Abstract
The peptidyl-prolyl isomerase Pin1 is frequently upregulated in human cancers in which Rel/NF-κB is constitutively activated but its role in these cancers remains to be determined, and evidence is still lacking to show that Pin1 contributes to cell transformation by Rel/NF-κB. Rel/NF-κB’s transcriptional and oncogenic activities are modulated by several post-translational modifications and co-regulatory proteins, and previous studies showed that cytokine treatment induces binding of Pin1 to the RelA subunit of NF-κB, thereby enhancing RelA’s nuclear localization and stability. Here we show that Pin1 associates with the Rel subunits of NF-κB that are implicated in leukemia/lymphomagenesis, and modulates their transcriptional and oncogenic activities. Pin1 markedly enhanced transformation of primary lymphocytes by the human c-Rel protein and also increased cell transformation by the potent viral Rel/NF-κB oncoprotein v-Rel, in contrast to a Pin1 mutant in the WW domain involved in interaction with NF-κB. Pin1 promoted nuclear accumulation of Rel proteins in absence of activating stimuli. Importantly, inhibition of Pin1 function with the small molecule inhibitor juglone or with Pin1-specific shRNA led to cytoplasmic relocalization of endogenous c-Rel in human lymphoma-derived cell lines, markedly interfered with lymphoma cell proliferation, and suppressed endogenous Rel/NF-κB-dependent gene expression. Together these results demonstrate that Pin1 is an important regulator of Rel/NF-κB’s transforming activity and suggest that Pin1 may be a potential therapeutic target in Rel/NF-κB-dependent leukemia/lymphomas.
Keywords: Rel, NF-κB, transformation, Pin1, Juglone
INTRODUCTION
The Rel/NF-κB family of inducible transcription factors plays pivotal roles in innate and adaptive immunity, inflammation and oncogenesis and includes the c-Rel, RelA, RelB, NF-κB1 (p50/p105) and NF-κB2 (p52/p100) proteins (1). In most cells NF-κB exists as latent cytoplasmic homo-/heterodimers, bound to inhibitory IκB proteins. Stimuli that activate the classical NF-κB cascade trigger the IKK kinase complex that mediates phosphorylation of IκBα, resulting in proteasomal degradation of IκBα, nuclear translocation of NF-κB dimers and their binding to κB DNA sites. This commonly results in transcriptional activation of genes important for the immune and inflammatory response, cell proliferation, adhesion, angiogenesis and inhibition of apoptosis (1–6). Binding of NF-κB to the promoter for the IκBα gene triggers a negative feedback loop that terminates NF-κB activation (1). Hence under normal conditions, activation of the classical Rel/NF-κB cascade is transient due to tight regulation of NF-κB’s subcellular localization by IκBα. Interference with this process can have severe consequences, as sustained activation of Rel/NF-κB is seen in many cancers where it promotes tumor cell survival, pathogenesis and chemoresistance (7).
The peptidyl-prolyl isomerase (PPIase) Pin1 interacts specifically and exclusively with certain phospho-Serine/Threonine-Proline (pSer/Thr-Pro) motifs in target proteins via its N-terminal WW domain and catalyzes rapid cis/trans isomerization of proline amide bonds through its C-terminal domain (8–10). This commonly alters the conformation and biological function of substrates and can have profound physiological relevance. Interaction of Pin1 with target proteins like p53, p73, cyclin D1, p66shc, tau, APP and IRF-3 has been implicated in cell cycle control, cellular stress, neuronal degeneration and tumor progression (11).
Pin1 was shown to associate with the p65/RelA subunit of NF-κB and to promote RelA’s nuclear translocation and extend its half-life by blocking its inhibition by IκBα and its SOCS-1-dependent degradation (12). Interestingly, Pin1 is upregulated in human breast cancer specimens and mouse mammary tumors, and is correlated with nuclear accumulation of RelA (12, 13). Oncomine database analysis revealed markedly elevated Pin1 levels in breast carcinoma and in human B-cell chronic lymphocytic leukemia (B-CLL), diffuse large B-cell lymphoma (DLBCL), mantle cell lymphoma (MCL) and multiple myeloma (MM) in which constitutive nuclear activation of NF-κB is necessary for cell survival and proliferation (Supplementary Fig. 1) (14–24). These observations suggest that Pin1 may contribute to Rel/NF-κB’s function in cancer. However the role of Pin1 in oncogenesis has been a subject of controversy (25), and there is no direct evidence that Pin1 plays a role in Rel/NF-κB-dependent tumor cells. Here we show that Pin1 associates with Rel proteins and demonstrate that it significantly potentiates their transforming activity in primary lymphocytes, coincident with increased Rel nuclear accumulation. We show that knockdown of Pin1 prompts the cytoplasmic relocalization of endogenous c-Rel in multiple Rel/NF-κB-dependent human lymphoma-derived cell lines, resulting in inhibition of tumor cell proliferation and decreased Rel/NF-κB-dependent gene expression. These results identify Pin1 as a critical regulator of Rel’s transforming activity and point to Pin1 as a potential therapeutic target in Rel/NF-κB-dependent tumors.
MATERIAL AND METHODS
Plasmids and cell culture
v-Rel and hc-Rel were expressed from pJDCMV19SV for GST pull-downs, or in the spleen necrosis virus (SNV) retroviral vector pUC19-pJD214 for immunofluorescence and transformation assays (26). Glutathione S-transferase (GST)-Pin1 and GST-v-Rel were expressed in pGEX-4T-1 (GE Healthcare). Pin1 tagged to green fluorescent protein (Pin1-GFP) or its mutants Pin1(S16E)-GFP or Pin1(S16A)-GFP were expressed from an internal ribosome entry site (IRES) in bicistronic retroviral vectors pUC19-pJD214-vRel or pUC19-pJD214-hcRel. Human 293T cells and primary chicken embryonic fibroblasts (CEF) were maintained as described (26). The human non-Hodgkin’s primary mediastinal B cell lymphoma cell line Karpas 1106 (a gift from Dr. A. Karpas, University of Cambridge, Cambridge, UK) (27) and Hodgkin’s lymphoma cell line KM-H2 (DSMZ, Braunschweig, Germany) were maintained in RPMI-1640 with 10% FBS and 1% antibiotics. KM-H2 cells were supplemented with 2 mM glutamine. Primary mediastinal large B-cell lymphoma MedB-1 cells (a gift from Drs. P. Moller and S. Bruderlein, Institute of Pathology, Ulm, Germany) were maintained in Iscove:RPMI-1640 medium (4:1) with 10% FBS, 2 mM glutamine and antibiotics. Rel-transformed chicken spleen cells (CSC) were maintained as described (26).
GST pull-down assays and co-immunoprecipitation
Extracts from 293T cells transfected with pJDCMV19SV-Rel, hc-Rel or hRelA, or with pPin1-GFP, pPin1(S16E)-GFP or pPin1(S16A)-GFP, from v-Rel-transformed CSC, or Karpas 1106 cells, were quantitated for equal amounts of total protein (1 mg) and used in pull-downs with GST-Pin1, GST-vRel or GST as described (12). Antibodies were against the v-Rel transactivation domain (vTAD) (#1691) (26), human c-Rel TAD (#265, a gift from M.K. Ernst and N.R. Rice, NCI-Frederick, Frederick, MD), GFP (TP401, Torrey Pines Biolab), PARP (#9542; Cell Signaling) or actin (Sigma). Endogenous Pin1 was detected with a monoclonal anti-Pin1 generated in the laboratory of KPL. Where indicated, 293T cells were treated with human tumor necrosis factor alpha (hTNF-α; 0 ng/ml, Roche) for 40 min. Endogenous hc-Rel was coimmunoprecipitated with endogenous Pin1 in extracts (1 mg) from lymphoma cells pre-treated with hTNFα, followed by immunoprecipitation with polyclonal anti-Pin1 (sc-15340, Santa Cruz), and immunoblotting with anti-hc-Rel (#265). The membrane was stained with Ponceau S.
