Abstract
We report a genome-wide association study of melanoma conducted by the GenoMEL consortium based on 317k tagging SNPs for 1650 genetically-enriched cases (from Europe and Australia) and 4336 controls and subsequent replication in 1149 genetically-enriched cases and 964 controls and a population-based case-control study of 1163 cases and 903 controls. The genome-wide screen identified five regions with genotyped or imputed SNPs reaching p < 5×10−7; three regions were replicated: 16q24 encompassing MC1R (overall p=2.54×10−27 for rs258322), 11q14-q21 encompassing TYR (p=2.41×10−14 for rs1393350) and 9p21 adjacent to MTAP and flanking CDKN2A (p=4.03×10−7 for rs7023329). MC1R and TYR are associated with pigmentation, freckling and cutaneous sun sensitivity, well-recognised melanoma risk factors, while the 9p21 locus is novel for common variants associated with melanoma. Despite wide variation in allele frequency, these genetic variants show notable homogeneity of effect across populations of European ancestry living at different latitudes and contribute independently to melanoma risk.
Cutaneous melanoma is almost entirely a disease of fair-skinned individuals. Increased risk for melanoma is associated with a family history of the disease1, pigmentation phenotypes, the number of melanocytic nevi2,3 and iatrogenic immuno-suppression4. Pigmentation risk factors include fair skin, blue or green eyes, blond or red hair, sun sensitivity or inability to tan5–8, each associated with an approximate doubling of risk. Variants of the melanocortin-1 receptor (MC1R) gene are associated with the combination of red hair, freckling and sun sensitivity, but MC1R may also have a non-pigmentation effect7–10. A detailed genetic screen of pigmentation loci found that tyrosinase (TYR) gene variants and a haplotype spanning the agouti signalling protein (ASIP) locus were also associated with melanoma11. Increased intermittent exposure to ultraviolet radiation, rather than chronic exposure, is thought to be responsible for the continued increase in incidence in many populations12.
The most common high-penetrance melanoma susceptibility gene (CDKN2A) maps to 9p2113. Germline CDKN2A mutations are carried by about 2% of all melanoma cases across populations14. The more cases of melanoma in a family, the more prevalent are CDKN2A germline mutations 15, which appear to be associated with increased numbers of naevi in at least some families16.
This study was carried out by the GenoMEL Consortium (see Supplementary Note), which focuses on genetic susceptibility to melanoma. The current genome-wide association study (GWAS) is based on population samples collected by GenoMEL participants to identify common genetic variants contributing to melanoma risk across populations of European ancestry living at different latitudes. In total, 10 GenoMEL groups contributed 1650 cases and 1065 controls of European ethnicity for this GWAS (Supplementary Table 1). Cases were selected in an effort to enrich for genetic susceptibility (see Methods). Genotyping results were also available from 1824 French controls held by Centre National de Génotypage (CNG) and 1447 UK controls from the 1958 Birth Cohort genotyped by the Wellcome Trust Case Control Consortium (WTCCC)17 (Supplementary Figure 1).
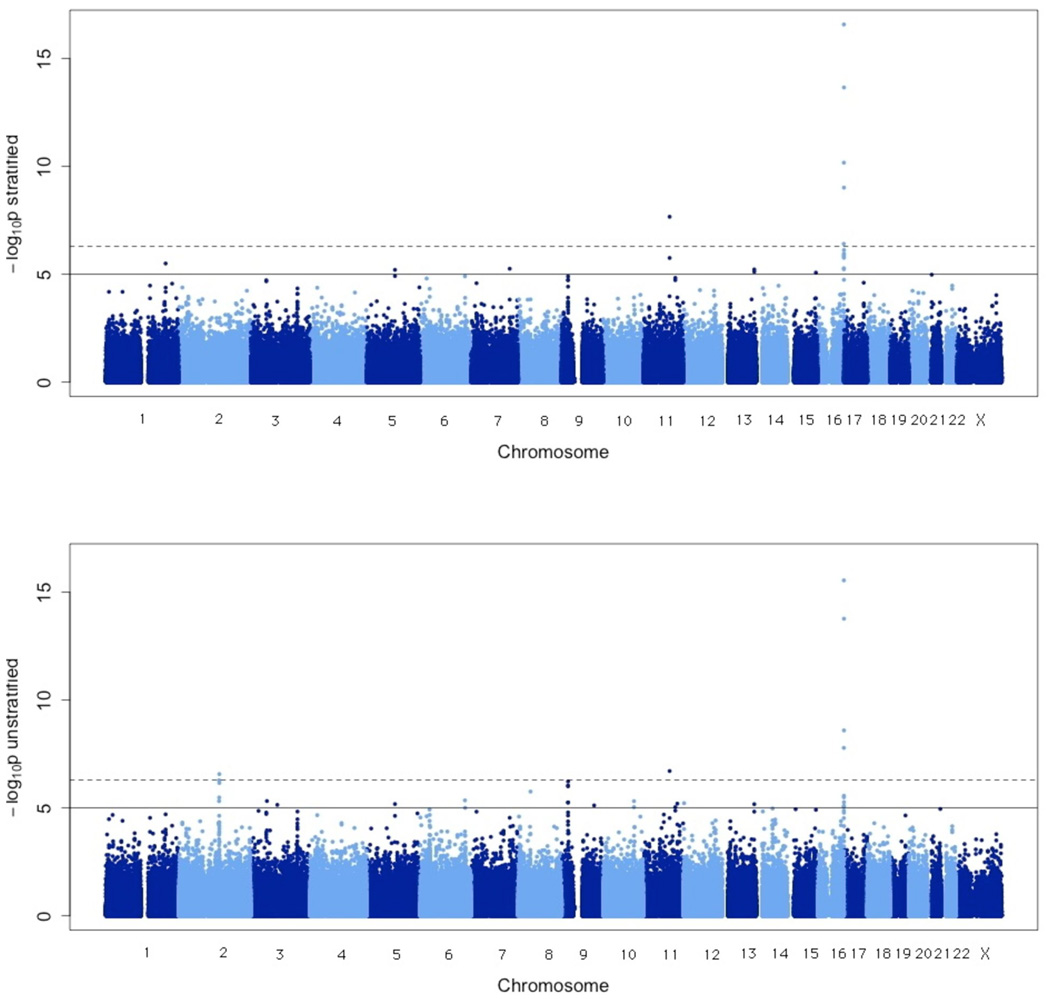
Following quality control and exclusion of persons of apparent non-European ancestry through principal components analysis (Supplementary Figure 2), case-control analyses were conducted both (i) stratified (the primary analysis) and (ii) unstratified by geographical region (Figure 1); the unstratified analysis should provide greater power for associations common to multiple sample sets, while the stratified analysis protects against population stratification but is likely conservative in the absence of stratification. To confirm the effectiveness of our methods for dealing with stratification we produced quantile-quantile plots (Supplementary Note). We selected regions with multiple SNPs (within 50kb) where each had a p-value less than 1O−5 from either the stratified or unstratified analysis and conducted imputation on these regions. This led to the identification of five regions on chromosomes 2, 5, 9,11 and 16 in which at least one imputed or genotyped SNP achieved genome-wide significance (p<5×10−7, Table 1, Supplementary Table 2, Figure 2). The chromosome 2 region showed strong evidence of association in the unstratified analysis only and was not followed up as it contains the lactase (LCT) gene, whose SNPs are known to vary in frequency across Europe (Supplementary Table 2)18. To replicate the other loci harbouring genome-wide significant SNPs, we analysed an independent set of 1149 cases and 964 controls from GenoMEL. Cases were selected to be genetically enriched as previously (see Methods and Supplementary Note). A further replication set comprised 1163 cases and 903 controls from population-based studies from Leeds, UK. Samples were genotyped at the most significant SNPs from the GWAS plus any imputed SNPs more significantly associated. If assays could not be produced or analysed, alternative SNPs were genotyped.
