Abstract
The structure and dynamics of proteins underlies the workings of virtually every biological process. Existing biophysical methods are inadequate to measure protein structure at atomic resolution, on a rapid time scale, with limited amounts of protein, and in the context of a cell or membrane. FRET can measure distances between two probes, but depends on the orientation of the probes and typically works only over long distances comparable with the size of many proteins. Also, common probes used for FRET can be large and have long, flexible attachment linkers that position dyes far from the protein backbone. Here, we improve and extend a fluorescence method called transition metal ion FRET that uses energy transfer to transition metal ions as a reporter of short-range distances in proteins with little orientation dependence. This method uses a very small cysteine-reactive dye monobromobimane, with virtually no linker, and various transition metal ions bound close to the peptide backbone as the acceptor. We show that, unlike larger fluorophores and longer linkers, this donor–acceptor pair accurately reports short-range distances and changes in backbone distances. We further extend the method by using cysteine-reactive metal chelators, which allow the technique to be used in protein regions of unknown secondary structure or when native metal ion binding sites are present. This improved method overcomes several of the key limitations of classical FRET for intramolecular distance measurements.
Keywords: fluorescence, peptides, FRET
Fluorescence resonance energy transfer (FRET) occurs when excitation energy is transferred from a donor fluorophore to an acceptor dye through weak dipole–dipole resonance interactions (1, 2). FRET is extremely sensitive to the distance between the two dyes, with the transfer rate inversely proportional to the sixth power of the distance between them. This steep distance dependence has suggested that FRET could be used as a spectroscopic ruler on the molecular scale (3, 4). In addition to distance, five other factors influence the rate of energy transfer: (i) the overlap of the donor's and acceptor's excitation and absorbance spectra, respectively; (ii) the refractive index of the solvent; (iii) the relative orientation of the dyes; (iv) the quantum yield of the donor; and (v) the extinction coefficient of the acceptor.
Although FRET is sensitive to the distance between probes, several factors have prevented the widespread use of FRET to easily map backbone distances within proteins. First, most commonly used FRET pairs have R0 values (distance at which FRET efficiency is 50%) between ≈30 and 60 Å and are only able to measure distances between ≈20 and 80 Å. Thus, to accurately measure distances, the probes must be separated by large distances relative to the size of many proteins. Also, the large size of many dyes and their long attachment linkers position fluorophores far from the backbone of the protein. These flexible linkers also increase the conformational space sampled by the dye (4, 5). Because of the strongly nonlinear dependence of FRET on distance, fluorophores can appear in a different position in FRET measurements than the average center position of the dye in space (4, 6, 7). This effect can be further exaggerated by the long excited-state lifetimes of some probes (8). Because of these issues, changes in backbone position might not be accurately represented by changes in FRET signals. Last, the difficulty in determining the relative orientation of the two probes can add uncertainty to the interpretation of FRET (9).
We previously developed a fluorescence method called transition metal ion FRET that improves on classical resonance energy transfer (10). Specifically, we used colored transition metal ions as ultrasmall, ultrashort-range FRET acceptors for the organic dye fluorescein. The metal ion acceptors were bound to engineered dihistidine sites in α-helices very close to the peptide backbone. This technique (i) has the ability to measure distances at close range (≈5–20 Å), (ii) maintains the steep distance dependence of FRET, (iii) exhibits a low orientation dependence, (iv) uses smaller dyes with little or no linkers and minimal perturbation to the protein backbone, (v) has the ability to independently label two sites within one protein, (vi) has the ability to add and remove the acceptor to measure absolute FRET efficiency by donor quenching/dequenching, and (vii) affords the possibility of using multiple acceptors in the same experiment.