Transformation of primary chicken lymphoid cells
Primary chicken spleen cells (CSC) were infected as described (3) with virus co-expressing v-Rel or hc-Rel along with wild type or mutant Pin1. Transformed colonies in soft agar were scored after two weeks. Animals were used according to the Institutional Animal Care and Use Committee guidelines under an approved protocol.
Immunofluorescence and cell fractionation
Cells (105) fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 were analyzed by immunofluorescence with anti-hc-Rel (sc-6955, Santa Cruz Biotechnology) and a rhodamine-conjugated secondary antibody (Jackson Lab), followed by staining with Hoechst 33258 (Sigma). Where indicated, lymphoma cells were pre-treated with juglone (0.1–10 µM; Calbiochem) for 2 hrs or with DMSO as control.
Karpas 1106 cells treated with juglone (0.1–10 µM) for 2 hrs were resuspended in hypotonic buffer (20 mM HEPES [pH 7.6], 20% glycerol. 10 mM KCl, 0.2 mM EDTA, 0.1% Triton X-100, 5 mM NaF, 10 mM glycerophosphate, 1 mM Na3VO4, 1 mM DTT, 1 mM PMSF, 1 × PIC) on ice for 10 min, followed by centrifugation at 10,000 × g to recover the cytosolic fraction. Nuclear pellets resuspended in hypertonic buffer containing 600 mM KCl, were centrifuged at 10,000 × g. Extracts (40 µg) were analyzed by western blotting with anti-Rel #265 and reprobed with anti-actin or anti-PARP (#9542, Cell Signaling) as cytosolic and nuclear markers.
RNA interference and reverse transcription-PCR
Pin1 shRNA in pSuper or a scrambled non-targeting shRNA control (NS) from KPL were delivered into human lymphoma cells by Amaxa nucleofection using Kit V (Amaxa AG, Gaithersburg, MD) and programs optimized for each cell line for transfection efficiencies >90% with pMaxGFP (Karpas 1106: X-001; KM-H2 and MedB-1: T-001). Reverse transcription–PCR (RT-PCR) was performed within the linear range of the PCR cycle using primers specific for pin1 (tgatcaacggctacatccag, caaacgaggcgtcttcaaat), cyclin D1 (ctggagcccgtgaaaaagagc, ctggagaggaagcgtgtgagg), vegf (ccctgatgagatcgagtacatctt, aacgctccaggacttataccg), Bfl-1 (caggctggctcaggactatc, cccagttaatgatgccgtct), and actin (tgacccacccagcacattta, tgaaagcagggcctgagg).
Cell proliferation and survival assays
The effects of Juglone (0.1 µM) or Pin1 shRNA on the proliferation and survival of lymphoma cells was determined by cell counting in the presence of trypan blue exclusion dye during a time course (0–24 hrs for Juglone vs. DMSO control; or 0–144 hrs for Pin1 shRNA vs. scrambled shRNA control).
RESULTS
Pin1 associates with the v-Rel and c-Rel subunits of NF-κB
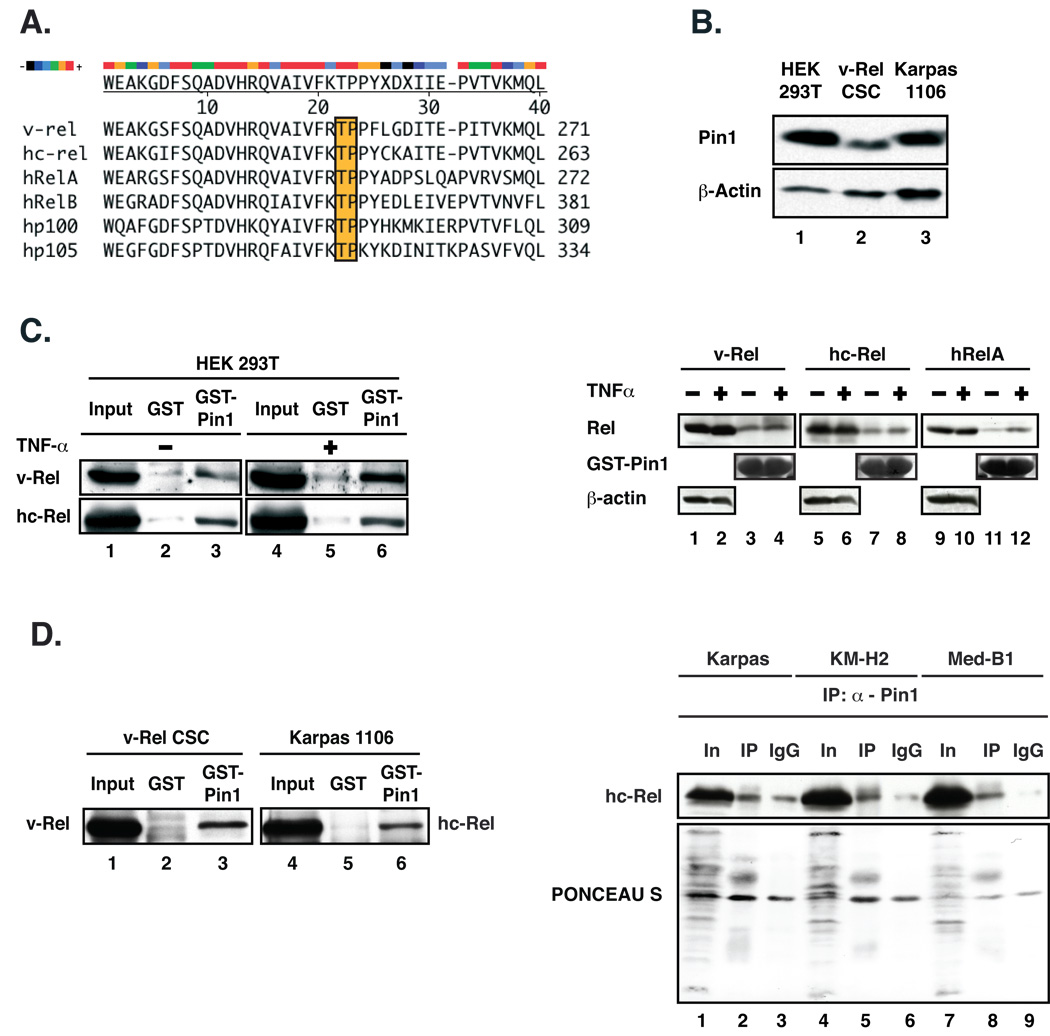
Human RelA was shown to interact with Pin1 via the Thr254-Pro motif in its Rel-homology domain (RHD) (12). Sequence alignment revealed conservation of this motif among vertebrate Rel/NF-κB family members. N-terminal sequences flanking this motif share a high degree of similarity and are in some cases identical, whereas those flanking its C-terminal end are significantly more divergent (Fig. 1A). Since overexpression of the Rel proteins can malignantly transform primary lymphocytes in vitro and induce fatal leukemia/lymphoma and mammary adenocarcinomas in animal models, unlike RelA (26, 28–31), we directly addressed the role of Pin1 in Rel/NF-κB-mediated oncogenesis by investigating its effects on the transforming activity of the viral (v-Rel) and cellular human c-Rel (hc-Rel) proteins.