Figure 1.
Results of Cochran-Armitage (CA) trend test for all SNPs passing quality control reported as −log10 p-values. The analysis stratified by geographic region is on the top with the unstratified analysis on the bottom (see Methods for details). The solid horizontal line indicates p-value of 10−5 and the dashed line indicates p-value of 5×10−7.
Table 1.
Genome-wide results for the most significant regions together with follow-up results. SNPs in the most significant regions are listed with their chromosome and position (in base-pair range) and the minor allele. Minor allele frequencies (MAF) are shown for the all controls genotyped genome-wide and in the combined replication sets, where appropriate.
| GENOME-WIDE PHASE | REPLICATION PHASE: GenoMEL + Leeds Case- control |
OVERALL: GENOME-WIDE + REPLICATION |
||||||
|---|---|---|---|---|---|---|---|---|
| SNP with base-pair range |
Minor allele |
MAF | P | OR [Conf. Int.] | P | OR [Conf. Int.] | P | OR [Conf. Int.] |
| Chromosomal Regions Considered for Followup on Basis of This Study | ||||||||
|
Chromosome 9: 21737803-21806528 |
||||||||
| rs4636294 | G | 0.51 | 1.77E-05 | 0.83 [0.76, 0.90] | 0.032 | 0.90 [0.83, 0.99] | 1.97E-06 | 0.86 [0.81, 0.92] |
| rs2218220 | T | 0.51 | 1.85E-05 | 0.83 [0.76, 0.90] | 0.046 | 0.91 [0.83, 1.00] | 6.40E-06 | 0.87 [0.82, 0.92] |
| rs1335510 | G | 0.41 | 1.14E-04 | 0.84 [0.77, 0.92] | - | - | - | - |
| rs935053* | A | 0.49 | 4.72E-06 | 0.81 [0.74, 0.89] | - | - | - | - |
| rs10757257 | A | 0.41 | 3.39E-05 | 0.83 [0.76, 0.91] | - | - | - | - |
| rs7023329 | G | 0.50 | 1.14E-05 | 0.82 [0.75, 0.90] | 0.023 | 0.90 [0.82, 0.99] | 4.03E-07 | 0.85 [0.80, 0.91] |
|
Chromosome 11: 88551344-88667691 |
||||||||
| rs1042602 (A) | A | 0.39 | 1.13E-01 | 0.93 [0.85, 1.02] | 0.054 | 0.91 [0.83, 1.00] | 1.07E-02 | 0.92 [0.87, 0.98] |
| rs1393350 | A | 0.27 | 4.28E-08 | 1.30 [1.19, 1.43] | 1.38E-07 | 1.30 [1.18, 1.44] | 2.41E-14 | 1.29 [1.21, 1.38] |
| rs1126809 (B) | A | 0.27 | - | - | 1.17E-06 | 1.27 [1.16, 1.40] | 1.17E-06 | 1.27 [1.16, 1.40] |
| rs1847142 (C )* | A | 0.32 | 8.35E-09 | 1.33 [1.21, 1.47] | 6.48E-04 | 1.26 [1.10, 1.44] | 2.24E-11 | 1.31 [1.21, 1.41] |
| rs1806319 | C | 0.35 | 2.49E-06 | 1.24 [1.13, 1.35] | - | - | - | - |
| rs10830253 (D) | G | 0.31 | - | - | 2.81E-06 | 1.26 [1.14, 1.39] | 2.81E-06 | 1.26 [1.14, 1.39] |
|
Chromosome 16: 87913062-88607035 |
||||||||
| rs2353033 | C | 0.41 | 3.32E-07 | 1.26 [1.15, 1.38] | - | - | - | - |
| rs352935 | C | 0.46 | 5.78E-06 | 1.22 [1.12. 1.33] | - | - | - | - |
| rs164741 | A | 0.29 | 1.50E-06 | 1.26 [1.15, 1.39] | - | - | - | - |
| rs7188458 (E) | A | 0.42 | 7.99E-11 | 1 34 [1.23, 1.46] | 1.20E-03 | 1.25 [1.09, 1.42] | 1.16E-12 | 1.30 [1.21, 1.40] |
| rs459920 | C | 0.47 | 5.41E-06 | 0.82 [0.75, 0.89] | - | - | - | - |
| rs12918773* | A | 0.08 | 1.34E-16 | 1.87 [1.62, 2.16] | - | - | - | - |
