regulation of intracellular calcium levels by actively pumping calcium out of the cell using energy derived from ATP hydrolysis (Strehler & Zacharias, 2001). The PMCAs are an important part of the calcium signalling toolkit (Berridge et al., 2003) and perform their role in a wide variety of different cell types. The family consists of four PMCA isoforms (PMCA1-4), all encoded by different genes, and all undergo alternative splicing (reviewed in (Strehler & Zacharias, 2001)). The pumps are regulated by a number of mechanisms, including the binding of Ca2+/calmodulin at the C-terminus to relieve the autoinhibition of the pump in the resting state (Enyedi et al., 1996). Alternative splicing at the carboxy(c)-terminal splice site produces two functionally distinct PMCA isoforms, a shorter “a” form and a longer `b' variant that contains a PDZ (PSD 95/Dlg/ZO-1) binding domain (DeMarco & Strehler, 2001).
The PMCA isoforms exhibit a differential pattern of expression across cell types. Whilst PMCAs 1 and 4 are ubiquitously expressed, PMCAs 2 and 3 are limited almost exclusively to excitable cells such as those in the central nervous system. In the brain, PMCA 2 exhibits particularly high levels of expression in the cerebellum (Stauffer et al., 1995).
Growing evidence supports the location and function of PMCAs at synapses, both at pre- and post-synaptic sites where control of intracellular calcium is critical for synapse function. It is likely that careful control of synaptic [Ca2+]i at both sites, aided by the extrusion kinetics of the PMCAs, will contribute to the timing of transmitter release and signalling dependent synaptic plasticity.
PMCAs have been found at pre-synaptic sites (Fujii et al.,1996; Morgans et al., 1998; Juhaszova et al., 2000, Jensen et al., 2007) where they provide one of the routes for pre-synaptic Ca2+ removal along with the Na+/Ca2+ exchanger (Kim et al., 2005, Usachev et al., 2002, Empson et al., 2007). Removal of PMCA2 function at both hippocampal and cerebellar synapses also enhances glutamate release (Jensen et al., 2007) and a form of short term plasticity called paired pulse facilitation as a consequence of elevated residual calcium within the pre-synaptic terminals (Jensen et al., 2007; Empson et al., 2007). At the post-synaptic side, PMCA2 is heavily expressed in the dendritic spines of the main output cells of the cerebellum, the Purkinje neurones (Stauffer et al., 1997; Burette & Weinberg, 2007 Sepúlveda et al., 2007) where it may be especially suited to controlling local calcium rises within this restricted compartment (Bloodgood & Sabatini, 2007). The location of PMCA isoforms 2b and 4b at synaptic spines is also supported by their molecular interaction with synapse-associated PDZ-domain containing proteins. These include members of the synapse-associated protein (SAP) family of proteins such as SAP90/PSD95, SAP97, chapsyn110/PSD93 and SAP102 (DeMarco & Strehler, 2001; Kim et al., 1998). Many members of the SAP family reside at or close to post-synaptic sites to fulfil a scaffolding role for the recruitment and stabilisation of signalling molecules at the post-synaptic density.
Given the wide array of interactions of PMCAs with signalling molecules (Strehler et al., 2007) and the potential for some or all of these to influence synapse function, we initially focussed on identifying PMCA2 interactions within a synapse enriched preparation in cerebellum. To do this we employed a proteomics screen followed by verification of positive hits with GST pull down, reciprocal immunoprecipitation, Western blot and immunohistochemistry. We confirm that PMCA2b interacts with PSD95 via the PDZ-domain binding motif in vivo and importantly, that this interaction also recruits the NMDA type glutamate receptor subunits, NR1 and NR2a. In addition we have identified a new in vivo interaction of PMCA2 with the pre-synaptic protein syntaxin-1A. Although their physiological impact is yet to be identified, these specific molecular interactions of PMCA2 at living cerebellar synapses provide the opportunity for PMCA2 to closely control local calcium dynamics during pre- and post-synaptic signalling events.
MATERIALS AND METHODS
Animals
6-8 week old Wistar rats were used for all tissue preparations in this study. Animals were euthanised by intra-peritoneal injection of pentobarbital followed by rapid decapitation, or by carbon dioxide asphyxiation. All experiments were carried out in accordance with the UK animals (Scientific Procedures) Act 1986 and within local ethical guidelines at the University of Otago.
Preparation of homogenates, synaptosomes, PSD fractions and pre-synapse web fractions
The cerebella or forebrains of Wistar rats were removed into 10mM Trizma® buffer, pH7.4, containing protease inhibitors (Sigma), and homogenised using a Dounce homogeniser. Synaptosomes, PSD and pre-synapse web fractions were prepared from cerebella removed from adult (6-8 weeks) Wistar rats as described by Phillips et. al. 2001. Briefly, cerebella from 10 rats were homogenised in homogenisation solution (0.32M sucrose, 0.1mM CaCl2, 1mM MgCl2 containing protease inhibitors) using a Dounce homogeniser. The final sucrose concentration was adjusted to 1.25M by the addition of 2M sucrose and 0.1mM CaCl2 in a total volume of 100ml. 15ml aliquots of homogenate were overlaid with 10ml 1.0M sucrose, 0.1mM CaCl2 and 5ml homogenisation solution and centrifuged at 100,000xgav for 3 hours at 4°C. Following the centrifugation a band at the 1.0M/1.25M interface representing synaptosomes was collected. For synaptosome preparation this material was diluted in 0.32M sucrose, 0.1mM CaCl2 and centrifuged at 100,000xgav for 30 minutes. The pellet was then resuspended in 10mM Tris, pH7.4 containing protease inhibitors. For synapse web preparation a 1ml aliquot of the synaptosome band was retained and the remainder was diluted 1:10 with ice-cold 0.1mM CaCl2. The solution was split into three equal aliquots. Two aliquots were solubilised for 30 minutes at 4°C in a final concentration of 20mM Tris, pH 6.0 and 1% triton X-100 and the third aliquot was solubilised for 30 minutes at 4°C in 20mM Tris, pH 8.0 and 1% Triton X-100. Insoluble material was then pelleted by centrifugation at 40,000xgav for 30 minutes at 4°C. For the pH 8 extraction and one pH 6 extraction the pellet was solubilised in 500ml 5% SDS and proteins in the supernatant were precipitated through the addition of 10 volumes of acetone and overnight incubation at -20°C. The precipitates were then solubilised in 5% SDS. The pellet from the other pH 6 extraction (serial extraction) was solubilised in 10ml 20mM Tris, pH8.0, 1% Triton X-100 for 30 minutes at 4°C. The insoluble and soluble material was then separated by centrifugation as above and the pellet and precipitated proteins from the supernatant were solubilised in 5% SDS.
Immunoprecipitations
SDS was added to synaptosome aliquots containing 1mg protein to give a final concentration of 1% SDS and the synaptosomes were incubated at room temperature for 15 minutes to solubilise. The synaptosomes were then diluted in 10 volumes of a solubilisation buffer containing 10mM Tris (pH 7.4), 1% Triton X-100, 10mM EDTA, 10mM EGTA and protease inhibitor cocktail (Sigma). The synaptosomes were incubated on ice for 30 minutes and then centrifuged at 10,000xgav for 5 minutes to pellet any insoluble material. Whilst the initial solubilisation step used a low (1%) concentration of SDS at room temperature, in order to avoid continuous solubilisation (and so potentially disrupt the protein-protein interactions of interest) the SDS was then diluted 10-fold during the remainder of the incubation and this was also done on ice. This 10x dilution aimed to minimise any excessive solubilisation. Should any SDS precipitation occur it was removed by the subsequent centrifugation. 1% of the supernatant was retained for analysis of the input material and the remainder was split into two equal aliquots. 2mg of precipitating antibody was added to one aliquot and 2mg of mouse IgG was added to the other. These were incubated at 4°C overnight with rotation. 25ml of protein G Dynabeads (Invitrogen) were then added to the tubes and these were rotated for a further 2 hours at 4°C. The beads were washed six times in the solubilisation buffer and then bound proteins were eluted from the beads by the addition of Sample buffer and then boiled for 5 minutes. The entire eluate and the input were then analysed by SDS-PAGE and Western blotting.
GST-fusion protein preparation
Three PMCA2 sequences were used in the pull down experiments, the PMCA2wb carboxy terminal domain, the PMCA2wa carboxy terminal domain and the more N-terminal splice site A region (Figure 4). PCR amplification of these sequences was accomplished using primers incorporating restriction sites for directional cloning into the pGEX-4T1 GST fusion expression vector (GE Healthcare). Templates for PCR were the full length PMCA2wa and PMCA2wb DNAs (Chicka & Strehler, 2003).