Figure 1. The c-Rel and v-Rel subunits of NFκB associate with Pin1.
(A) Sequence alignment of the vertebrate Rel/NF-κB family proteins, highlighting conservation of N-terminal sequences flanking the Thr254-Pro Pin1 recognition motif in RelA, whereas sequences flanking its C-terminal end are more divergent. (B) Immunoblots showing endogenous expression of Pin1 in 293T cells, v-Rel-transformed CSC and in the primary mediastinal B cell lymphoma cell line Karpas 1106. The blot was probed with anti-Pin1 and reprobed for actin. (C) Left panel. Pull-down of v-Rel or hc-Rel in extracts from transiently transfected 293T cells with GST-Pin1 or GST as control, followed by immunoblotting with anti-Rel. Where indicated, cells were treated with hTNFα prior to harvest (lanes 4–6). Right panel. Pull-down of v-Rel, hc-Rel or hRelA transfected in 293T cells with GST-Pin1, followed by immunoblotting with anti-Rel. Where indicated, cells were treated with hTNFα. Input (1/10 of lysate; 1–2, 5–6 and 9–10). The blot was reprobed for actin and Pin1. (D) Left panel. Pull-down of endogenous v-Rel from v-Rel-transformed CSC (lanes 1–3) or hc-Rel from human lymphoma-derived Karpas 1106 cells (lanes 4–6) with GST-Pin1 or GST, followed by immunoblotting with anti-Rel (v-Rel: #1691 and hc-Rel: #265). Right panel. Co-immunoprecipitation of endogenous hc-Rel with Pin1 in extracts from human lymphoma cell lines using anti-Pin1, followed by immunoblotting with anti-hc-Rel. Input (1/10 of lysate). The membrane was stained with Ponceau S (bottom panel).
We used GST-pull downs to determine if Pin1 could engage in physical interaction with Rel proteins. As expected Pin1 is ubiquitously expressed in human 293T cells, in primary chicken spleen cells (CSC) transformed by v-Rel, and in human lymphoma Karpas 1106 cells in which NF-κB is markedly activated due to nuclear accumulation of hc-Rel (32) (Fig. 1B). GST-Pin1 interacted both with v-Rel and its cellular homologue hc-Rel in human 293T cells, compared to the GST control (Fig. 1C left panel, lane 3 vs. 2). Interestingly while association of RelA with Pin1 depends on cell stimulation with cytokine TNFα that triggers phosphorylation and activation of RelA (12), TNFα treatment had little or no effect on the interaction of either v-Rel or hc-Rel with GST-Pin1 (Fig. 1C left panel, lane 6 vs. 3; right panel, lanes 4, 8 12 vs. 3, 7, 11). GST-Pin1 also interacted with endogenous v-Rel from v-Rel-transformed chicken spleen cells (CSC), and with endogenous hc-Rel in extracts from the human lymphoma cell line Karpas 1106 (Fig. 1D left panel, lanes 3, 6 vs. 2, 5). Importantly, coimmunoprecipitation assays verified association between endogenous Pin1 and endogenous hc-Rel in lymphoma-derived cell lines, particularly evident in KM-H2 and MedB-1 cells (Fig. 1D right panel, lanes 2 vs. 3, 5 vs. 6 and 8 vs. 9). These results demonstrate that v-Rel and hc-Rel can specifically associate with Pin1.
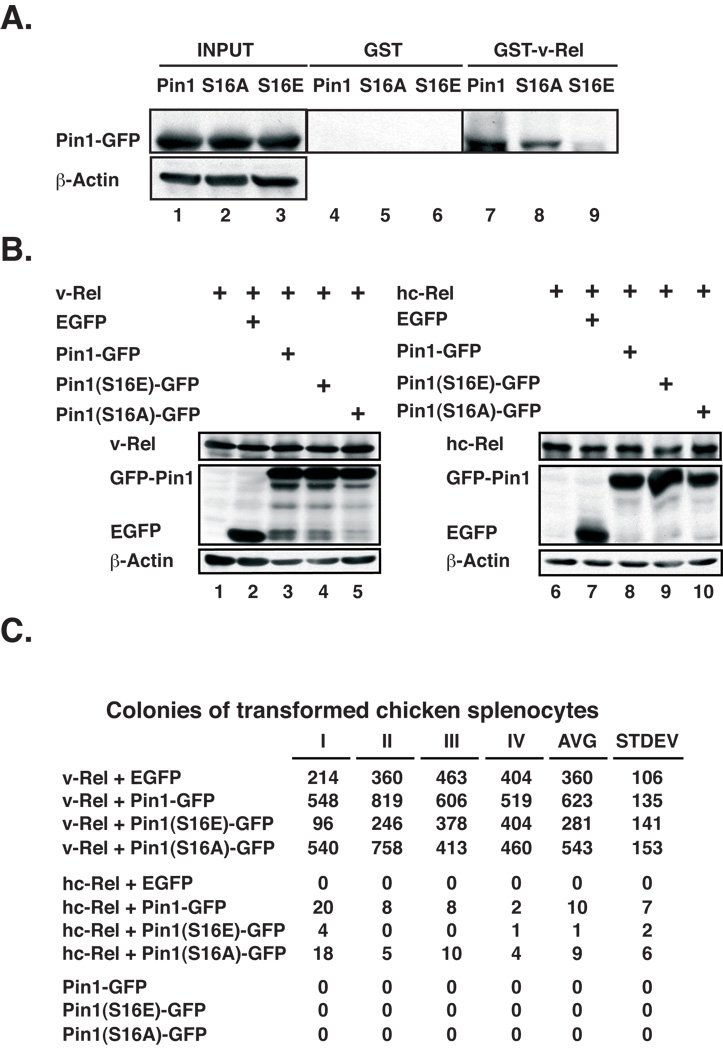
Conversely Pin1 transfected in 293T cells was pulled-down with GST-v-Rel, confirming their interaction (Fig. 2A, lane 7 vs. 4). In contrast, a Pin1 mutant with a serine to glutamate substitution at position 16 (S16E) in the N-terminal WW domain that interacts with pSer/Thr-Pro motifs showed significantly less interaction with GST-v-Rel (lane 9). This is consistent this mutant’s failure to efficiently associate with RelA (12). A mutant with substitution of serine 16 to alanine (S16A) retained the ability to interact with GST-v-Rel, although with somewhat reduced efficiency (Fig. 2A, lane 2). Consistent with previous findings that Pin1(S16A) constitutively bind to Pin1 substrates, in contrast to S16E (33), our data show that v-Rel preferentially associates with Pin1 and Pin1(S16A).
Figure 2. Pin1 markedly increases the transforming activity of Rel proteins.