| rs258322 | A | 0.09 | 7.54E-17 | 1.81 [1.58, 2.08] | 1.07E-10 | 1.55 [1.36, 1,77] | 2.54E-27 | 1.67 [1.52, 1.83] |
| rs1800359 | A | 0.42 | 1.41E-06 | 0.80 [0.74, 0.88] | - | - | - | - |
| rs11861084 | A | 0.43 | 1.64E-06 | 0.81 [0.74, 0.88] | - | - | - | - |
| rs4408545 | T | 0.51 | 1.29E-06 | 0.81 [0.74, 0.88] | - | - | - | - |
| rs4238833 | G | 0.36 | 1.04E-09 | 1.32 [1.21, 1.44] | - | - | - | - |
| rs4785763 | A | 0.32 | 2.84E-14 | 1.42 [1.30, 1.56] | 5.13E-08 | 1.30 [1.18, 1.43] | 5.96E-22 | 1.36 [1.28, 1.45] |
| rs8059973 | A | 0.17 | 6.81E-07 | 0.74 [0.65, 0.83) | - | - | - | - |
| Chromosomal Regions Considered for Followup on Basis of Other Studies | ||||||||
|
Chromosome 20: 32635433-33040650 |
||||||||
| rs910873 | A | 0.07 | - | - | 1.92E-08 | 1.55 [1.33, 1.81] | 1.92E-08 | 1.55 [1.33, 1.81] |
| rs17305573 | C | 0.09 | - | - | 3.95E-05 | 1.53 [1.25, 1.87] | 395E-05 | 1.53 [1.25, 1.87] |
| rs4911442 | G | 0.12 | - | - | 1.80E-05 | 1.48 [1.24, 1,77] | 1.80E-05 | 1.48 [1,24, 1.77] |
| rs1885120 | C | 0.07 | - | - | 1.25E-08 | 1.58 [1.35, 1.85] | 1.25E-08 | 1.58 [1.35, 1.85] |
|
Chromsome 22: 36874244-36875565 |
||||||||
| rs2284063 | G | 0.37 | 5.60E-05 | 0.83 [0.76, 0.91] | 6.45E-05 | 0.82 [0.75, 0.90] | 2.40E-09 | 0.83 [0.78, 0.88] |
| rs6001027 | G | 0.37 | 4.50E-05 | 0.83 [0.76, 0.91] | 2.26E-03 | 0.86 [0.78, 0.95] | 1.94E-08 | 0.83 [0.78, 0.89] |
P-values and ORs (stratified by geographic region) are given for the genome-wide data, the replication data and the two stages combined. Imputed SNPs are marked with an asterisk (*) after the SNP number. Other SNPs are genotyped in the genome-wide dataset. The replication data sets consist of the Leeds case-control study and GenoMEL case-control samples. The following SNPs were also examined because of published literature: (A) rs1042602 was reported to be associated with hair colour and skin phenotype, so was genotyped in both replication sets11, (B) rs1126809 is a functional SNP in TYR, so was genotyped in both replication sets. Also, (C) rsl87142 was genotyped in the genome-wide dataset and in the Leeds case-control samples but not genotyped in the GenoMEL replication set, (D) rsl0830253 is a replacement for rsl806319 and (E) rs7188458 was genotyped in the genome-wide dataset and in the GenoMEL replication set but was not genotyped in the Leeds case-control samples. The table also includes two other regions implicated to contain melanoma susceptibility genes. The chromosome 20 region contains ASIP, a pigmentation gene, identified by an analysis of pigmentation genes11 which is in proximity to a region identified in a pooled genome-wide analysis21 (none of these SNPs were on the genome-wide array); the chromosome 22 region was identified in an adjoining paper involving a GWAS of nevus count variation22.
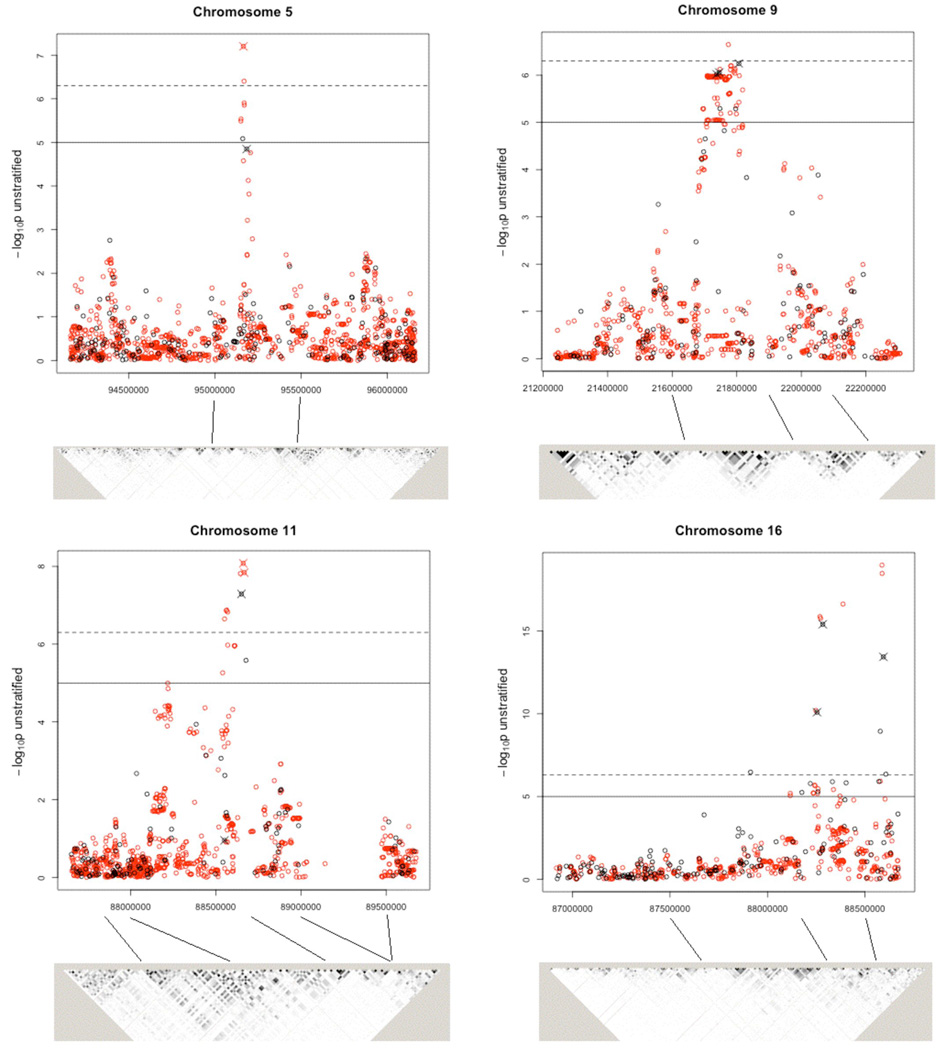
Figure 2.