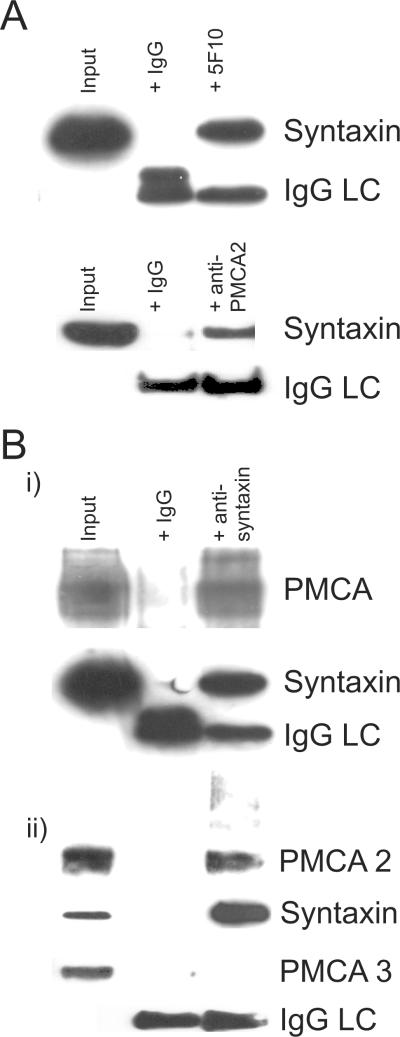
Figure 4. Reciprocal co-immunoprecipitation of syntaxin with PMCA and PMCA2.
(A) shows representative results from immunoprecipitation experiments in cerebellar synaptosomes using the pan-PMCA antibody (5F10) and the PMCA2 specific antibody NR-2 (lower panel) with Western blotting to identify syntaxin as indicated. Input represents 1% of the synaptosomes used in the immunoprecipitation and was analysed alongside on the same gel and blot. Control IgG was also used to immunoprecipitate and the absence of any bands in this lane confirms that precipitation by both 5F10 and anti PMCA2 antibodies was specific. The signal from the IgG light chain (LC) in both lanes is also shown.
In (B) a representative reciprocal immunoprecipitation using the anti syntaxin antibody shows that PMCA, as detected using the 5F10 antibody (Bi), and in a separate experiment PMCA2 (Bii) were both co-immunoprecipitated, with the same controls as in A. Note that syntaxin was detected in these experiments as expected. Also shown in the lower part of (Bii) is an example of one of four separate experiments where the anti syntaxin antibody was unable to co-immunoprecipitate PMCA3, despite this isoform being readily detectable in cerebellar tissue (albeit at lower levels than PMCA2), see input lane. IgG light chain (LC) signal is also shown for the PMCA3 experiment.
Primer sequences for the PMCA2wb carboxy terminal domain were 5'-CTGAATCGGATCCGTCGACAGATCCGCGTC and 5'-CTTGGATCCTCTAGAGCGGCCGCCTAAAG, for the PMCA2wa domain 5'-CCTGAATCGGATCCGTCGACAGATTGAAG and 5'-CTGAACTCTGTGCGGCCGCTTCTCTAGCC and for the splice site A variant 5'-GTGAACTCTCAGAGTCGACTCATCTTTACC and 5'-CATCTCCATGGCGGCCGCCTAGTCCTGTTG. Following amplification and restriction digestion, fragments were cloned into pGEX-4T1 using standard molecular biology procedures.
The mouse Syntaxin 1A-GST construct was a gift from Prof R. D. Burgoyne (Graham et al., 2004). GST-fusion constructs and the empty pGEX-4T-1 vector were expressed in E.coli, BL21DE3 cells (Novagen) with 1mM IPTG (Sigma). The cells were lysed by sonication and the soluble material was rotated with glutathione-sepharose 4B beads (GE Healthcare) for 30 minutes at room temperature. The beads were washed in PBS and immobilised fusion proteins quantified by electrophoresis and Coomassie staining. Equivalent amounts (50mg) were used in subsequent pull down experiments.
GST pull down
Synaptosomes containing 1mg protein were solubilised in 1% SDS and solubilisation buffer as described for immunoprecipitations above. The solubilised synaptosomes were rotated overnight at 4°C with GST or GST-fusion protein bound beads. The beads were washed six times in solubilisation buffer and bound proteins were eluted from the beads by the addition of 25μl of sample buffer followed by boiling for five minutes. Samples were then analysed by SDS-PAGE and Western blotting.
SDS-PAGE and Western blotting
SDS-PAGE and Western blotting were carried out as previously described (Buckby et al., 2006). In brief, for analysis of synapse web preparations, samples of equalised protein content (15μg) were loaded onto 7.5% polyacrylamide gels, separated by electrophoresis and transferred to a nitrocellulose membrane using standard Western blotting techniques. For analysis of immunoprecipitation and pull down experiments the entire eluates were separated by SDS-PAGE on 4-12% NuPAGE bis-tris gels (Invitrogen). Protein levels were also controlled by post-hoc analysis of Ponceau stains of transfers and Coomassie stained gels to ensure equal protein loading. Transferred proteins were probed using primary antibodies specific for N-terminal epitopes of total PMCA1, 2, 3 or 4 (NR-1-3 and JA9 respectively, Abcam, Cambridge, UK), PSD95 (monoclonal, Abcam), PSD93 (Abcam), SAP102 (Antibodies Inc.), NR1 (mouse monoclonal, BD Pharmingen), NR2a (mouse monoclonal, Chemicon) Syntaxin (mouse monoclonal, Sigma) and visualised on Kodak Biomax film using HRP-conjugated secondary antibodies (Dako Ltd, Glostrup, Denmark) with ECL substrates (Pierce, Illinois, USA).
Proteomics analysis
Immunoprecipitations using the 5F10 antibody and in some cases the specific anti PMCA2 NR-2 antibody as well as control IgG were carried out as described above and the eluted material was separated by SDS-PAGE on a 4-12% NuPAGE bis-tris gel (Invitrogen). The gel was silver stained using the Proteosilver Plus silver stain kit (Sigma). Bands of approximately 1mm in width were cut from both the IgG control and the 5F10 lanes of the gel, cut into 1mm cubes and destained. These were then processed for trypsin digestion and mass spectrometric analysis using standard protocols. Briefly, the bands were reduced in 75mM DTT in 25mM ammonium bicarbonate for an hour at 56°C and then treated with 110mM iodoacteamide in 25mM ammonium bicarbonate for 45 minutes at room temperature in the dark. The gel pieces were equilibrated in 25mM ammonium bicarbonate and dried in 100% acetonitrile before being rehydrated in 25mg/ml sequencing-grade porcine trypsin (Promega) in 25mM ammonium bicarbonate and incubated overnight at 37°C. Following digestion the supernatant was removed and the gel pieces washed in 50% acetonitrile/0.1% TFA. The supernatant and washes were combined and dried down. The peptides were resuspended in 0.1% TFA, desalted using C18 zip-tips (Millipore) and eluted in 10μl 70% acetonitrile/0.1% TFA. The peptides were then combined with HCCA matrix and loaded onto an Anchorchip plate for analysis using the Bruker Reflex III MALDI-TOF mass spectrometer and for subsequent MS2 using the Thermofinnigan LCQ Deca Plus coupled to a Surveyor LC system. Tryptic peptides were separated on a micro C18 column over 40 min with an acetonitrile gradient running to 60% in 0.1% formic acid, and then electrosprayed into the ion-trap. MS2 data was collected, and searches were performed at MatrixScience (www.matrixscience.com).
Immunohistochemistry
Rat brains were removed into ice-cold aCSF containing in (mM) NaCl 126, KCl 2.5, NaH2PO4 1.2, MgCl2 1.3, CaCl2 2, NaHCO3 26, glucose 10, pre-bubbled with 95% oxygen and 5% carbon dioxide. Sagittal slices (150μm thick) were cut from the cerebella using a Vibroslice (Campden Instruments, Loughborough, UK) and transferred into fresh ice-cold aCSF. These were then fixed for 15 minutes at 4°C in 4% formaldehyde. The slices were washed three times in PBS before being permeabilised and blocked for 4 hours at 37°C in 0.5% Triton X-100 in blocking solution (PBS containing 0.5% BSA and 1% goat serum). The slices were then incubated overnight at 4°C with primary antibodies raised against PMCA2 (NR-2, Abcam, 1:200) PSD95 (Abcam, 1:100) NR2a (1:100 Chemicon) and syntaxin (1:500) in blocking solution. Following three washes in PBS the slices were incubated in secondary antibodies (goat anti mouse Alexa-488 and goat anti rabbit Alexa-555, Molecular Probes, Invitrogen) in blocking solution for 4 hours at room temperature. The slices were then extensively washed in PBS before being mounted in Vectashield mounting medium onto microscope slides. The slices were viewed using a 60x oil immersion objective on an upright light microscope (Eclipse model E600FN, Nikon, Japan) attached to a laser scanning confocal (Bio-Rad Radiance 2100). The microscope was controlled and images collected with the BioRad Lasersharp 2000 software.