(A) Pin1 and mutant S16A associate with v-Rel in GST pull down assays significantly more than mutant S16E. Pull downs were carried out as in Figure 1C with extracts from 293T cells transfected with Pin1-GFP, Pin1(S16E)-GFP or Pin1(S16A)-GFP and GST-v-Rel or GST, followed by western blot with anti-Pin1. Input (1/10 of lysate). (B) Immunoblots showing efficient co-expression of v-Rel or hc-Rel with GFP-tagged Pin1, Pin1(S16E), Pin1(S16A) or GFP control in CEFs used as the source of virus to infect primary CSCs. The blots were probed with antibodies to v-Rel (#2716) or hc-Rel (#265) (top panels) or GFP (middle), and reprobed for actin as control (bottom). (C) Co-expression of Pin1 or mutant S16A markedly increases the transforming activity of hc-Rel and also enhances that of v-Rel in primary CSC, in contrast to Pin1(S16E), as detected by colony formation in soft agar. The results of four independent experiments are shown along with averages and standard deviations.
Pin1 markedly enhances the transforming activities of hc-Rel and v-Rel
We investigated Pin1’s ability to modulate the oncogenic activity of Rel proteins in primary lymphocytes, using a bicistronic retroviral vector to co-express v-Rel or hc-Rel along with GFP-tagged Pin1 or mutants S16A and S16E. Western blots confirmed equivalent expression of all proteins (Fig. 2B). Co-expression of Pin1-GFP significantly increased the already potent transforming activity of v-Rel in primary lymphocytes nearly 2-fold, as seen by colony formation in soft agar (Fig. 2C). In contrast Pin1(S16E) failed to do so, consistent with its very weak association with v-Rel (Fig. 2C). Coexpression of Pin1(S16A) generally enhanced the efficiency of v-Rel-mediated transformation, albeit less than wild type Pin1. Importantly, the effects of Pin1 on lymphocyte transformation by the cellular hc-Rel protein were even more pronounced, despite variability in the efficiency of individual transformation assays that is commonly observed in these assays with primary chicken splenocytes, particularly with more weakly transforming proteins, like hc-Rel. While hc-Rel co-expressed with GFP failed to transform cells, Pin1 and Pin1(S16A) markedly boosted hc-Rel’s transforming activity, yielding up to twenty colonies that could be picked and propagated in liquid culture, in contrast to Pin1(S16E) (Fig. 2C). Enhancement of Rel-mediated transformation did not result from Pin1 acting as an oncogene, as neither Pin1-GFP nor its mutants could transform splenocytes on their own (Fig. 2C). These results demonstrate that Pin1 markedly increases Rel-mediated transformation. Given the weak transforming activity of hc-Rel (26, 31), these data suggest that Pin1 may play a significant role in enhancing hc-Rel’s oncogenic activity.
Pin1 increases the nuclear distribution of v-Rel and hc-Rel
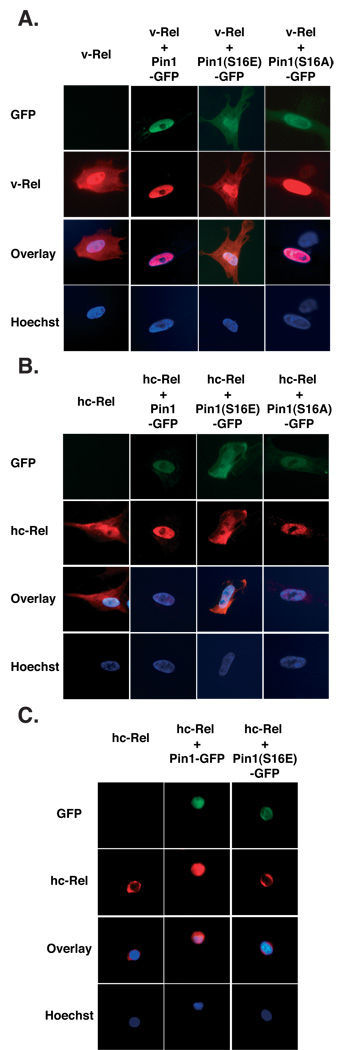
Pin1’s ability to enhance the transforming activity of hc-Rel and v-Rel led us to investigate its effect on their subcellular localization. Previous studies showed that Pin1 and Pin1(S16A) localize to the nucleus, whereas Pin1(S16E) is diffused throughout the cell (33). Immunofluorescence showed that Pin1-GFP localizes to the nucleus of infected CEFs and its coexpression with either v-Rel or hc-Rel prompted their relocalization to the nucleus (Fig. 3A, B). This is consistent with Pin1’s ability to enhance their transforming potential. In contrast Pin1(S16E) expressed alone was found in both the cytoplasm and the nucleus, and was unable to induce nuclear accumulation of v-Rel or hc-Rel. Pin1(S16A) was primarily nuclear, with low-level residual localization to the cytoplasm, and effectively prompted nuclear accumulation of v-Rel and hc-Rel in infected CEFs. This agrees with the capacity of Pin1(S16A) to retain association with Rel proteins and enhance their transforming activity. These results indicate that Pin1 can promote nuclear localization of Rel factors and that this is correlated with enhanced Rel transforming activity.
Figure 3. Pin1 and Pin1(S16A) promote relocalization of v-Rel and hc-Rel to the nucleus in the absence of stimuli.
(A) Immunofluorescence showing that co-expression of Pin1 or Pin1(S16A) provokes nuclear translocation of v-Rel in CEFs infected with bicistronic retroviral vectors, in contrast to Pin1(S16E). (B) Pin1 or Pin1(S16A) similarly induce accumulation of hc-Rel in the nucleus of infected CEFs. (C) Immunofluorescence showing relocalization of hc-Rel to the nucleus of transformed CSCs coexpressing hc-Rel proteins with Pin1, but not in those co-expressing Pin1(S16E).
We investigated the relevance of these observations in the physiological setting of transformed primary lymphocytes by probing the effects of Pin1 or its mutants on the subcellular localization of hc-Rel in CSCs. As anticipated, hc-Rel was predominantly cytoplasmic in primary splenocytes transformed by hc-Rel alone, as previously reported (Fig. 3C left panel) (26, 31). However hc-Rel was predominantly nuclear in splenocytes transformed by its co-expression with Pin1, consistent with their increased transforming efficiency. In contrast Pin1(S16E), that showed very weak association with Rel, was unable to promote nuclear localization of hc-Rel, in agreement with its inability to enhance hc-Rel-mediated transformation. These data suggest that Rel proteins are substrates for the peptidyl-prolyl isomerase activity of Pin1, and that Pin1 may significantly enhance their oncogenic activity by changing the equilibrium between their localization to the nucleus vs. cytoplasm.