Stratified CA trend tests. The −log10 p-values are from the CA trend test stratified (by geographical region) for genotyped and imputed SNPs in candidate regions for follow-up. Genotyped SNPs are shown in black and imputed SNPs in red; those SNPs that were followed up in the replication stage are indicated by a cross. The solid horizontal line indicates a p-value of 10−5 and the dashed line indicates a p-value of 5×10−7. The grey scale plots indicate extent of pairwise linkage disequilibrium (measured by r2) between SNPs, estimated from our data using Haploview23. Imputed data were not used for this. The darkest shading (black) indicates r2≥0.9, the next darkest 0.8≥ r2 > 0.9, etc. The position of the SNPs relative to the graph is indicated by the lines between the two.
Of the four regions that we attempted to replicate, only one (chromosome 5) showed no evidence of replication (Table 1 and Supplementary Table 3). The strongest association was with a region near the telomere of chromosome 16q: the three SNPs chosen for replication covered a 340kb region with p-values ranging between 7.99×10−11 and 7.54×10−17 in the GWAS sample and between 1.16×l0−12 and 2.54×10−27 in the overall (genome-wide plus replication) sample. This region includes several candidate genes, notably MC1R, CDK10 (involved in cell cycle regulation) and FANCA (which regulates genomic stability). A number of the 16q SNPs were reported earlier by Han et al19 who in a GWAS examining hair colour and skin pigmentation identified rs258322, which showed the strongest effect in both their and our study. Han et al 19 showed that the rs258322 signal was explained by functional MC1R variants. In this study, stepwise regression analysis of 30 tag SNPs covering 1.4Mb on 16q24 (Supplementary Table 4) showed evidence for association with three SNPs (rs258322, rs4785763, rs8059973); one of these (rs8059973) was not reported by the Han et al study22 and appears to have an association independent of the other two on melanoma risk. The magnitude of the association with rs258322 (per-allele odds ratio (OR) of 1.67, 95% confidence interval (CI) (1.52, 1.83), Table 1) is notable, since it is comparable to that found by a recent meta-analysis of MC1R variants 9 even though this SNP is distant from MC1R.
The chromosome 11 region (overall strongest evidence for rsl393350, p = 2.41×10−14) includes the TYR gene, which has previously been associated with melanoma11; the coding variant (rsll26809) is included in the replication panel and shows a strong association in terms of OR for genotyped SNPs (OR = 1.27, 95% CI (1.16, 1.40), p = 1.17×10−6; Table 1 and Supplementary Table 3). Stepwise regression analysis indicates that, after accounting for rsl393350, no other SNPs in the region were independently associated with melanoma (Methods and Supplementary Table 4).
The third locus replicated is on 9p21, with the overall p-value reaching 4.03×l0−7 for rs7023329 which lies within the MTAP gene (Table 1 and Supplementary Table 3). Stepwise regression analysis of 11 tag SNPs covering a 2 Mb region on 9p21 showed evidence for the independent associations with one replicated SNP (rs7023329) plus another one, rsl011970, that is 246kb centromeric and is within ANRIL, an antisense non-coding mRNA starting in the promoter of p14/ARF gene and overlapping CDKN2B20 (Supplementary Table 4, Supplementary Figure 3). Interestingly, antisense transcripts in the MTAP region have been suggested for CDKN2A and CDKN2B3.
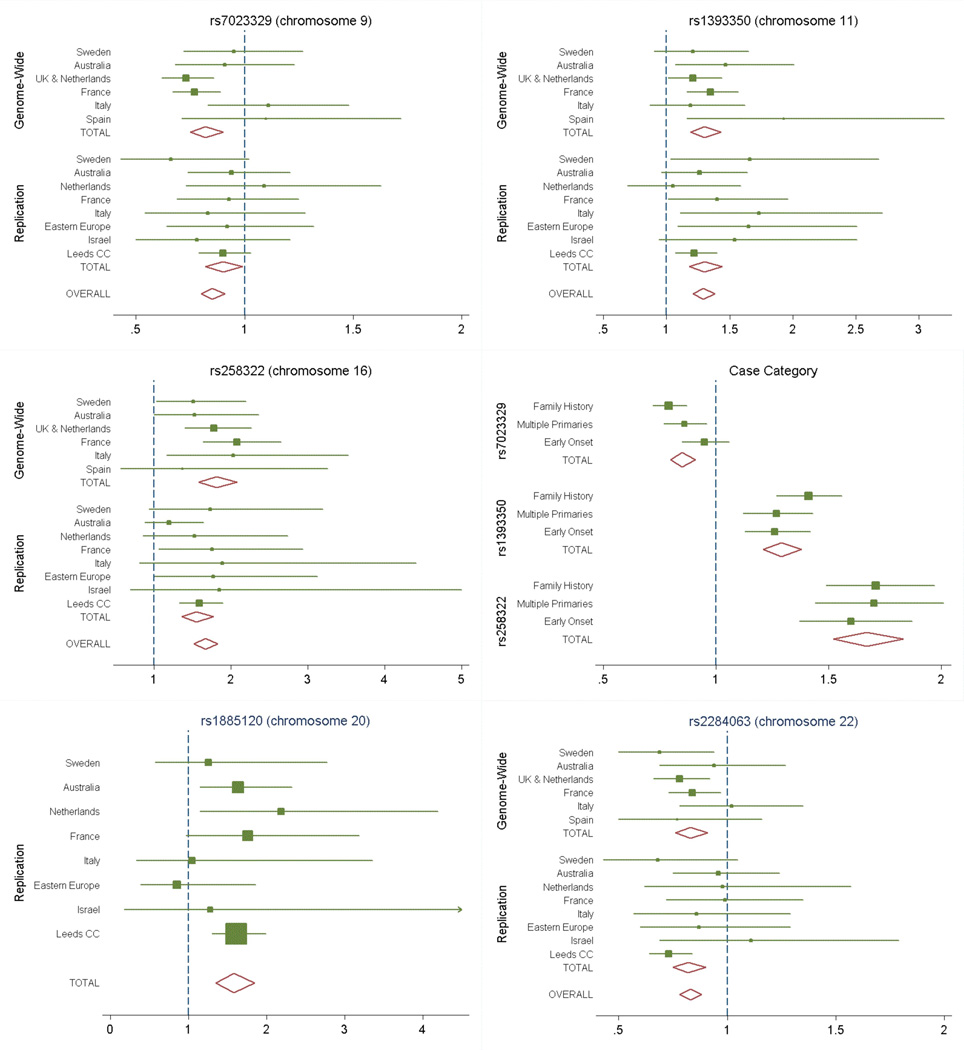
We also attempted to replicate findings from two other related genome-wide studies; SNPs in these regions did not achieve the critical significance level for follow-up in our GWAS study, but the available evidence suggested these regions as strong candidates for containing melanoma susceptibility genes. A further pigmentation gene has been reported to be associated with melanoma (ASIP11), adjacent to a reported melanoma locus recently identified by a pooling study21; this chromosomal region is confirmed by the present data (Table 1 and Supplementary Table 3). In an accompanying article a GWAS of nevus count variation based on volunteer twins from the UK and replicated in an Australian population found evidence for two regions, one of which overlaps with the chromosome 9 region identified here (see Supplementary Note). The second nevus locus reported in this paper is on chromosome 22 and was also associated with risk of melanoma in our study (Table 1 and Supplementary Table 3). Figure 3 shows the estimated ORs by geographical region.