Statistical Analysis
Colocalisation analysis used the BioRad Lasersharp 2000 software and ImageJ (http://rsbweb.nih.gov/ij/) with the JACoP colocalisation analysis plug-in (http://rsb.info.nih.gov/ij/plugins/track/jacop.html) to calculate Pearson's correlation values from the raw images. Similar Pearson's correlation values were obtained with images processed with similar settings using either software. Prism 3.03 (Graphpad Software) calculated mean and standard error of the mean (SEM) Pearson's correlation values from several slices from at least 3 animals and used unpaired t-test comparisons.
RESULTS
An initial screen to identify PMCA interactions within rat forebrain synaptosomes revealed PSD95 (post-synaptic) and syntaxin (pre-synaptic) as potential partners
Given the multiplicity of interactions of PMCAs within a wide range of different tissues we initially focussed our search for PMCA interactions at synapses using an immunoprecipitation approach from forebrain synaptosomes, where PCMA binding partners had previously been identified (de Marco & Strehler, 2001). We also chose to use the pan-PMCA antibody, 5F10, rather than the anti PMCA2 antibody NR-2, in this initial screen, as 5F10 is recommended for immunoprecipitations and we encountered early technical difficulties using the NR-2 antibody with this technique. Precipitated material was separated using SDS PAGE on a one dimensional gel (see Supplementary Fig. 1) and silver stained bands were excised, digested and processed for peptide identification using either MALDI-ToF alone or separation followed by electrospray and subsequent MS2 detection. A number of bands containing material precipitated by 5F10 (distinct from those immunoprecipitated by the IgG control) were observed and positive identifications of the proteins in these bands revealed PMCAs1, 2, 3 and 4 (expected to be precipitated by the 5F10) followed by the PSD95/PSD93 members of the MAGUK family, the NR2a subunit of the NMDA receptor and syntaxin. PMCAs, PSD95 and PSD93 were all identified with a high confidence level (p<0.05, Mowse scores > 77 in Mascot) whereas both NR2a and syntaxin were identified with lower confidence (Mowse score of 70 in Mascot).
PMCA2 locates to pre- and post-synaptic subfractions of synaptosomes
To provide confidence to the results from the initial screen, we screened subfractions of cerebellar synaptosomes for the presence of PMCA2. We decided to focus on PMCA2 as it is the most highly enriched of the PMCAs in the cerebellum (Filoteo et al., 1997). A protocol based on Philips et al., 2001 separates the post-synaptic density (PSD) from the pre-synapse web and vesicle fractions as shown in Fig. 1A. Verification of successful subfractionation used the separation of known pre- and post-synaptic proteins, as detected by Western blot and shown in Fig. 1B. Post-synaptic fractions were free of pre-synaptic proteins such as syntaxin or the synaptic vesicle protein synaptophysin and also enriched with PSD95 and the NMDA receptor NR1 subunit (pH8 pellet and serial pH8 pellet). Using the PMCA2 specific antibody NR-2, PMCA2 could also be detected in the serial pH 8 pellet PSD fraction, supporting the proposal that in vivo PMCA2 exists in close proximity to PSD95 as suggested by previous work (DeMarco & Strehler, 2001) and the results from our initial screen above, although why there was little PMCA2 in the single Triton extracted pH 8 PSD material is unclear. However, since it was a consistent finding, the result may represent the looser attachment of PMCA2 retained by the two step PSD preparation. It is also possible that pre-synaptic PMCA2 might contaminate the serial pellet, but this seems less likely since other potentially contaminating pre-synaptic proteins were not detected in the pellet.
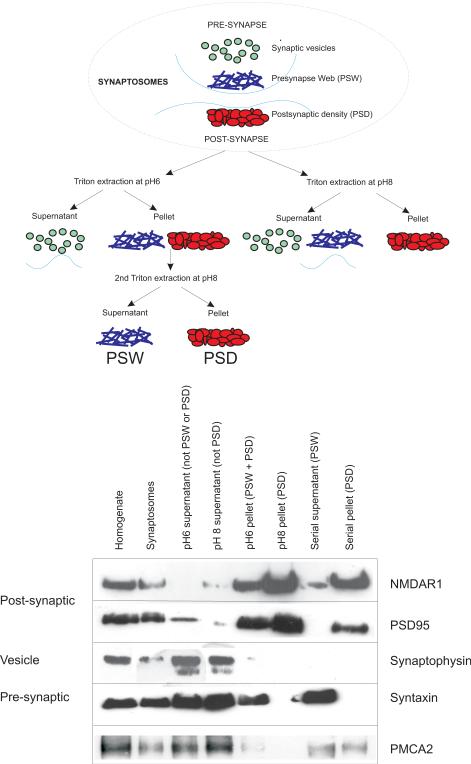
Figure 1. PMCA2 is present in pre- and post-synaptic fractions of cerebellar synaptosomes.
The scheme on top shows the fractionation protocol used to separate preand post-synaptic components based upon the method of Philips et al., 2001. The bottom panel shows a representative western blot to identify pre- and post-synaptic fractions. Note in particular that the serial supernatant, or pre-synaptic web (PSW) does not contain PSD95, but is enriched for syntaxin and contains PMCA2, as detected by the PMCA2 specific antibody NR-2. Also note that the PSW does not contain the synaptic vesicle protein synaptophysin, since vesicle membrane was removed during the first Triton extraction at pH 6. The serial pellet, or post-synaptic density (PSD) is enriched for NMDA receptor subunits NR1 and also for PSD95 and whilst it does not contain pre-synaptic marker proteins syntaxin or synaptophysin, it does contain PMCA2. Note that some PSD95 is in the pH 6 supernatant and may represent extra-synaptic or pre-synaptic membrane associated PSD95 (Castejon et al., 2004) or incomplete separation by the pH 6 Triton extraction. Note, as described in the methods that protein loading was equalised across the different samples. The labels on the left of the bottom panel broadly indicate the membrane specificity of the proteins identified by the Western blots.
Of note, the pre-synaptic fraction, or pre-synapse web (PSW) serial supernatant (characterized by the abundance of the pre-synaptic “marker” protein syntaxin) contained a significant amount of PMCA2, as detected by the NR-2 PMCA2 specific antibody. This PSW was free of the post-synaptic proteins and the synaptic vesicle protein synaptophysin and supports the pre-synaptic location for PMCA2. Interestingly, NR1 was also detected in the PSW and may indicate the presence of pre-synaptic NMDA receptors in the cerebellum (Fiszmann et al., 2005). Since PMCA2 was enriched in the other membrane fractions, particularly the initial pH6 supernatant not thought to contain either the PSD or the PSW, we suggest that PMCA2 also locates to extra synaptic neuronal membranes.
Reciprocal immunoprecipitation confirms PMCA2 and PSD95 interactions in cerebellar synaptosomes
GST fusion protein constructs of PMCA2b and PMCA4b have previously identified the interaction of these PMCAs with PSD95 and other MAGUK family proteins (Kim et al., 1998; de Marco & Strehler, 2001), and our proteomics confirmed this in the forebrain (see Supplementary Fig. 1). We used an immunoprecipitation approach to determine if similar partnerships took place in cerebellar synapse tissue. First, using the pan-PMCA antibody 5F10 we immunoprecipitated PMCA protein from cerebellar synaptosomal preparations. Precipitated material was then immunoblotted using an antibody against PSD95. As shown in Fig. 2A PSD95 co-precipitated with the PMCAs, suggesting an interaction between PSD95 and PMCAs in vivo in the cerebellum. The same result was also obtained using forebrain synaptosomes to confirm the MALDI-ToF result from Supplementary Fig. 1 (data not shown). To further corroborate the evidence for the PMCA2-PSD95 interaction in cerebellar tissue, we carried out a reciprocal immunoprecipitation, using the antibody against PSD95 as the precipitating antibody. Immunoblot analysis with the PMCA2 specific antibody NR-2, revealed that the PSD95 precipitated material contained PMCA2 (Fig. 2B) but did not contain PMCA3 as evidenced by the lack of any detection by the NR-3 PMCA3 specific antibody (see also lower panel Fig. 2B). A previous report (DeMarco & Strehler, 2001) showed that PMCA2b interacts with other members of the MAGUK family, including PSD93 and SAP97, via the same C-terminal PDZ motif. However, these partnerships could not be detected in cerebellar synaptosomes although they were consistently seen with forebrain synaptosomes (data not shown) indicating that these interactions are weaker in the cerebellum. A possible explanation might be a lower abundance of these other members of the MAGUK family of proteins in cerebellum compared with forebrain.