Juglone induces cytoplasmic relocalization of endogenous hc-Rel in lymphoma cells, coincident with lymphoma cell death and/or growth inhibition
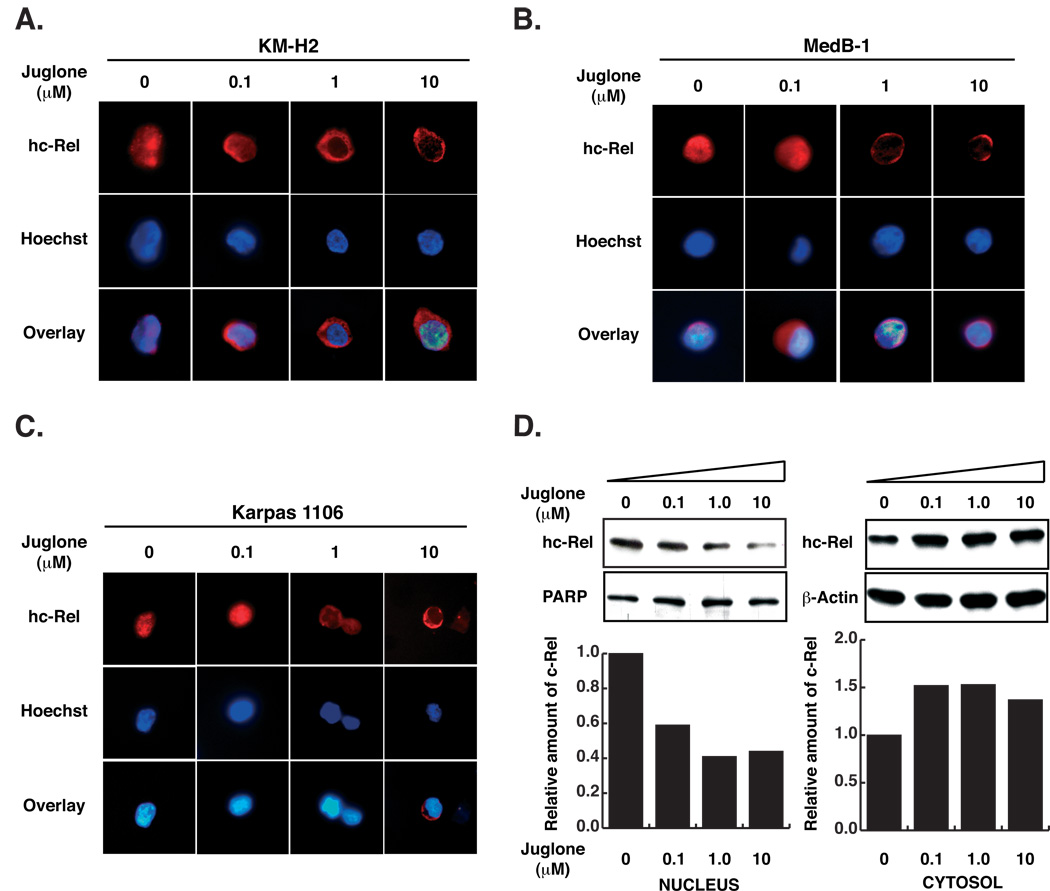
Since Pin1 significantly increased the transforming activity of hc-Rel, we asked if endogenous Pin1 plays a role in the subcellular distribution of endogenous hc-Rel in human lymphoma-derived cells, since Pin1 is highly expressed in lymphoma cells in which Rel/NF-κB is activated (Fig. 1B, lane 3 and Supplementary Fig. 1). Juglone (5-hydroxy-1, 4-naphthoquinone) is a natural and irreversible inhibitor of the parvulin family of PPIases that blocks interaction of Pin1 with its substrates by covalently modifying its only two cysteine residues (Cys 57, 113) (34). Juglone is frequently used to study the relevance of Pin1 function in vivo, since it can give rise to a similar phenotype as Pin1 dominant negative mutants or Pin1 knockdown (35–37). We analyzed juglone’s effects on the distribution of endogenous hc-Rel in human lymphoma cell lines in which Rel/NF-κB is constitutively activated including KM-H2, MedB-1 and Karpas 1106 cells (32, 38, 39). Treatment with increasing concentrations of juglone for 2 hours provoked rapid redistribution of predominantly nuclear hc-Rel to the cytoplasm in a dose-dependent manner, compared to DMSO control (Fig. 4A–C). hc-Rel was efficiently excluded from the nucleus with 1µM juglone, and its cytoplasmic relocalization was detected with as little as 0.1 µM juglone in KM-H2 cells. Fractionation of Karpas 1106 cells confirmed the significant redistribution of hc-Rel upon treatment with juglone in a dose dependent manner (Fig. 4D). These data suggest that Pin1 may play a role in modulating the localization of endogenous hc-Rel in human lymphoma cells.
Figure 4. Juglone induces cytoplasmic relocalization of endogenous hc-Rel in human lymphoma cells.
Immunofluorescence analysis of endogenous hc-Rel subcellular localization in presence of increasing concentrations of Juglone (0.1, 1 or 10 µM) for 2hrs in human lymphoma-derived KM-H2 (A), MedB-1 (B) or Karpas 1106 (C) cell lines, compared to DMSO control, using anti-hc-Rel and a rhodamine-conjugated secondary antibody. Nuclei were stained with Hoechst 33258 dye. (D) Fractionation of Karpas 1106 cells treated with juglone for 2hrs confirmed rapid redistribution of endogenous hc-Rel from the nucleus to the cytoplasm. The blots were probed with anti-hc-Rel and reprobed for uncleaved poly-ADP-ribose polymerase (PARP; ~115 kDa) or actin as nuclear and cytosolic markers. Histograms show quantitation of hc-Rel band intensities in the nucleus and cytoplasm normalized to those of PARP and actin.
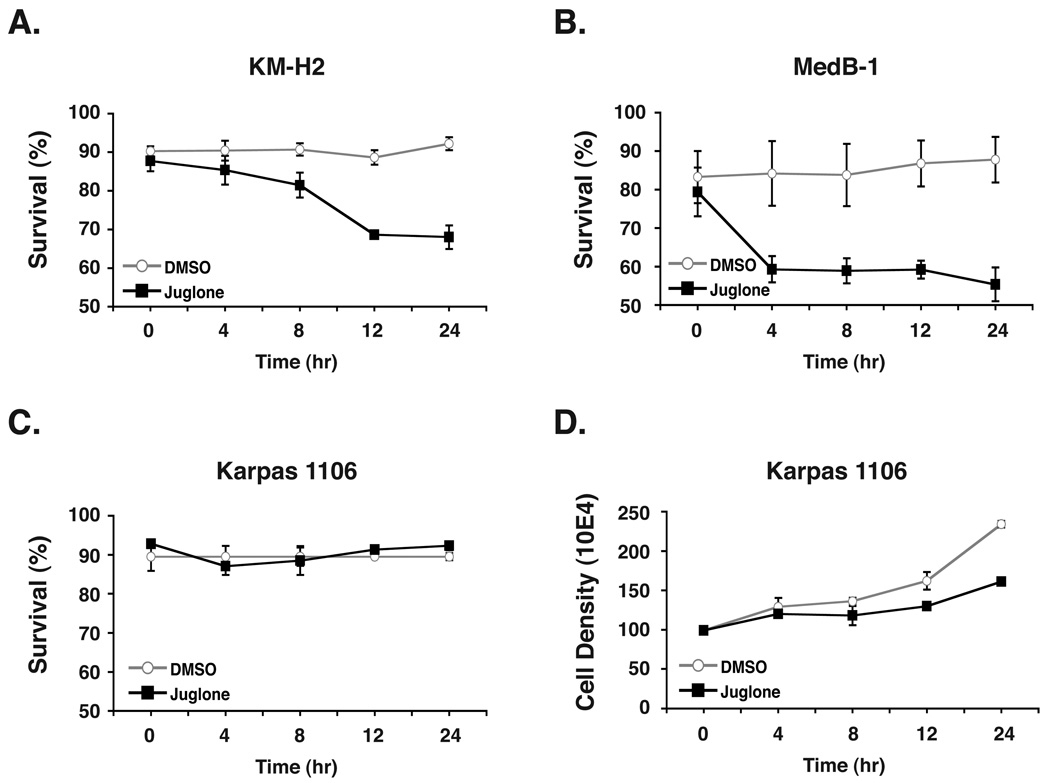
Given the important implications of our finding that Pin1 can markedly enhance the transforming activity of hc-Rel, we investigated the impact of inhibiting Pin1 function with juglone on the biological properties of human lymphoma cells in which Rel/NF-κB is constitutively activated. We found that significant detrimental effects on tumor cell survival and/or proliferation accompanied the rapid relocalization of hc-Rel to the cytoplasm of human lymphoma cell lines treated with juglone. Treatment of either KM-H2 or MedB-1 cells with 0.1 µM juglone, the lowest concentration that provoked cytoplasmic relocalization of hc-Rel within 2 hrs of treatment, triggered 20–30% cell death within 4 hrs of treatment compared to DMSO (Fig. 5A and B). While there was no significant difference in the survival for Karpas 1106 cells subjected to the same treatment (Fig. 5C), these cells displayed a noticeable decrease in growth rate starting at 6 hrs post-treatment. By 24 hrs, the growth of Karpas 1106 cells was severely blunted compared to those treated with DMSO (Fig. 5D). These data indicate that juglone restricts hyperproliferation and compromises the survival of tumor-derived cells.