Figure 3.
Forest plot of associations by geography and case category. The forest plot of the estimated per-allele OR from the CA trend test across geographical regions and overall for (i) chromosome 9 (rs7023329, top left), (ii) chromosome 11 (rsl393350, top right), (iii) chromosome 16 (rs258322, middle left), (iv) by case-category for these three SNPs (middle right), (v) chromosome 20 (rsl885120, only genotyped in replication set, bottom left) and (vi) chromosome 22 (rs2284063, bottom right). Overall the associations are consistent across populations. The case categories show some limited variation for both chromosome 9 and chromosome 16 with family history and multiple primaries being associated with more extreme risk than early onset.
Multiple logistic regression analysis of the three loci on chromosomes 9, 11 and 16 showed significant evidence for six SNPs being associated with melanoma risk (Supplementary Table 4). No pairwise interactions between the most significant SNPs at the three loci were significant (all p>0.19), showing that the pattern of association with the three loci is consistent with a multiplicative model (Supplementary Table 4). Furthermore, examination of the associations by case phenotype category showed similar associations for family history, multiple primaries and early onset (Figure 3), although for some SNPs at the 9q21 and 16q241oci the effect size was marginally weaker for early onset melanoma.
The strength of our study, which combines cases and controls from geographically distinct sites, lies in its ability to identify genetic variants that affect disease risk within diverse populations of European origin and to estimate the effects by population. Interestingly despite the variation in allele frequencies between groups (Supplementary Table 2) and the large differences in sun exposure between sites, the effect sizes for all three identified regions show remarkable homogeneity across the groups (Figure 3).
Given that this study has good power to detect well-tagged variants with OR>1.5 there are unlikely to be many other common SNPs with a large association with melanoma risk. Some of the genetic variants found to date to be associated with melanoma risk have also been found to be associated with pigmentation phenotypes11–15 and nevus numbers22, but other variants may be involved. Further, Falchi et al22 found that adjustment of the SNP-melanoma association for nevus count weakened that association with the SNPs, suggesting that the SNP and nevus count are measuring in part the same disease-associated factor(s). Further studies are required to assess whether the loci characterized by the present study act mainly through melanoma-associated phenotypes and/or have independent associations with melanoma risk. Our findings have important implications for understanding melanoma etiology.
Methods
Overview
This is a GWAS of melanoma cases and controls, contributed by GenoMEL participating groups. Supplementary Figure 1 contains an overview of the study. Groups were asked to identify, preferentially, melanoma cases either with a family history (but confirmed as not having a germline CDKN2A mutation), multiple primaries or onset before age 40 years. We argued that these criteria were all likely associated with an increased genetic risk of melanoma so that the cases would be “enriched” for genetic susceptibility, thereby increasing power to identify germline variation increasing risk 24. Family history was restricted to three cases for families from European populations and four cases for Australian participants to reduce the chance of including individuals with high-penetrance mutations. Controls were recruited by the same research groups from the same populations. Initially the study design involved minimally 100 cases and 100 controls from each of the 10 centres, but this was expanded subsequently. Genotyping was conducted in two phases. The first phase involved genotyping through ServiceXS in Leiden, The Netherlands, using the Illumina HumanHap300 BeadChip version 2 duo array (with 317k tagging SNPs). Subsequent genotyping of cases was conducted at CNG in Paris using the Illumina Humancnv370k array. Finally control data were available from two further groups: French controls from CNG genotyped on the Illumina HumanHap300 BeadChip version 2 duo array and UK controls from the WTCCC (described in 17 but based for this analysis on Illumina HumanHap300 BeadChip version 2 duo array genotyping).
We categorised the research groups by their geographical locations, but to enhance power identified six geographical regions within which the data from individual groups were pooled. These geographical regions were: Sweden (Lund and Stockholm), Australia (Brisbane, Sydney and Australian Melanoma Family Study (AMFS) sites), Italy (Genoa and Emilia-Romagna), UK/Leiden, France and Spain.
Replication was carried out in a further independent set of genetically-enriched cases and controls supplemented by a population-based case-control study conducted in Leeds, UK (Supplementary Figure 1).
Sample Sets
The cases and controls were selected from sets of samples accrued by the various contributing groups; these samples were recruited in different ways as described in the relevant publications. The contributing groups for the genome-wide genotyping were: Barcelona25, Brisbane (cases26, controls27), Emilia-Romagna28, Genoa29, Leiden, Leeds16,30, Lund31, Paris 32, Stockholm33 and Sydney34,35. The samples from Sydney included population-based early-onset cases and matching controls from the Australian Melanoma Family Study21 (AMFS, recruited in Sydney, Brisbane and Melbourne). Genotyping within the replication studies included samples from Poland36, Slovenia37 and Tel Aviv. Ethical consent was obtained from all participants.
The Leeds-based case-control study recruited 1274 population-based incident melanoma cases diagnosed between September 2000 and December 2006 from a geographically defined area of Yorkshire and the Northern region of the UK (63% response rate). Cases were identified by clinicians, pathology registers and via the Northern and Yorkshire Cancer Registry and Information Service to ensure overall ascertainment. For all but 18 months of the study period, recruitment was restricted to patients with Breslow thickness of at least 0.75mm. Some of these cases were genotyped genome-wide if they satisfied the case inclusion criteria for genetic enrichment; the reminder (1163) were genotyped in the replication set.
Controls were ascertained by contacting general practitioners to identify eligible individuals. These controls were frequency matched with cases for age and sex from general practitioners who had also had cases as part of their patient register. Overall there was a 55% response for controls (496 subjects); a subset of these controls was genotyped genome-wide and hence is excluded from the replication set. Controls were supplemented by a population-based group of 574 women who, following informed consent, agreed to participate in a study recording their history of sun exposure and sun bathing and for whom various measures of skin aging were recorded. DNA samples were also provided. A total of 903 controls were genotyped in the replication phase.