Figure 2. Co-immunoprecipitation of PMCA2 with PSD95 in cerebellar synaptosomes.
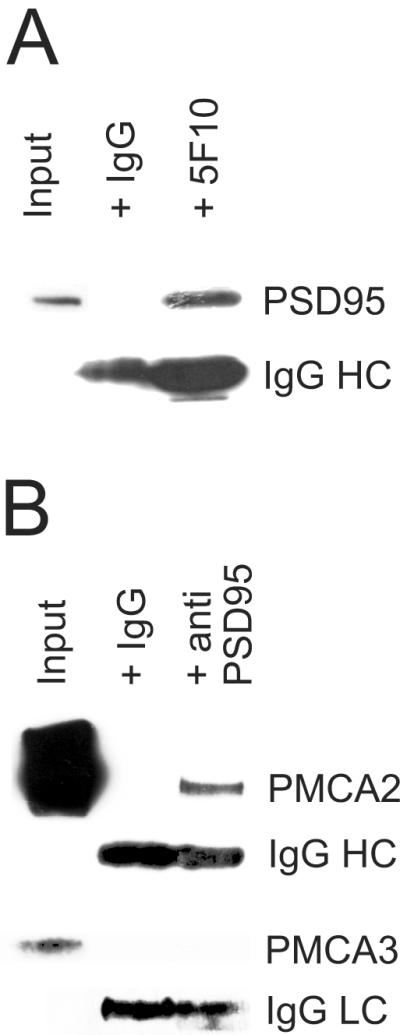
(A) Representative results from immunoprecipitation experiments from cerebellar synaptosomes using the pan-PMCA antibody (5F10) using Western blotting to identify PSD95. Input represents 1% of the synaptosomes used in the immunoprecipitation and was analysed alongside on the same gel and blot. Control IgG was also used to immunoprecipitate and the absence of any bands in this lane confirms that the 5F10 mediated precipitation was specific. The signal from the IgG heavy chain (HC) in both lanes is also shown.
In (B) a representative reciprocal immunoprecipitation from cerebellar synaptosomes using the PSD95 antibody shows that PMCA2 was immunoprecipitated and detected with the PMCA2 specific antibody NR-2, with the same controls as in A. Also shown in the lower panel is the absence of immunoprecipitation of PMCA3 by the anti PSD 95 antibody, from one of four separate experiments that showed this negative result, even though PMCA3 was readily detected in the input lane. The signal from the IgG light chain (LC) is also shown in both lanes for the PMCA3 experiment.
Immunoprecipitation and GST-pull down identifies PMCA2b interactions with PSD95 and the NMDA receptor subunits NR1 and NR2a
Whilst the reciprocal immunoprecipitation of PMCA2b with PSD95 provided strong evidence for this interaction in vivo at cerebellar synapses, a functional pointer for this interaction was still lacking. PSD95 is a classical adaptor protein with three PDZ domains that provide the opportunity to link several different molecules via their PDZ-binding motif (Funke et al., 2005). Our initial proteomics screen identified NR2a as a potential partner for PMCA and since this subunit of the NMDA subtype of glutamate receptor possesses a C-terminus that can interact with PSD95 (Niethammer et al., 1996) our result raised the possibility that all three of PMCA, PSD95 and the NMDA receptor subunit could form a complex at synapses. As shown in Fig. 3, only a GST-PMCA2b fusion protein containing the PDZ domain-binding sequence pulled down PSD95 from the forebrain (Fig. 3B), whilst other GST-PMCA2 fusion proteins containing the alternatively spliced PMCA2a C-terminal tail or the internal splice site “A” sequence (Fig. 3A) were completely ineffective. GST-2b also reliably pulled down PSD95 in the cerebellum, as well as both the NR1 and NR2a receptor subunits (Fig. 3C). Importantly, only the GST fusion protein that encoded the C-terminal PDZ domain-interacting region of PMCA2, GST-2b, was effective at pulling down all three of PSD95, NR1 and NR2a. Immunoprecipitation experiments, using the pan-PMCA antibody 5F10, also confirmed these interactions, as shown in Fig. 3D and E. This result provides further evidence that PMCA2b can form a close association with calcium permeable NMDA receptors at cerebellar post-synaptic sites quite possibly via the PSD95 adaptor protein. Whilst PMCA4b could also make the same interaction with PSD95 (DeMarco & Strehler, 2001), the lower abundance of PMCA4 in cerebellar tissue (Filoteo et al., 1997) and synapse preparations (MG & RME, unpublished) may reduce the likelihood of a PMCA4-NMDA receptor interaction.
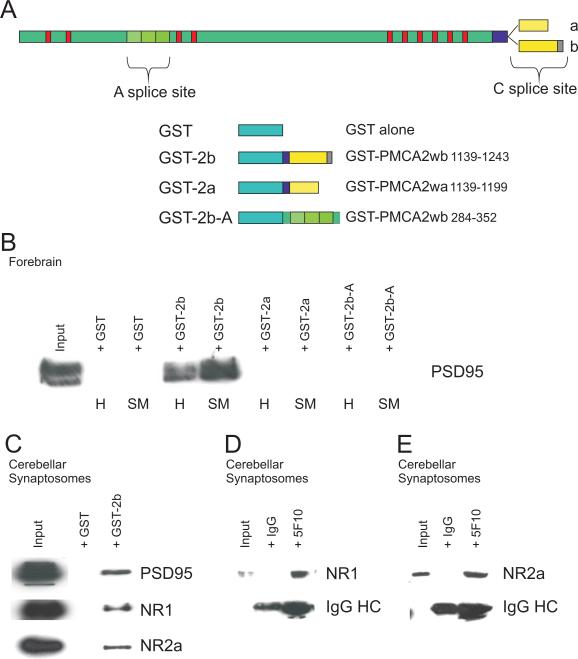
Figure 3. GST pull down confirms the PMCA2b-PSD95 and PMCA2-NR2a and NR1 interactions in cerebellar synaptosomes.
(A) shows a schematic of the GST-PMCA2 fusion protein sequences that encompass the PMCA2 “A” and “C” splice sites and the shortened “a” and longer “b” PMCA2 C-terminal tail regions, with the latter containing the PDZ domain-binding sequence (grey box). (B) shows the outcome of pull down experiments with the different GST-fusion proteins using forebrain samples. Only GST-PMCA2wb (GST-2b) was able to pull down PSD95, and this was evident whether using homogenate (H) or synapse enriched synaptosomes (SM), although, as expected, it was more efficient in the latter. GST alone as a control was ineffective. None of the other GST fusion proteins were successful at pulling down PSD95. (C) GST-PMCA2wb (GST-2b) was also effective in cerebellar synaptosomes and pulled down PSD95, as well as NR2a and NR1, whilst GST alone used as control was ineffective in these experiments.
(D) and (E) show representative results from immunoprecipitation experiments from cerebellar synaptosomes using the pan-PMCA antibody (5F10) and Western blotting to identify NMDA receptor subunits NR1 and NR2a, respectively. Input represents 1% of the synaptosomes used in the immunoprecipitation and was analysed alongside on the same gel and blot. Control IgG was also used to immunoprecipitate and the absence of any bands in this lane confirms that the 5F10 mediated precipitation was specific. The signal from the IgG heavy chain (HC) in both lanes is also shown.
Reciprocal immunoprecipitation confirms the PMCA2 and syntaxin interaction in cerebellar synaptosomes
We next sought to confirm the other intriguing positive hit from the proteomics study, which had revealed syntaxin as a putative partner of the PMCA (see Supplementary Fig. 1). As shown in Fig. 4A, material immunoprecipitated from cerebellar synaptosomes (and also from forebrain synaptosomes; data not shown) by the pan PMCA antibody 5F10 contained syntaxin. A reciprocal experiment using an anti syntaxin antibody also immunoprecipitated material recognised by the pan PMCA antibody 5F10 (Fig. 4B i), and the PMCA2-specific antibody NR-2 (Fig. 4B ii). Together these results strongly support an association between syntaxin and PMCA2 within cerebellar synaptosomes. In a separate set of experiments none of the GST- PMCA2 fusion proteins used in Fig. 3 could pull down syntaxin (data not shown). This result indicates that the PMCA2-syntaxin interaction requires sites distinct from the C-terminus of PMCA2b or PMCA2a, or the PMCA2 A-splice region. Similar immunoprecipitation experiments with the anti syntaxin antibody also did not immunoprecipitate PMCA3, as shown in the lower panel of Fig. 4B (ii) so providing further support for the specificity of the syntaxin interaction with PMCA2.