Figure 5. Juglone interferes with cell survival and/or growth in Rel-dependent human lymphoma cells.
Treatment of human lymphoma-derived KM-H2 (A) or MedB-1 (B) cells with 0.1 µM juglone during a time course induces cell death compared to the DMSO control, as determined by live cell count following Trypan blue exclusion staining. Whereas this treatment did not affect the survival of Karpas 1106 cells (C), juglone significantly interfered with Karpas 1106 cell growth compared to the control (D). The average of three independent experiments is shown.
Pin1 knockdown prompts cytoplasmic relocalization of hc-Rel, interferes with lymphoma cell growth and suppresses Rel/NF-κB-dependent gene expression
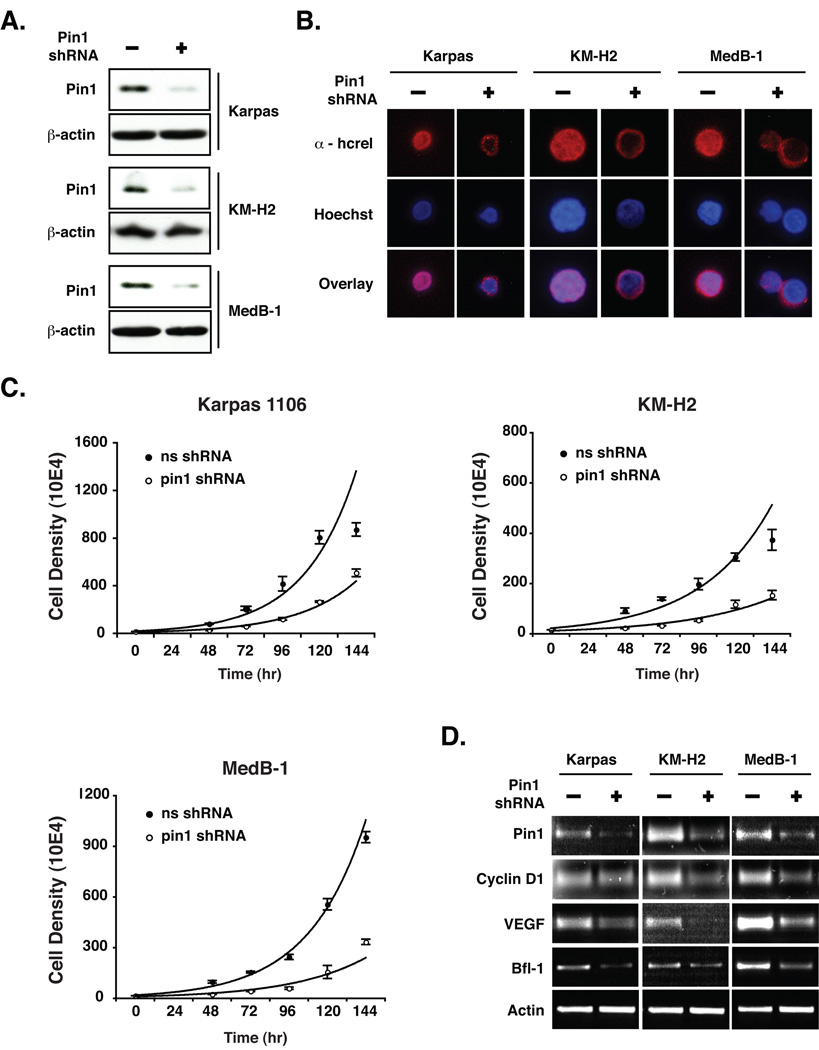
Since pharmacological inhibition of Pin1 with juglone could have some indirect and/or off-target effects, we used a Pin1-specific shRNA to verify our hypothesis. Nucleofection of Pin1 shRNA significantly reduced Pin1 protein levels at 96 hrs in all lymphoma cell lines tested, compared to the non-targeting shRNA control (Fig. 6A, lane 2 vs. 1). Consistent with our findings with juglone, Pin1 knockdown prompted redistribution of endogenous hc-Rel to the cytoplasm compared to the shRNA control, as seen by immunofluorescence (Fig. 6B). Furthermore Pin1 shRNA significantly interfered with the proliferation of all lymphoma cell lines compared to the shRNA control (Fig. 6C), but did not affect their viability (data not shown). This indicates that juglone has some off-target effects. Importantly, Pin1 knockdown with shRNA significantly reduced the expression of known endogenous Rel/NF-κB target genes including cyclin D1, VEGF and Bfl-1 (Fig 6D). Together with our finding that Pin1 promotes lymphocyte transformation by v-Rel and hc-Rel, these results demonstrate that Pin1 is an important modulator of Rel/NF-κB function in transcription and oncogenesis.
Figure 6. Pin1 knockdown prompts cytoplasmic relocalization of hc-Rel, interferes with lymphoma cell growth and suppresses expression of Rel/NF-κB target genes.
(A) Immunoblot of lymphoma-derived Karpas 1106, KM-H2 or MedB-1 cells harvested at 96hrs post-nucleofection with Pin1 shRNA or a scrambled shRNA control, using monoclonal anti-Pin1 or anti-actin. (B) Immunofluorescence of endogenous hc-Rel subcellular localization in lymphoma cells lines transfected with Pin1 shRNA or shRNA control, using anti-hc-Rel and a rhodamine-conjugated secondary antibody. Nuclei were stained with Hoechst 33258 dye. (C) Pin1 knockdown with shRNA (○) significantly delays the growth of lymphoma cell lines, compared to a scrambled shRNA control (●). The average of three independent experiments is shown. (D) RNA was analyzed by RT-PCR at 48hrs post-nucleofection with Pin1 shRNA or a scrambled shRNA control using primers specific for the Rel/NF-κB-regulated genes cyclin D1, VEGF or Bfl-1. Actin mRNA was amplified as a control.