Sample Handling and DNA preparation
Genome-wide genotyping for this study was conducted in Leiden by ServiceXS (SXS in Supplementary Table 1) and in Paris by Centre National de Génotypage (CNG in Supplementary Table 1) (Supplementary Figure 1). Samples genotyped at ServiceXS were processed by the Department of Human and Clinical Genetics, Leiden University Medical Centre (LUMC). Sample lists were provided by the contributing GenoMEL centres and sample tubes and barcodes were returned to the centres from LUMC. Samples returned from the GenoMEL centres were then crosschecked against the manifest list. At the same time, phenotypic information was sent to the analysis centre in Leeds. Processed samples were then checked for QC by performing a single PCR test, and examining DNA concentration and quantities; a round of sample replacement proceeded for samples that were considered to have failed QC. A similar process applied to samples genotyped at CNG.
QC was performed for all samples before processing on the Illumina arrays. DNAs with a minimum concentration of 30 ng/ul (measured by NanoDrop(r) ND-1000) were subjected to a test PCR and checked on an agarose gel.
Replication genotyping was carried out using Taqman™ SNP genotyping assay (Applied Biosystems, Foster City, USA), with the exception of rs4911442, which was genotyped using the KASPar™ competitive allele specific PCR system (KBiosciences, Hoddesdon, UK) for the Leeds case-control study.
Quality Control (QC)
Genotypes were called using the proprietary software supplied by Illumina (BeadStudio, version 3.2), with imported cluster centres based on HapMap samples (supplied by Illumina) and call threshold set at 0.15 as recommended by Illumina. Initially no SNP was excluded from the analysis on the basis of QC, but SNPs showing some evidence of association were screened intensively for signs of poor QC. Some problems with poor chip quality were identified, and where possible samples with low (<97%) call rates were re-genotyped.
Sample exclusions
Samples were excluded for any of the following reasons: a) call rate of less than 97% (of the total number of SNPs on the array); b) evidence of non-European origin from principal components analysis (PCA); c) sex as inferred from genotyping not matching reported sex; d) evidence of first degree relationship or genetic identity with another sample. Sex was ascertained from genotype data by calculating the heterozygosity rate on the 9035 X-chromosome markers within Beadstudio; persons with > 10% heterozygosity were classified as female, otherwise male. Relationship analysis was carried out in PLINK 38 using estimated IBD sharing.
SNPQC
SNP QC was assessed by considering a range of measures: a) test of Hardy-Weinberg equilibrium for the different control groups; b) call rate; c) minor allele frequency (MAF); d) differences in MAF between genotyping centres; e) test for homogeneity of ORs across regions; f) concordance of results with neighbouring SNPs; g) where possible review of clusters from Bead Studio. Rather than using fixed thresholds, these measures were considered for all SNPs showing evidence of association at 10−5. We had no formal exclusion QC criteria for SNPs; most SNPs either clearly failed QC on multiple measures or raised no concern on any measure.
PCA and Population Stratification
To identify individuals of non-European ethnicity the SNPs were thinned to reduce linkage disequilibrium (LD) to a set of 67315 SNPs such that no pair had r2>0.2. The data were then combined with the HapMap data from 270 individuals of European, Asian and African origin. Before thinning, a QC step was implemented; SNPs with callrate less than 97% were excluded as were SNPs with a Hardy-Weinberg p-value < 0.0001 in any case-by-genotyping-laboratory (SXS or CNG) or control-by-genotyping-laboratory analysis. PCA was applied to the remaining SNPs using EIGENSTRAT39,40 with the first two principal components (PCs) clearly separating the HapMap data into three distinct clusters according to ethnicity. The majority of our samples were clustered with the HapMap European samples (Supplementary Figure 2B) but those that weren't were excluded from subsequent analysis. The first two PCs captured 72% of the variation in the first 20 PCs (and 83% of the variation in the first 10 PCs).
The remaining European ethnicity-only data were analysed similarly by PCA implemented in EIGENSTRAT. After applying QC the SNPs were thinned such that pairwise r2 never exceeded 0.5 (167517 SNPs). Despite the fact that PCA makes no use of the geographical origin of the samples, plotting the first two PCs clearly grouped the samples by the centre from which they were collected, with surprisingly little overlap between centres (Supplementary Figure 2A). The first PC corresponded roughly to latitude and the second PC to longitude as has been predicted elsewhere 18. The third PC separated those groups from the NE or SW from those groups from the NW or SE (also predicted18). This demonstrated that the majority of individuals were ethnically from the regions they had been collected from, while a few were clearly outliers from elsewhere in Europe.
For instance, while the majority of the Australian samples were clustered with the UK samples, several appeared to be more similar to the Italian samples, as did several of the French samples, suggesting that some had a southern European origin. Here the first three PCs capture 77% of the variation in the first 20 PCs (87% of the variation from the first 20 PCs is in the first 10). We were thus able to confirm that our groupings were roughly correct and we could account for stratification within Europe by adjusting for the first two or three PCs. Furthermore, by examining the loadings on the PCs we could identify particular SNPs or regions with particularly strong loadings suggesting that these showed considerable variation across Europe. The regions with notably high loading were around Lactase, OCA2 and HLA.
Association analysis
The primary analysis was a CA trend test, stratified by region (see Overview). In addition an unstratified CA trend test was carried out. Two further analyses were conducted to assist in interpretation of results: logistic regression analysis adjusted for region and the first three PCs (to adjust for any residual within-region population stratification) and a CA trend test stratified by study group defined by geographical centre and genotyping laboratory. Equivalent 1 degree of freedom stratified and unstratified trend tests were carried out for the X chromosome, in which males are treated as equivalent to homozygous females and a variance estimate that allows for the different variance of male and female contributions is used41.
Subgroup analyses of the different subtypes of case against all controls were also conducted for the most interesting SNPs.
Imputation
Imputation of ungenotyped SNPs was conducted using IMPUTE42, which predicts the genotypes of unobserved SNPs by means of a hidden Markov model using the genotype data at our observed markers and a set of known haplotypes (in this case the HapMap European samples). As the method is computationally intensive we applied it only to those regions in which at least one SNP reached a p-value of <10−5 either in the CA trend test, the test stratified by group or the test stratified by region and further SNPs within 50kb supported this result. We imputed 1Mb either side of any SNPs that reached the required p-value as well as a 250kb buffer either side to avoid end effects. We assumed an effective population size of 11400 and used a calling threshold of 0.9. We then applied both the CA trend test and a logistic regression adjusting for region.