Syntaxin-1A interacts specifically with PMCA2
The syntaxin antibody used in the reciprocal immunoprecipitations is able to recognize syntaxin-1A and syntaxin-1B, and although there is more than 80% homology between these two isoforms, the syntaxin family of proteins is rather large (Teng et al., 2001). We therefore further tested the specificity of the interaction between PMCA2 and syntaxin with a GST pull down approach. As shown in Fig. 5, a full length syntaxin-1A GST construct was able to pull down material that contained PMCA2. In the same experiments the GST construct did not pull down PMCA1, 3 or 4, demonstrating the specificity of the interaction between syntaxin-1A and PMCA2 in cerebellar synaptosomes.
Figure 5. GST-Syntaxin-1A specifically pulls down PMCA2, and not PMCA1, 3 or 4 from cerebellar synaptosomes.
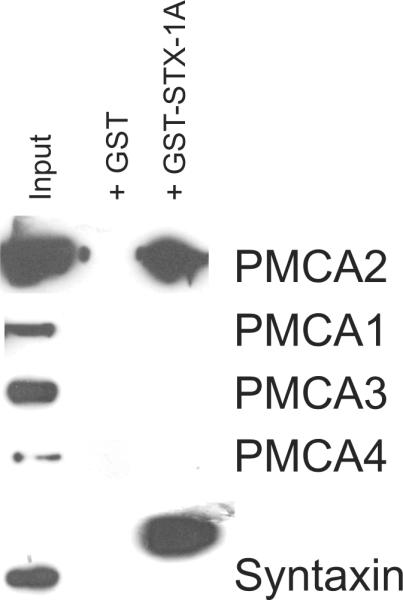
Full length GST-syntaxin-1A was able to pull down PMCA2, whereas GST alone was ineffective. Anti syntaxin antibodies detected syntaxin in the input and detected the higher molecular weight GST-syntaxin-1A present in the pull down lane. No signal was evident in either the GST-syntaxin-1A or GST alone lanes when probed with antibodies to PMCA1, 3 and 4. Note, however, that PMCA4, 1 and 3 were less abundant than PMCA2, as revealed by the input lane.
PMCA2 co-localises with its interacting partners PSD95, NR2a and syntaxin within the layers of the cerebellar cortex
Having established that a molecular interaction occurs between PMCA2 and PSD95/NMDA receptor subunits and PMCA2 and syntaxin-1A at cerebellar synapses, we wanted to know if these interactions could be detected within the structure of the cerebellum. Therefore, we used immunohistochemistry to co-localise PMCA2 (red) and syntaxin (green), PMCA2 (red) and PSD95 (green) and PMCA2 (red) and the NMDA receptor subunit NR2a (green) within the layers of the cerebellar cortex, as illustrated in Fig. 6A, B and C respectively. Immunolabelling for PMCA2, PSD95, NR2a and syntaxin showed discrete puncta reminiscent of the location of these proteins at synaptic sites. Colocalisation of syntaxin with PMCA2 was detected within both the molecular and granule cell layers (white arrows in Fig. 6A). Puncta of PMCA2 alone, without syntaxin present could also be detected and many of these are likely present at post-synaptic sites. Indeed, discrete puncta of PSD95/PMCA2 colocalisation (yellow puncta indicated by white arrows in the left panel of Fig. 6B), were also observed in both the molecular and granule cell layers. Quantitatively, colocalisation of PSD95 with PMCA2 was significantly greater in the granule cell layer compared with the molecular layer, with a mean Pearson's correlation value of 0.58 ± 0.03 compared with 0.22 ± 0.04 (n=12, p<0.0001, t-test; Fig. 6B). The NR2a expression was similar to that seen in previous studies (Piochon et al., 2007) and colocalisation between PMCA2 and NR2a in the granule cell layer gave a mean Pearson's correlation of 0.38 ± 0.06 (n=5), lower than that seen between PSD95 and PMCA2. Like the PMCA2 / PSD95 colocalisation the PMCA2/NR2a colocalisation was also much lower in the molecular layer, with a mean Pearson's correlation of 0.11 ± 0.01 (n=5, p<0.01, t-test, Fig. 6C)). This latter result may reflect the lower expression of functional NMDA receptors in the Purkinje neurone dendrites of rats less than 3 months old (Piochon et al., 2007).
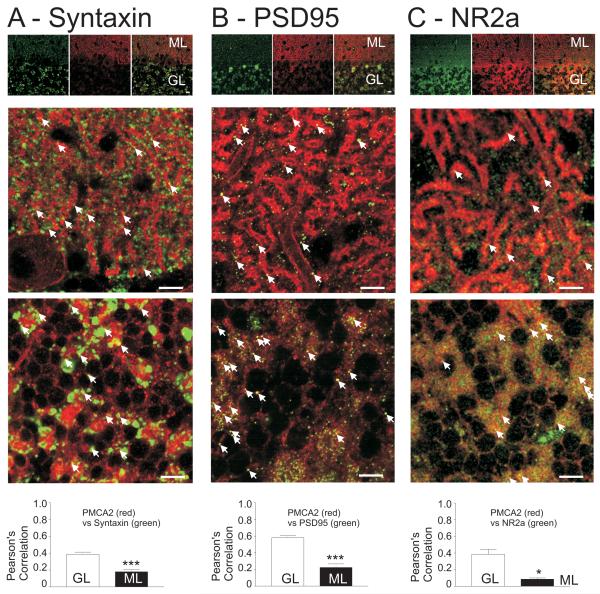
Figure 6. Syntaxin, PSD95 and the NMDA receptor subunit NR2a colocalise with PMCA2 within the different layers of the cerebellar cortex.
(A) Low power (top panel) and high power (bottom two panels) fluorescence images of syntaxin (green) and PMCA2 (red) localisation. Scale bars represent 10mm. In the high power images of the molecular layer (ML) and the granule cell layer (GL) arrow heads point to regions where staining for the two proteins colocalised. Note the red outline of the Purkinje neurone cell body membrane and the punctate nature of the PMCA2 expression in both the molecular and granule cells layers. Pearson's correlations show the extent of colocalisation in the two regions as a bar graph in the lower panel. Pearson's correlation uses a plot of the intensities of all green pixels (x-axis) versus red pixels (y-axis) within the image, so the more often a green pixel colocalises with a red pixel of similar intensity the better the correlation (r-value) of the plot. A value of 1 in a Pearson's correlation indicates 100% colocalisation. Pearsons correlation values less than 1 therefore indicate incomplete colocalisation, where bright green pixels do not have any (or very little) corresponding red intensity (and vice versa).
(B) Low power (upper panel) and high power (bottom two panels) fluorescence images of PSD95 (green) and PMCA2 (red) staining, with the average Pearson's correlations for colocalisation shown below in the bar graph. Note the PMCA2 signal in the membrane of the Purkinje neuron dendrite. Error bars represent SEM and *** represents p<0.0001, unpaired t-test.
(C) Low power (upper panel) and high power (bottom two panels) fluorescence images of NR2a (green) and PMCA2 (red) staining, with the average Pearson's correlations for colocalisation shown below in the bar graph. Note the very few instances of colocalisation of NR2a in the molecular layer, reflecting the very low Pearson's correlation value <0.1 in this region. Error bars represent SEM and * represents p<0. 01, unpaired t-test.
The syntaxin and PMCA2 localisation was also greater in the granule cell layer than the molecular layer, with a mean value of 0.39 ± 0.02 compared with 0.18 ± 0.03 (n=12, p<0.0001, t-test; Fig. 6A). Interestingly, the correlation of expression between PMCA2 and an alternative pre-synaptic marker protein, synaptophysin, revealed a similar pattern between the molecular layer and the granule cell layer, but with a greater Pearson's correlation than with syntaxin. The mean Pearson's correlations for the PMCA2-synaptophysin colocalisation were 0.84 ± 0.01 in the granule cell layer compared with 0.64 ± 0.04 in the molecular layer (n=11, p<0.0001, t-test, data not shown). This might indicate that the PMCA2-syntaxin interaction only occurs at a subset of terminals even though syntaxin is present in all the terminals.