DISCUSSION
Accumulating evidence shows that Pin1 is upregulated in many human cancers including breast, prostate, lung, hepatic, cervical and colon cancer, and that increased expression of Pin1 is correlated with poor prognosis in prostate cancer (13, 40–42). Since ablation of Pin1 could prevent development of mammary carcinoma induced by oncogenic Neu or Ras in mice, this suggests an important role for Pin1 in cancer (43). Others suggested that it plays a tumor suppressor role (25). Indeed loss of Pin1 can reportedly lead to deregulation of cyclin E and c-Myc thereby increasing genomic instability, and is believed to sensitize cells to oncogenic transformation (44, 45). Here we provide evidence that Pin1 functionally associates with the oncogenic Rel subunits of NF-κB and that this interaction plays an important role in promoting the nuclear localization and the transcriptional, pro-proliferative and transforming properties of Rel proteins. We show that Pin1 markedly enhances the weak transforming activity of hc-Rel in primary lymphocytes. We also demonstrate that inhibition of Pin1 severely compromises proliferation of Rel/NF-κB dependent human lymphoma cells and is accompanied by suppression of Rel/NF-κB-dependent gene expression. These findings are consistent with accumulating evidence supporting an important role for Pin1 deregulation during tumorigenesis and the pro-proliferative capacity of tumor cells (46), and emphasize an important role in Rel’s oncogenic activity. This may be particularly relevant since upregulation of Pin1 is seen in many human leukemia and lymphoma specimens in which Rel/NF-κB is known to be constitutively activated (Supplementary Fig. 1).
While Pin1 failed to transform primary lymphocytes on its own, it significantly enhanced Rel’s transforming activity dependent on its ability to alter the dynamics of Rel protein nuclear import/export to tip the equilibrium in favor of increased nuclear accumulation. This most likely results from Pin1’s ability to induce proline isomerization, thereby preventing Rel inhibition by IκBα (12).In this regard it is not surprising that Pin1 had a more dramatic effect in enhancing the transforming activity of the weakly transforming hc-Rel protein compared to the potently oncogenic v-Rel, since v-Rel is known to be significantly more resistant to inhibition by IκBα than c-Rel (47). Upregulation of Pin1 may thus emerge as novel means to enhance the contribution of hc-Rel in human cancer by helping to dampen its negative feedback inhibition. Additionally since the 14-3-3 proteins can facilitate efficient nuclear export of IκBα-p65 complexes by binding to both RelA/p65 and IκBα, and 14-3-3 binds to RelA (amino acids 278–283) in close proximity of the Thr254-Pro recognition motif for Pin1 (48), this raises the possibility that Pin1 might also preclude export of NF-κB/IκB complexes by interfering with NF-κB regulation by 14-3-3. In both scenarios, altered nucleo-cytoplasmic shuttling of Rel/NF-κB following upregulation Pin1 would contribute to sustained activation of Rel/NF-κB signaling and oncogenesis.
The N-terminal sequences that flank RelA’s Thr254-Pro motif are highly conserved among Rel/NF-κB family members, but greater variability is seen in those that flank its C-terminus. Although the kinase responsible for phosphorylation of Thr254 in RelA’s Thr-Pro motif has yet to be identified, efficient association of RelA with Pin1 is dependent on cell stimulation with cytokine TNFα (12 and Fig 1C right panel). In contrast, TNF had little effect on the interaction of v-Rel or hc-Rel with Pin1 (Fig 1C). This, together with the sequence divergence at the C-terminus of the Pin1 recognition motif in Rel/NF-κB proteins, suggests that different regulatory mechanisms may dictate interaction of Pin-1 with Rel and RelA proteins. Further studies will be needed to address this issue.
Our finding that Pin1 knockdown compromised the growth of Rel-dependent lymphoma cells and the expression Rel-dependent target genes points to Pin1 as a possible therapeutic target in these and other Rel-dependent tumors. However since pharmacological inhibition of Pin1 with juglone compromised both the growth and the survival of lymphoma cells, it appears that juglone shows some off-target effects. Indeed juglone has been shown to affect the activity of RNA polymerase II through Pin1 and to also prevent postmitotic protein dephosphorylation (49, 50), although its effects on the growth and survival of lymphoma cells was observed at a concentration 10- to 75-fold lower than those reported to inhibit the activity of RNA polymerase II (49) or to block dephosphorylation of mitotic phosphoproteins (50). Nevertheless our results with Pin1 shRNA revealed off-target effects for juglone, and suggest that more specific inhibitors of Pin1 will need to be identified for possible therapeutic application. Overall, our studies demonstrate that Pin1 plays a crucial role in promoting Rel-mediated transcription and transformation, and that inhibition of Pin1 can significantly compromise the proliferation of Rel-dependent lymphoma cells. Thus Pin1 may be an attractive therapeutic target in Rel/NF-κB-dependent leukemia/lymphomas.
Supplementary Material
ACKNOWLEDGEMENTS
Grant Support: This work was supported by Public Health Service grant CA054999 from the National Cancer Institute to CG.
We thank Drs. Abraham Karpas (Univ. of Cambridge, Cambridge, United Kingdom), Silke Bruderlein and Peter Moller (Institute of Pathology, Ulm, Germany) for the gifts of lymphoma cell lines, and J Dutta, MJ Simmons, G Xiao, K Madura and E White for fruitful discussions and suggestions during the course of this work.
Footnotes
Contributors’ statement: All contributors declare that they have no conflict of interest.
REFERENCES
- 1.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Campbell KJ, Rocha S, Perkins ND. Active repression of antiapoptotic gene expression by RelA(p65) NF-kappa B. Mol Cell. 2004;13:853–865. doi: 10.1016/s1097-2765(04)00131-5. [DOI] [PubMed] [Google Scholar]
- 3.Gupta N, Delrow J, Drawid A, Sengupta AM, Fan G, Gelinas C. Repression of B-cell linker (BLNK) and B-cell adaptor for phosphoinositide 3-kinase (BCAP) is important for lymphocyte transformation by rel proteins. Cancer Res. 2008;68:808–814. doi: 10.1158/0008-5472.CAN-07-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majid SM, Liss AS, You M, Bose HR. The suppression of SH3BGRL is important for v-Rel-mediated transformation. Oncogene. 2006;25:756–768. doi: 10.1038/sj.onc.1209107. [DOI] [PubMed] [Google Scholar]
- 5.Dutta J, Fan Y, Gupta N, Fan G, Gelinas C. Current insights into the regulation of programmed cell death by NF-κB. Oncogene. 2006;25:6800–6816. doi: 10.1038/sj.onc.1209938. [DOI] [PubMed] [Google Scholar]
- 6.Rocha S, Martin AM, Meek DW, Perkins ND. p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NF-kappaB subunit with histone deacetylase 1. Mol Cell Biol. 2003;23:4713–4727. doi: 10.1128/MCB.23.13.4713-4727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: balancing life and death--a new approach to cancer therapy. J Clin Invest. 2005;115:2625–2632. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 9.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 10.Yaffe MB, Schutkowski M, Shen M, et al. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 11.Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 12.Ryo A, Suizu F, Yoshida Y, et al. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 2003;12:1413–1426. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 13.Wulf GM, Ryo A, Wulf GG, et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 2001;20:3459–3472. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sovak MA, Bellas RE, Kim DW, et al. Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. J Clin Invest. 1997;100:2952–2960. doi: 10.1172/JCI119848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS., Jr Selective activation of NF-κB subunits in human breast cancer: potential roles for NF-κB2/p52 and for Bcl-3. Oncogene. 2000;19:1123–1131. doi: 10.1038/sj.onc.1203412. [DOI] [PubMed] [Google Scholar]