Multiple Regression Analysis
Stepwise logistic regression was performed to identify those SNPs independently associated with melanoma within each of the validated regions (chromosomes 9, 11 and 16). We identified all SNPs that passed the QC for imputation within the genome-wide analysis and that were within 1Mb of the SNP giving the strongest evidence within each region. (The region covered on chromosome 16 is 1.4Mb long since it is close to the telomere). Any SNP not associated with melanoma (p>0.01) was eliminated, and SNPs were thinned such that no pair had r2>0.8. This left 50 SNPs from a total set of 480. The model assumed an additive effect on the logistic scale at the locus of interest, and analyses were adjusted for geographical region (as previously defined). Chromosomal regions were analysed separately and in combination, and the analysis included either every individual or was restricted to individuals with complete genotyping at the markers being analysed. At each step of the forward-selection procedure, the global significance of the model was evaluated, as was the significance of the new marker. The criteria for accepting a new marker were p <0.001 for each marker included in the model from the likelihood ratio test and a decrement in the global p-value of the model. Of the 5456 individuals in the dataset, 4959 (90.9%) had complete genotyping. The stepwise regression was conducted using Stata. Including individuals without complete data or considering regions separately made no qualitative difference to the results. Therefore results are presented for analyses restricted to those with complete genotyping and a combined analysis of all three genetic regions.
Case category analysis
Cases were classified as having a family history if at least one first degree relative had melanoma; individuals with multiple primary melanoma but no family history were classified in the “multiple melanoma” category, and cases with onset before age 40 without either a family history or multiple primary melanoma were classified as “early onset” (Supplementary Table 1). Using these categories we performed classical logistic regression adjusted for geographical region to estimate ORs for a given case category (e.g. cases with family history vs controls) and to test for homogeneity of the SNP effect across regions using a likelihood ratio test. The homogeneity of ORs by case category was tested using trichotomous regression (e.g. controls, cases with family history, cases without family history) and a likelihood-ratio test with 1 df (i.e. comparing a model where the ORs with and without family history are equal to a model where the two ORs are estimated).
Power
We calculated the power to detect disease-associated regions in our initial genome-wide analysis at a p-value of 5×10−7 and a sample size of 1539 cases and 3917 controls. We used effect sizes estimated in the replication stage in order to avoid bias, although this may be conservative since the cases from Leeds were not genetically enriched. We assumed the marker being tested was in complete LD with the causative locus and that the baseline risk of disease was 0.05.
Our calculations show that we have 97% and 84% power to detect the most significant SNPs on chromosomes 16 and 11 respectively. However, we have very low power (~1%) to detect the region on chromosome 9. While power is good (>95%) to detect the previously-identified region found on chromosome 2021, the low frequency led to it not being tagged by our array. Considering the range of values we have the power to detect, we have at least 80% power to detect any SNP with an OR of 1.6 if MAF>0.05 (assuming sufficient coverage), an OR of 1.5 if MAF>0.08, an OR of 1.4 if MAF>0.12 or an OR of 1.3 if MAF>0.25. The lowest OR we have 80% to detect is 1.27 (when MAF=0.5). Thus, it is unlikely that we have missed any common variants with effect sizes >1.5 (unless coverage is poor in these areas). Power is not very good to detect effects below 1.27 - thus there may well be other regions with SNPs of similar effect to those we found on chromosome 9 still to be discovered.
Replication Analysis and Israeli Samples
The genome-wide analysis involved European populations, and PCA was employed to identify participants apparently not of European ancestry. Within the replication set, all samples were again derived from European populations with the exception of one population from Israel. Without genome-wide analyses to confirm comparability with the European genetic profile, we have taken the approach of analysing the replication data both with and without the Israeli samples. The analyses presented here include the Israeli samples but analyses excluding these samples are not qualitatively different from these.
Supplementary Material
Acknowledgements
The Principal Investigators of Q-MEGA would like to thank Dixie Statham, Monica de Nooyer, Isabel Gardner and Barbara Haddon for project management, Anjali Henders, Megan Campbell and Mitchell Stark for managing sample processing and preparation, David Smyth and Harry Beeby for data management, Jane Palmer and Judy Simmons for ascertainment of clinical records. We also thank the numerous interviewers who collected questionnaire data. Most of all we thank the melanoma patients and their families for their cooperation.
The Sydney and AMFS investigators are grateful to all members of the recruitment, data collection and laboratory team, especially Caroline Watts, Robyn Dalziell, Kate Mahendran, Gayathri St George and Chantelle Agha-Hamilton and AMFS coordinators Judi Maskiell, Megan Ferguson and Jodi Jettan.
This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data is available from the Wellcome Trust website. Funding for the project was provided by the Wellcome Trust under award 076113
The authors thank the EGEA cooperative group for having given access to data on the EGEA (Epidemiological study on the Genetics and Environment of Asthma) study. Biological specimens of the French Family study group were obtained from IGR Biobank.
The authors are grateful for the contributions of Patricia A. Van Belle to the work of GenoMEL.