PMCA3 did not colocalise well with syntaxin in either the granule cell or molecular layers, mean Pearson's correlations were 0.19±0.02 and 0.15±0.03 (n=4). This finding was consistent with our inability to detect a molecular interaction between PMCA3 and syntaxin using immunoprecipitation (see above).
Additional immunohistochemistry (not shown) also could not detect any substantial colocalisation of PSD93 and PMCA2 puncta, or between PMCA3 and PSD95 in either the granule or molecular layers of cerebellar slices, with mean correlation values of 0.09 ± 0.02 and 0.11 ± 0.02, (n=11, PSD93 and PMCA2) and 0.22±0.02 and 0.1±0.01 (n=5, PMCA3 and PSD95) respectively. These findings were consistent with our inability to detect a molecular interaction between these two sets of proteins using immunoprecipitation from cerebellar synaptosomes (see above).
DISCUSSION
Using a variety of approaches, and building upon previous work, we show that PMCA2 locates to pre- and post-synaptic sites in synapse-enriched rat cerebellar preparations. PMCA2 also makes molecular interactions with specific partners, syntaxin-1A at the pre-synapse, and with PSD95 and NMDA receptor subunits at the post-synapse.
PMCA2 and NMDA receptors as part of a functional post-synaptic signalling complex
It has been previously established that PMCA b-splice variants can interact with a variety of PDZ-domain containing proteins via their C-terminal domains. These include members of the SAP family of proteins such as SAP90/PSD95, chapsyn 110/PSD93, SAP97 and SAP102 (DeMarco & Strehler, 2001; Kim et al., 1998) as well as other molecules including nNOS (Schuh et al., 2001), the Na+/H+ exchange regulatory factor 2 (DeMarco et al., 2002), CASK (Ca2+/calmodulin-dependent serine protein kinase) (Schuh et al., 2003), PISP (PMCA-interacting single-PDZ protein) (Goellner et al., 2003) and ania-3 (Sgambato-Faure et al., 2006). Indeed, some of these interactions have important functional consequences, such as the down-regulation of nNOS activity (Williams et al., 2006).
The available evidence therefore strongly supports the idea that PMCAs form part of one or several multi-protein signalling complexes at the cell membrane (Strehler et al., 2007). Here, in synapse enriched tissue from cerebellum, we show that the specific interaction of PMCA2 with PSD95 occurs in vivo. Furthermore, we demonstrate that this interaction may also be capable of recruiting the NMDA type glutamate receptor into close proximity with the PMCA2 isoform. Since both the NR1 and the NR2a subunits of the NMDA receptor were immunoprecipitated by the anti PMCA antibody and also pulled down by the recombinant GST-PMCA2b, we propose that functionally complete and synaptically located NMDA receptors, consisting of these two subunits, form a close partnership with PMCA2 in vivo. Previous work has identified molecular and functional interactions between PSD95 and the NMDA receptor in cerebellar neurones (Niethammer et al., 1996; Losi et al., 2003) and more recently PSD95 has been shown to bring NR2a-containing NMDA type glutamate receptors into functional synaptic signalling complexes (Elias et al., 2008). Hence, the possibility exists that PMCA2b, PSD95 and the NMDA receptor exist within a trimeric post-synaptic signalling complex. These interactions would bring the PMCA2 mediated Ca2+ extrusion mechanism into close proximity with Ca2+ entry through the NMDA receptor, perhaps as an efficient way to limit the spread of local calcium.
Based on our data, the simplest model proposes a direct PMCA2-PSD95-NMDA receptor interaction, where PSD95 uses one or both of its first two PDZ binding domains to grab the NR2a subunit (Niethammer et al.,1996) and then one or more of its remaining PDZ binding domains to bind the PMCA2b C-terminal tail. However, the potential for additional partners and alternative indirect interactions may also explain our findings. The promiscuity of the PMCA2b tail allows it to interact with other PDZ domain containing proteins, such as SAP97 and PSD93 (DeMarco & Strehler, 2001), and these too can interact with the subunits of the NMDA receptor (Muller et al., 1996; Cousins et al., 2008). However, our inability to detect PMCA2b-PSD93 interactions here suggests that this is less likely, at least in the cerebellum. More recently, PMCA2b was shown to interact with the ania3-Homer complex within hippocampal neurones (Sgambato-Faure et al., 2006) and with Homer 3 in the cerebellum (Kurnellas et al., 2007) and since NMDA receptor NR2a subunits can also co-immunoprecipitate Homer (Husi et al., 2000; Al-Hallaq et al., 2007) this presents an alternative basis for the NMDA receptor-PMCA2 interaction. Furthermore, the NMDA receptor, Homer and PSD95 could all interact via the shank scaffolding protein within a four-way partnership (Tu et al., 1999) as a way to bring PMCA2b into a synapse associated complex. However, irrespective of the partners, the functional significance of the PMCA2b-NMDA receptor interaction is to provide the potential, along with other calcium “off” mechanisms (Berridge et al, 2003), to efficiently control local post-synaptic calcium levels. The high level of colocalisation between PSD95 and PMCA2 and NR2a and PMCA2 in the cerebellar granule cell layer where fully functional calcium permeable NMDA receptors (containing NR1 and NR2a subunits) drive the plasticity of the mossy fibre to granule cell pathway (D'Angelo et al., 1999), supports the idea that the partnership functions during sustained depolarisation and NMDA receptor activation. More recently, in hippocampal pyramidal neurones, where PMCA2b is also expressed, NMDA receptor mediated calcium entry was shown to slow the extrusion kinetics of the PMCA (Scheuss et al., 2006) through a protease mediated mechanism (Ferragamo et al., 2009). This provides supporting evidence for a close partnership between the NMDA receptor and PMCA2b.
PMCA2 and Syntaxin partner at cerebellar pre-synaptic sites
There is anatomical and functional evidence to support a location and role for PMCA2 to control calcium extrusion from pre-synaptic terminals (Kim et al., 2005, Jensen et al., 2007, Empson et al., 2007), and thereby influence calcium dependent transmitter release kinetics. We now show the presence of PMCA2 in the pre-synapse web and a molecular interaction that could underlie its functional contribution at this site. Immunoprecipitation of syntaxin by anti PMCA and anti PMCA2 antibodies from synapse-enriched cerebellar tissue supported the original finding from the proteomics screen and added weight to the proposal that PMCA2 and syntaxin co-exist at synapses both in cerebellum and forebrain.
The syntaxins are a diverse family of proteins (Teng, et al., 2001), but in the context of synapse function we expected syntaxin-1A to be the most likely partner for PMCA2, and indeed this was the isoform identified by the proteomics screen. Syntaxin-1A contains a single membrane-spanning domain and together with the membrane-associated protein SNAP25 and synaptobrevin forms part of the vesicle fusion complex known as the SNARE complex. The SNARE complex is responsible for the fusion and exocytosis of neurotransmitter-containing synaptic vesicles at the pre-synaptic membrane in response to the calcium trigger (Sorensen, 2005). As the most efficient way to confirm whether syntaxin-1A was the partner for PMCA2 we employed a GST-syntaxin-1A fusion protein that efficiently pulled-down PMCA2; importantly none of PMCA1, 3 or 4 were detected though they are all expressed in the cerebellum (albeit at lower levels than PMCA2). Although PMCA3 has been detected at pre-synaptic parallel fibre terminals in cerebellum (Eakin et al., 1995; Burette & Weinberg, 2007) our results indicate that it may use an alternative interaction partner to syntaxin-1A.
Some concerns exist about the use of immunoprecipitation experiments to demonstrate interactions between syntaxin-1A and reported binding partners (Fletcher et al., 2003) since syntaxin-1A can bind directly to the sepharose and agarose resins commonly used in immunoprecipitation experiments. However, we are confident that our results are not false positives for a number of reasons: (i) we carried out immunoprecipitations using magnetic beads rather than agarose or sepharose resins, (ii) if the syntaxin was binding to the beads directly we would have detected it in control immunoprecipitations using IgG in place of the precipitating antibody, and (iii) the reciprocal immunoprecipitation using anti syntaxin to co-precipitate PMCA would have yielded negative results had the syntaxin simply bound to the beads.
At a functional level we could detect the PMCA2-syntaxin interaction in all regions of the cerebellar cortex characterized by a high density of synaptic terminals, although it was greatest in the granule cell layer. At the present time we can only speculate about how this interaction might work. Syntaxin-1A binds to L-, N- and P/Q-type voltage-gated Ca2+ channels and influences the location and gating of these important channels; with obvious importance for the regulation of Ca2+ influx and subsequent neurotransmitter release (Sheng et al., 1994; Martin-Moutot et al., 1996; Keith et al., 2007). Syntaxin-1A binds to these Ca2+ channels via a synaptic protein interaction (synprint) site. Since the PMCAs do not possess this domain, or indeed any sequence similarity to this region of Ca2+ channels, the molecular basis or stoichiometry of the PMCA2-syntaxin-1A interaction requires further investigation. However, the association with syntaxin may provide an anchor for PMCA2 close to the site of neurotransmitter release where precise control of pre-synaptic Ca2+ levels via extrusion can influence residual calcium to enhance and prolong paired pulse facilitation (Jensen et al., 2007; Empson et al., 2007).