- 16.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 18.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 20.Haslinger C, Schweifer N, Stilgenbauer S, et al. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J Clin Oncol. 2004;22:3937–3949. doi: 10.1200/JCO.2004.12.133. [DOI] [PubMed] [Google Scholar]
- 21.Zhan F, Barlogie B, Arzoumanian V, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109:1692–1700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kern C, Cornuel JF, Billard C, et al. Involvement of BAFF and APRIL in the resistance to apoptosis of B-CLL through an autocrine pathway. Blood. 2004;103:679–688. doi: 10.1182/blood-2003-02-0540. [DOI] [PubMed] [Google Scholar]
- 23.Pham LV, Tamayo AT, Yoshimura LC, Lo P, Ford RJ. Inhibition of constitutive NF-kappa B activation in mantle cell lymphoma B cells leads to induction of cell cycle arrest and apoptosis. J Immunol. 2003;171:88–95. doi: 10.4049/jimmunol.171.1.88. [DOI] [PubMed] [Google Scholar]
- 24.Hideshima T, Chauhan D, Richardson P, et al. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem. 2002;277:16639–16647. doi: 10.1074/jbc.M200360200. [DOI] [PubMed] [Google Scholar]
- 25.Yeh ES, Means AR. PIN1, the cell cycle and cancer. Nat Rev Cancer. 2007;7:381–388. doi: 10.1038/nrc2107. [DOI] [PubMed] [Google Scholar]
- 26.Fan Y, Rayet B, Gélinas C. Divergent C-terminal transactivation domains of Rel/NF-κB proteins are critical determinants of their oncogenic potential in lymphocytes. Oncogene. 2004;23:1030–1042. doi: 10.1038/sj.onc.1207221. [DOI] [PubMed] [Google Scholar]
- 27.Nacheva E, Dyer MJ, Metivier C, et al. B-cell non-Hodgkin's lymphoma cell line (Karpas 1106) with complex translocation involving 18q21.3 but lacking BCL2 rearrangement and expression. Blood. 1994;84:3422–3428. [PubMed] [Google Scholar]
- 28.Zhang JY, Olson W, Ewert D, Bargmann W, Bose HR., Jr The v-rel oncogene of avian reticuloendotheliosis virus transforms immature and mature lymphoid cells of the B cell lineage in vitro. Virology. 1991;183:457–466. doi: 10.1016/0042-6822(91)90975-h. [DOI] [PubMed] [Google Scholar]
- 29.Gilmore TD, Cormier C, Jean-Jacques J, Gapuzan ME. Malignant transformation of primary chicken spleen cells by human transcription factor c-Rel. Oncogene. 2001;20:7098–7103. doi: 10.1038/sj.onc.1204898. [DOI] [PubMed] [Google Scholar]
- 30.Romieu-Mourez R, Kim DW, Shin SM, et al. Mouse mammary tumor virus c-rel transgenic mice develop mammary tumors. Mol Cell Biol. 2003;23:5738–5754. doi: 10.1128/MCB.23.16.5738-5754.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starczynowski DT, Reynolds JG, Gilmore TD. Deletion of either C-terminal transactivation subdomain enhances the in vitro transforming activity of human transcription factor REL in chicken spleen cells. Oncogene. 2003;22:6928–6936. doi: 10.1038/sj.onc.1206801. [DOI] [PubMed] [Google Scholar]
- 32.Feuerhake F, Kutok JL, Monti S, et al. NFkappaB activity, function, and target-gene signatures in primary mediastinal large B-cell lymphoma and diffuse large B-cell lymphoma subtypes. Blood. 2005;106:1392–1399. doi: 10.1182/blood-2004-12-4901. [DOI] [PubMed] [Google Scholar]
- 33.Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem. 2002;277:2381–2384. doi: 10.1074/jbc.C100228200. [DOI] [PubMed] [Google Scholar]
- 34.Hennig L, Christner C, Kipping M, et al. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry. 1998;37:5953–5960. doi: 10.1021/bi973162p. [DOI] [PubMed] [Google Scholar]
- 35.Galas MC, Dourlen P, Begard S, et al. The peptidylprolyl cis/trans-isomerase Pin1 modulates stress-induced dephosphorylation of Tau in neurons. Implication in a pathological mechanism related to Alzheimer disease. J Biol Chem. 2006;281:19296–19304. doi: 10.1074/jbc.M601849200. [DOI] [PubMed] [Google Scholar]
- 36.Esnault S, Braun RK, Shen ZJ, et al. Pin1 modulates the type 1 immune response. PLoS ONE. 2007;2:e226. doi: 10.1371/journal.pone.0000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kesavapany S, Patel V, Zheng YL, et al. Inhibition of Pin1 reduces glutamate-induced perikaryal accumulation of phosphorylated neurofilament-H in neurons. Mol Biol Cell. 2007;18:3645–3655. doi: 10.1091/mbc.E07-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weniger MA, Gesk S, Ehrlich S, et al. Gains of REL in primary mediastinal B-cell lymphoma coincide with nuclear accumulation of REL protein. Genes Chromosomes Cancer. 2007;46:406–415. doi: 10.1002/gcc.20420. [DOI] [PubMed] [Google Scholar]
- 39.Hinz M, Loser P, Mathas S, Krappmann D, Dorken B, Scheidereit C. Constitutive NF-kappaB maintains high expression of a characteristic gene network, including CD40, CD86, and a set of antiapoptotic genes in Hodgkin/Reed-Sternberg cells. Blood. 2001;97:2798–2807. doi: 10.1182/blood.v97.9.2798. [DOI] [PubMed] [Google Scholar]
- 40.Pang RW, Lee TK, Man K, et al. PIN1 expression contributes to hepatic carcinogenesis. J Pathol. 2006;210:19–25. doi: 10.1002/path.2024. [DOI] [PubMed] [Google Scholar]
- 41.Pang R, Yuen J, Yuen MF, et al. PIN1 overexpression and beta-catenin gene mutations are distinct oncogenic events in human hepatocellular carcinoma. Oncogene. 2004;23:4182–4186. doi: 10.1038/sj.onc.1207493. [DOI] [PubMed] [Google Scholar]
- 42.Ayala G, Wang D, Wulf G, et al. The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer Res. 2003;63:6244–6251. [PubMed] [Google Scholar]
- 43.Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. 2004;23:3397–3407. doi: 10.1038/sj.emboj.7600323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeh E, Cunningham M, Arnold H, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 45.Yeh ES, Lew BO, Means AR. The loss of PIN1 deregulates cyclin E and sensitizes mouse embryo fibroblasts to genomic instability. J Biol Chem. 2006;281:241–251. doi: 10.1074/jbc.M505770200. [DOI] [PubMed] [Google Scholar]
- 46.Finn G, Lu KP. Phosphorylation-specific prolyl isomerase Pin1 as a new diagnostic and therapeutic target for cancer. Curr Cancer Drug Targets. 2008;8:223–229. doi: 10.2174/156800908784293622. [DOI] [PubMed] [Google Scholar]
- 47.Sachdev S, Hannink M. Loss of IkBa-mediated control over nuclear import and DNA binding enables oncogenic activation of c-Rel. Mol Cell Biol. 1998;18:5445–5456. doi: 10.1128/mcb.18.9.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguilera C, Fernandez-Majada V, Ingles-Esteve J, Rodilla V, Bigas A, Espinosa L. Efficient nuclear export of p65-IkappaBalpha complexes requires 14-3-3 proteins. J Cell Sci. 2006;119:3695–3704. doi: 10.1242/jcs.03086. [DOI] [PubMed] [Google Scholar]
- 49.Xu YX, Hirose Y, Zhou XZ, Lu KP, Manley JL. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 2003;17:2765–2776. doi: 10.1101/gad.1135503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fila C, Metz C, van der Sluijs P. Juglone inactivates cysteine-rich proteins required for progression through mitosis. J Biol Chem. 2008;283:21714–21724. doi: 10.1074/jbc.M710264200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.