Footnotes
URLs
GenoMEL www.genomel.org
EGEA (Epidemiological study on the Genetics and Environment of Asthma) study http://ifr69.vjf.inserm.fr/~egeanet
WTCCC (Wellcome Trust Case Control Consortium) www.wtccc.org.uk
References
- 1.Cannon-Albright LA, Bishop DT, Goldgar C, Skolnick MH. Genetic predisposition to cancer. Important Adv Oncol. 1991:39–55. [PubMed] [Google Scholar]
- 2.Bataille V, et al. Risk of cutaneous melanoma in relation to the numbers, types and sites of naevi: a case-control study. Br J Cancer. 1996;73:1605–1611. doi: 10.1038/bjc.1996.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang YM, et al. A pooled analysis of melanocytic nevus phenotype and the risk of cutaneous melanoma at different latitudes. Int J Cancer. 2009;124:420–428. doi: 10.1002/ijc.23869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollenbeak CS, et al. Increased incidence of melanoma in renal transplantation recipients. Cancer. 2005;104:1962–1967. doi: 10.1002/cncr.21404. [DOI] [PubMed] [Google Scholar]
- 5.Naldi L, et al. Cutaneous malignant melanoma in women. Phenotypic characteristics, sun exposure, and hormonal factors: a case-control study from Italy. Ann Epidemiol. 2005;15:545–550. doi: 10.1016/j.annepidem.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Titus-Ernstoff L, et al. Pigmentary characteristics and moles in relation to melanoma risk. Int J Cancer. 2005;116:144–149. doi: 10.1002/ijc.21001. [DOI] [PubMed] [Google Scholar]
- 7.Holly EA, Aston DA, Cress RD, Ahn DK, Kristiansen JJ. Cutaneous melanoma in women. I. Exposure to sunlight, ability to tan, and other risk factors related to ultraviolet light. Am J Epidemiol. 1995;141:923–933. doi: 10.1093/oxfordjournals.aje.a117359. [DOI] [PubMed] [Google Scholar]
- 8.Holly EA, Aston DA, Cress RD, Ahn DK, Kristiansen JJ. Cutaneous melanoma in women. II. Phenotypic characteristics and other host-related factors. Am J Epidemiol. 1995;141:934–942. doi: 10.1093/oxfordjournals.aje.a117360. [DOI] [PubMed] [Google Scholar]
- 9.Raimondi S, et al. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer. 2008;122:2753–2760. doi: 10.1002/ijc.23396. [DOI] [PubMed] [Google Scholar]
- 10.Kanetsky PA, et al. Population-based study of natural variation in the melanocortin-1 receptor gene and melanoma. Cancer Res. 2006;66:9330–9337. doi: 10.1158/0008-5472.CAN-06-1634. [DOI] [PubMed] [Google Scholar]
- 11.Gudbjartsson DF, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40:886–891. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 12.Elwood JM, Jopson J. Melanoma and sun exposure: an overview of published studies. Int J Cancer. 1997;73:198–203. doi: 10.1002/(sici)1097-0215(19971009)73:2<198::aid-ijc6>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 13.Kamb A, et al. Analysis of the pl6 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet. 1994;8:23–26. doi: 10.1038/ng0994-22. [DOI] [PubMed] [Google Scholar]
- 14.Berwick M, et al. The prevalence of CDKN2A germ-line mutations and relative risk for cutaneous malignant melanoma: an international population-based study. Cancer Epidemiol Biomarkers Prev. 2006;15:1520–1525. doi: 10.1158/1055-9965.EPI-06-0270. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein AM. Features associated with germline CDKN2A mutations: a Geno MEL study of melanoma-prone families from three continents. J Med Genet. 2007;44:99–106. doi: 10.1136/jmg.2006.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishop JA, et al. Genotype/phenotype and penetrance studies in melanoma families with germline CDKN2A mutations. J Invest Dermatol. 2000;114:28–33. doi: 10.1046/j.1523-1747.2000.00823.x. [DOI] [PubMed] [Google Scholar]
- 17.WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genets. 2008;4:el000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasmant E, et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 21.Brown KM, et al. Common sequence variants on 20q 11.22 confer melanoma susceptibility. Nat Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falchi M, et al. Loci at 9p21 and 22ql3 harbour alleles for development of cutaneous nevi and melanoma. (Submitted) [Google Scholar]
- 23.Barrett JC, Fry B, Mailer J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Antoniou AC, Easton DF. Polygenic inheritance of breast cancer: Implications for design of association studies. Genet Epidemiol. 2003;25:190–202. doi: 10.1002/gepi.10261. [DOI] [PubMed] [Google Scholar]
- 25.Puig S, et al. Role of the CDKN2A locus in patients with multiple primary melanomas. J Clin Oncol. 2005;23:3043–3051. doi: 10.1200/JCO.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 26.Baxter AJ, et al. The Queensland Study of Melanoma: environmental and genetic associations (Q-MEGA); study design, baseline characteristics, and repeatability of phenotype and sun exposure measures. Twin Res Hum Genet. 2008;11:183–196. doi: 10.1375/twin.11.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiteman DC, et al. Combined effects of obesity, acid reflux and smoking on the risk of adenocarcinomas of the oesophagus. Gut. 2008;57:173–180. doi: 10.1136/gut.2007.131375. [DOI] [PubMed] [Google Scholar]
- 28.Landi MT, et al. Genetic susceptibility in familial melanoma from northeastern Italy. J Med Genet. 2004;41:557–566. doi: 10.1136/jmg.2003.016907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pastorino L, et al. CDKN2A mutations and MC1R variants in Italian patients with single or multiple primary melanoma. Pigment Cell Melanoma Res. 2008 doi: 10.1111/j.1755-148X.2008.00512.x. [DOI] [PubMed] [Google Scholar]
- 30.Newton Bishop JA, et al. Mutation testing in melanoma families: INK4A, CDK4 and INK4D. Br J Cancer. 1999;80:295–300. doi: 10.1038/sj.bjc.6690354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borg A, et al. High frequency of multiple melanomas and breast and pancreas carcinomas in CDKN2A mutation-positive melanoma families. J Natl Cancer Inst. 2000;92:1260–1266. doi: 10.1093/jnci/92.15.1260. [DOI] [PubMed] [Google Scholar]
- 32.Chaudru V, et al. Influence of genes, nevi, and sun sensitivity on melanoma risk in a family sample unselected by family history and in melanoma-prone families. J Natl Cancer Inst. 2004;96:785–795. doi: 10.1093/jnci/djh136. [DOI] [PubMed] [Google Scholar]
- 33.Platz A, et al. Screening of germline mutations in the CDKN2Aand CDKN2B genes in Swedish families with hereditary cutaneous melanoma. J Natl Cancer Inst. 1997;89:697–702. doi: 10.1093/jnci/89.10.697. [DOI] [PubMed] [Google Scholar]
- 34.Holland EA, et al. Analysis of the pl6 gene, CDKN2, in 17 Australian melanoma kindreds. Oncogene. 1995;11:2289–2294. [PubMed] [Google Scholar]
- 35.Holland EA, Schmid H, Kefford RF, Mann GJ. CDKN2A (P16(INK4a)) and CDK4 mutation analysis in 131 Australian melanoma probands: effect of family history and multiple primary melanomas. Genes Chromosomes Cancer. 1999;25:339–348. [PubMed] [Google Scholar]
- 36.Debniak T, et al. Common variants of DNA repair genes and malignant melanoma. Eur J Cancer. 2008;44:110–114. doi: 10.1016/j.ejca.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Peric B, et al. Prevalence of variations in melanoma susceptibility genes among Slovenian melanoma families. BMC Med Genet. 2008;9:86. doi: 10.1186/1471-2350-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 41.Clayton D. Testing for association on the X chromosome. Biostatistics. 2008;9:593–600. doi: 10.1093/biostatistics/kxn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.