In summary we provide new evidence that PMCA2, which is enriched in the cerebellum, makes specific partner interactions with important pre- and post-synaptic signalling molecules in vivo to contribute to the functional physiology of cerebellar synapses. Further studies will also be required to tease out the mechanistic implications of these PMCA2 interactions for synapse pathology, such as during amyloid beta toxicity in human Alzheimer's disease (Berrocal et al., 2009), and will be aided by the availability of PMCA2 null mice that exhibit cerebellar, spinal and motor defects (Kozel et al., 1998; Kurnellas et al., 2005; Souayah et al., 2008).
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the support from the BBSRC (BBS/0338) and Royal Holloway College University of London to RME, PWB, MG, PRT, a University of Otago Research Grant to RME, and a grant from the NIH (NS51769) to EES. We thank Professor Robert Burgoyne, University of Liverpool, UK for the generous donation of GST-syntaxin-1A DNA. We also extend our thanks to the Biomics Centre at St George's for proteomics technical support and to Dr Vincent O'Connor, University of Southampton, UK for early help with the pre-synapse web preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Al-Hallaq RA, Conrads TP, Veenstra TD, Wenthold RJ. NMDA diheteromeric receptor populations and associated proteins in rat hippocampus. J Neurosci. 2007;27(31):8334–43. doi: 10.1523/JNEUROSCI.2155-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4(7):517–29. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 3.Berrocal M, Marcos D, Sepúlveda MR, Pérez M, Avila J, Mata AM. Altered Ca2+ dependence of synaptosomal plasma membrane Ca2+-ATPase in human brain affected by Alzheimer's disease. FASEB J. 2009 doi: 10.1096/fj.08-121459. (In Press) [DOI] [PubMed] [Google Scholar]
- 4.Bloodgood BL, Sabatini BL. Ca(2+) signaling in dendritic spines. Curr Opin Neurobiol. 2007;17(3):345–51. doi: 10.1016/j.conb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Buckby LE, Jensen TP, Smith PJ, Empson RM. Network stability through homeostatic scaling of excitatory and inhibitory synapses following inactivity in CA3 of rat organotypic hippocampal slice cultures. Mol Cell Neurosci. 2006;31(4):805–16. doi: 10.1016/j.mcn.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Burette A, Weinberg RJ. Perisynaptic organization of plasma membrane calcium pumps in cerebellar cortex. J Comp Neurol. 2007;500(6):1127–35. doi: 10.1002/cne.21237. [DOI] [PubMed] [Google Scholar]
- 7.Castejón OJ, Fuller L, Dailey ME. Localization of synapsin-I and PSD-95 in developing postnatal rat cerebellar cortex. Brain Res Dev Brain Res. 2004;151(12):25–32. doi: 10.1016/j.devbrainres.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 8.Chicka MC, Strehler EE. Alternative splicing of the first intracellular loop of plasma membrane Ca2+-ATPase isoform 2 alters its membrane targeting. J Biol Chem. 2003;278(20):18464–70. doi: 10.1074/jbc.M301482200. [DOI] [PubMed] [Google Scholar]
- 9.Cousins SL, Papadakis M, Rutter AR, Stephenson FA. Differential interaction of NMDA receptor subtypes with the post-synaptic density-95 family of membrane associated guanylate kinase proteins. J Neurochem. 2008;104(4):903–13. doi: 10.1111/j.1471-4159.2007.05067.x. [DOI] [PubMed] [Google Scholar]
- 10.D'Angelo E, Rossi P, Armano S, Taglietti V. Evidence for NMDA and mGlu receptor-dependent long-term potentiation of mossy fiber-granule cell transmission in rat cerebellum. J Neurophysiol. 1999;81(1):277–87. doi: 10.1152/jn.1999.81.1.277. [DOI] [PubMed] [Google Scholar]
- 11.DeMarco SJ, Strehler EE. Plasma membrane Ca2+-atpase isoforms 2b and 4b interact promiscuously and selectively with members of the membrane-associated guanylate kinase family of PDZ (PSD95/Dlg/ZO-1) domain-containing proteins. J Biol Chem. 2001;276(24):21594–600. doi: 10.1074/jbc.M101448200. [DOI] [PubMed] [Google Scholar]
- 12.DeMarco SJ, Chicka MC, Strehler EE. Plasma membrane Ca2+ ATPase isoform 2b interacts preferentially with Na+/H+ exchanger regulatory factor 2 in apical plasma membranes. J Biol Chem. 2002;277(12):10506–11. doi: 10.1074/jbc.M111616200. [DOI] [PubMed] [Google Scholar]
- 13.Eakin TJ, Antonelli MC, Malchiodi EL, Baskin DG, Stahl WL. Localization of the plasma membrane Ca(2+)-ATPase isoform PMCA3 in rat cerebellum, choroid plexus and hippocampus. Brain Res Mol Brain Res. 1995;29(1):71–80. doi: 10.1016/0169-328x(94)00231-3. [DOI] [PubMed] [Google Scholar]
- 14.Elias GM, Elias LA, Apostolides PF, Kriegstein AR, Nicoll RA. Differential trafficking of AMPA and NMDA receptors by SAP102 and PSD-95 underlies synapse development. Proc Natl Acad Sci U S A. 2008;105(52):20953–8. doi: 10.1073/pnas.0811025106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enyedi A, Verma AK, Filoteo AG, Penniston JT. Protein kinase C activates the plasma membrane Ca2+ pump isoform 4b by phosphorylation of an inhibitory region downstream of the calmodulin-binding domain. J Biol Chem. 1996;271(50):32461–7. doi: 10.1074/jbc.271.50.32461. [DOI] [PubMed] [Google Scholar]
- 16.Empson RM, Garside ML, Knöpfel T. Plasma membrane Ca2+ ATPase 2 contributes to short-term synapse plasticity at the parallel fiber to Purkinje neuron synapse. J Neurosci. 2007;27(14):3753–8. doi: 10.1523/JNEUROSCI.0069-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferragamo MJ, Reinardy JL, Thayer SA. Ca2+-dependent, stimulus-specific modulation of the plasma membrane Ca2+ pump in hippocampal neurons. J Neurophysiol. 2009 doi: 10.1152/jn.90774.2008. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filoteo AG, Elwess NL, Enyedi A, Caride A, Aung HH, Penniston JT. Plasma membrane Ca2+ pump in rat brain. Patterns of alternative splices seen by isoform-specific antibodies. J Biol Chem. 1997;272(38):23741–7. doi: 10.1074/jbc.272.38.23741. [DOI] [PubMed] [Google Scholar]
- 19.Fiszman ML, Barberis A, Lu C, Fu Z, Erdélyi F, Szabó G, Vicini S. NMDA receptors increase the size of GABAergic terminals and enhance GABA release. J Neurosci. 2005;25(8):2024–31. doi: 10.1523/JNEUROSCI.4980-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fletcher S, Bowden SE, Marrion NV. False interaction of syntaxin 1A with a Ca(2+)-activated K(+) channel revealed by coimmunoprecipitation and pull-down assays: implications for identification of protein-protein interactions. Neuropharmacology. 2003;44(6):817–27. doi: 10.1016/s0028-3908(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 21.Fujii JT, Su FT, Woodbury DJ, Kurpakus M, Hu XJ, Pourcho R. Plasma membrane calcium ATPase in synaptic terminals of chick Edinger-Westphal neurons. Brain Res. 1996;734(12):193–202. [PubMed] [Google Scholar]
- 22.Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–45. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 23.Graham ME, Barclay JW, Burgoyne RD. Syntaxin/Munc18 interactions in the late events during vesicle fusion and release in exocytosis. J Biol Chem. 2004;279(31):32751–60. doi: 10.1074/jbc.M400827200. [DOI] [PubMed] [Google Scholar]
- 24.Goellner GM, DeMarco SJ, Strehler EE. Characterization of PISP, a novel single-PDZ protein that binds to all plasma membrane Ca2+-ATPase b-splice variants. Ann N Y Acad Sci. 2003;986:461–71. doi: 10.1111/j.1749-6632.2003.tb07230.x. [DOI] [PubMed] [Google Scholar]
- 25.Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3(7):661–9. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 26.Juhaszova M, Church P, Blaustein MP, Stanley EF. Location of calcium transporters at presynaptic terminals. Eur J Neurosci. 2000;3:839–46. doi: 10.1046/j.1460-9568.2000.00974.x. [DOI] [PubMed] [Google Scholar]
- 27.Jensen TP, Filoteo AG, Knopfel T, Empson RM. Presynaptic plasma membrane Ca2+ ATPase isoform 2a regulates excitatory synaptic transmission in rat hippocampal CA3. J Physiol. 2007;579(Pt 1):85–99. doi: 10.1113/jphysiol.2006.123901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keith RK, Poage RE, Yokoyama CT, Catterall WA, Meriney SD. Bidirectional modulation of transmitter release by calcium channel/syntaxin interactions in vivo. J Neurosci. 2007;27(2):265–9. doi: 10.1523/JNEUROSCI.4213-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim E, DeMarco SJ, Marfatia SM, Chishti AH, Sheng M, Strehler EE. Plasma membrane Ca2+ ATPase isoform 4b binds to membrane-associated guanylate kinase (MAGUK) proteins via their PDZ (PSD-95/Dlg/ZO-1) domains. J Biol Chem. 1998;273(3):1591–5. doi: 10.1074/jbc.273.3.1591. [DOI] [PubMed] [Google Scholar]
- 30.Kim MH, Korogod N, Schneggenburger R, Ho WK, Lee SH. Interplay between Na+/Ca2+ exchangers and mitochondria in Ca2+ clearance at the calyx of Held. J Neurosci. 2005;25(26):6057–65. doi: 10.1523/JNEUROSCI.0454-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozel PJ, Friedman RA, Erway LC, Yamoah EN, Liu LH, Riddle T, Duffy JJ, Doetschman T, Miller ML, Cardell EL, Shull GE. Balance and hearing deficits in mice with a null mutation in the gene encoding plasma membrane Ca2+-ATPase isoform 2. J Biol Chem. 1998;273(30):18693–6. doi: 10.1074/jbc.273.30.18693. [DOI] [PubMed] [Google Scholar]
- 32.Kurnellas MP, Lee AK, Li H, Deng L, Ehrlich DJ, Elkabes S. Molecular alterations in the cerebellum of the plasma membrane calcium ATPase 2 (PMCA2)-null mouse indicate abnormalities in Purkinje neurons. Mol Cell Neurosci. 2007;34:178–88. doi: 10.1016/j.mcn.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurnellas MP, Nicot A, Shull GE, Elkabes S. Plasma membrane calcium ATPase deficiency causes neuronal pathology in the spinal cord: a potential mechanism for neurodegeneration in multiple sclerosis and spinal cord injury. FASEB J. 2005;19:298–300. doi: 10.1096/fj.04-2549fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Losi G, Prybylowski K, Fu Z, Luo J, Wenthold RJ, Vicini S. PSD-95 regulates NMDA receptors in developing cerebellar granule neurons of the rat. J Physiol. 2003;548(Pt 1):21–9. doi: 10.1113/jphysiol.2002.034918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Moutot N, Charvin N, Leveque C, Sato K, Nishiki T, Kozaki S, Takahashi M, Seagar M. Interaction of SNARE complexes with P/Q-type calcium channels in rat cerebellar synaptosomes. J Biol Chem. 1996;271(12):6567–70. doi: 10.1074/jbc.271.12.6567. [DOI] [PubMed] [Google Scholar]
- 36.Morgans CW, El Far O, Berntson A, Wässle H, Taylor WR. Calcium extrusion from mammalian photoreceptor terminals. J Neurosci. 1998;18(7):2467–74. doi: 10.1523/JNEUROSCI.18-07-02467.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16(7):2157–63. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips GR, Huang JK, Wang Y, Tanaka H, Shapiro L, Zhang W, Shan WS, Arndt K, Frank M, Gordon RE, Gawinowicz MA, Zhao Y, Colman DR. The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron. 2001;32(1):63–77. doi: 10.1016/s0896-6273(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 39.Piochon C, Irinopoulou T, Brusciano D, Bailly Y, Mariani J, Levenes C. NMDA receptor contribution to the climbing fiber response in the adult mouse Purkinje cell. J Neurosci. 2007;27(40):10797–809. doi: 10.1523/JNEUROSCI.2422-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheuss V, Yasuda R, Sobczyk A, Svoboda K. Nonlinear [Ca2+] signaling in dendrites and spines caused by activity-dependent depression of Ca2+ extrusion. J Neurosci. 2006;26(31):8183–94. doi: 10.1523/JNEUROSCI.1962-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuh K, Uldrijan S, Telkamp M, Rothlein N, Neyses L. The plasmamembrane calmodulin-dependent calcium pump: a major regulator of nitric oxide synthase I. J Cell Biol. 2001;155(2):201–5. doi: 10.1083/jcb.200104131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuh K, Uldrijan S, Gambaryan S, Roethlein N, Neyses L. Interaction of the plasma membrane Ca2+ pump 4b/CI with the Ca2+/calmodulin-dependent membrane-associated kinase CASK. J Biol Chem. 2003;278(11):9778–83. doi: 10.1074/jbc.M212507200. [DOI] [PubMed] [Google Scholar]
- 43.Sepúlveda MR, Hidalgo-Sánchez M, Marcos D, Mata AM. Developmental distribution of plasma membrane Ca2+-ATPase isoforms in chick cerebellum. Dev Dyn. 2007;236(5):1227–36. doi: 10.1002/dvdy.21131. [DOI] [PubMed] [Google Scholar]
- 44.Sgambato-Faure V, Xiong Y, Berke JD, Hyman SE, Strehler EE. The Homer-1 protein Ania-3 interacts with the plasma membrane calcium pump. Biochem Biophys Res Commun. 2006;343(2):630–7. doi: 10.1016/j.bbrc.2006.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sørensen JB. SNARE complexes prepare for membrane fusion. Trends Neurosci. 2005 Sep;28(9):453–5. doi: 10.1016/j.tins.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Souayah N, Sharovetskaya A, Kurnellas MP, Myerson M, Deitch JS, Elkabes S. Reductions in motor unit number estimates (MUNE) precede motor neuron loss in the plasma membrane calcium ATPase 2 (PMCA2)-heterozygous mice. Exp Neurol. 2008;214(2):341–6. doi: 10.1016/j.expneurol.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stauffer TP, Guerini D, Carafoli E. Tissue distribution of the four gene products of the plasma membrane Ca2+ pump. A study using specific antibodies. J Biol Chem. 1995;270(20):12184–90. doi: 10.1074/jbc.270.20.12184. [DOI] [PubMed] [Google Scholar]
- 48.Stauffer TP, Guerini D, Celio MR, Carafoli E. Immunolocalization of the plasma membrane Ca2+ pump isoforms in the rat brain. Brain Res. 1997;748:21–29. doi: 10.1016/s0006-8993(96)01282-6. [DOI] [PubMed] [Google Scholar]
- 49.Strehler EE, Zacharias DA. Role of alternative splicing in generating isoform diversity among plasma membrane calcium pumps. Physiol Rev. 2001;81(1):21–50. doi: 10.1152/physrev.2001.81.1.21. [DOI] [PubMed] [Google Scholar]
- 50.Strehler EE, Filoteo AG, Penniston JT, Caride AJ. Plasma membrane Ca(2+) pumps: structural diversity as the basis for functional versatility. Biochem Soc Trans. 2007;35(Pt 5):919–22. doi: 10.1042/BST0350919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teng FY, Wang Y, Tang BL. The syntaxins. Genome Biol. 2001;2(11) doi: 10.1186/gb-2001-2-11-reviews3012. REVIEWS3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23(3):583–92. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- 53.Usachev YM, DeMarco SJ, Campbell C, Strehler EE, Thayer SA. Bradykinin and ATP accelerate Ca(2+) efflux from rat sensory neurons via protein kinase C and the plasma membrane Ca(2+) pump isoform 4. Neuron. 2002;33(1):113–22. doi: 10.1016/s0896-6273(01)00557-8. [DOI] [PubMed] [Google Scholar]
- 54.Williams JC, Armesilla AL, Mohamed TM, Hagarty CL, McIntyre FH, Schomburg S, Zaki AO, Oceandy D, Cartwright EJ, Buch MH, Emerson M, Neyses L. The sarcolemmal calcium pump, alpha-1 syntrophin, and neuronal nitric-oxide synthase are parts of a macromolecular protein complex. J Biol Chem. 2006;281(33):23341–8. doi: 10.1074/jbc.M513341200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.