Here, we have improved on transition metal ion FRET by using a much smaller fluorescent donor (monobromobimane, mBBr), which is close to the size of a natural amino acid, and three different transition metal ion acceptors (11). mBBr was attached via cysteine residues to model α-helical peptides, and metal ions were bound to either dihistidine sites or small cysteine-attached metal chelators. These FRET pairs afforded accurate measurements of distance in the peptide backbone. When larger dyes or longer linkers were used, we observed smaller changes in distance than those expected from the structure of the peptides. This technique should open the door to the complete mapping of unknown protein structures or changes in protein structure with FRET.
Results
Transition Metal Ions as Distance-Dependent FRET Acceptors for Bimane.
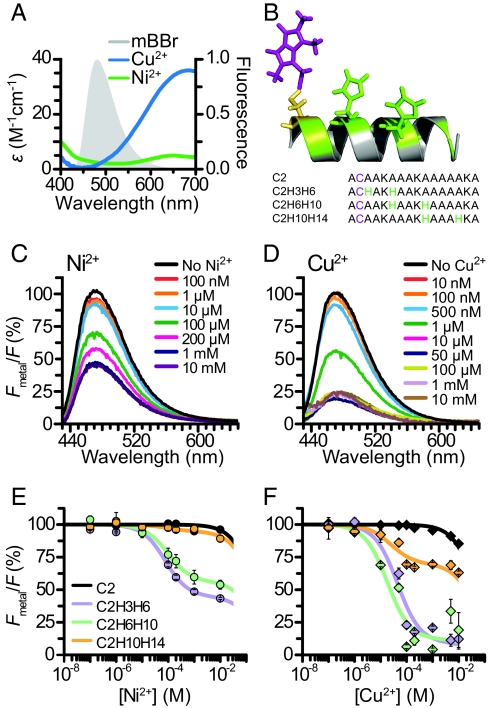
Some transition metal ions including Ni2+, Cu2+, and Co2+ absorb visible light, making them potential FRET acceptors for many fluorophores (12–15). The absorbance spectra of these metals are broad, and their extinction coefficients are very small, between 5- and 30-fold less than commonly used organic fluorophores (12). Fig. 1A shows the overlap of the absorbance of Ni2+ and Cu2+ bound to histidine residues in a helical peptide with the emission of the small organic fluorophore bimane. The extent of this overlap, low extinction coefficient of the metals, and low quantum yield of bimane suggest that these metals should work as efficient FRET acceptors for bimane only at very close distances. Indeed, the calculated R0 (distance where FRET efficiency is half-maximal) between bimane and these metals is 10 Å for Ni2+ and 12 Å for Cu2+. Also, the multiple transition dipoles of these metals virtually eliminate the orientation dependence normally associated with FRET (12, 13).
Fig. 1.
Measuring distances between transition metal ions bound to dihistidine motifs and mBBr in helical peptides. (A) Spectral overlap between the emission of mBBr reacted with glutathione and the absorbance of Cu2+ and Ni2+ bound to the peptide ACAAKAAAKHAAAHKA. (B) Sequences of model peptides. Diagram depicting C2H6H10, with mBBr reacted to the cysteine at position 2 (purple), and histidine residues at positions 6 and 10 (green). The positions of the other histidine residues used (3 and 14) are highlighted in green. (C) Spectra of the mBBr-reacted peptide C2mBBrH3H6 at increasing [Ni2+]. (D) Spectra of C2mBBrH3H6 at increasing [Cu2+]. (E) Average Ni2+ quenching of mBBr-reacted peptides. Fluorescence was normalized to the intensity before the addition of metal. Black trace is a single-site binding fit to the data. All other solid lines are two-site binding curves fit to the data. (F) Average Cu2+ quenching of the peptides in E.
To explore close-range FRET between fluorophores and transition metal ions, we created a set of short 16-aa α-helical model peptides (Fig. 1B). The sequences of our peptides were based on previously described alanine-containing peptides, which are strongly helical in aqueous solution (16). CD spectroscopy confirmed that our peptides were α-helical (Fig. S1). This reduced chemical scaffold simulates larger proteins and allowed us to easily manipulate several aspects of the system, including the structure of the probes, their mechanism of attachment, and the molecular distances between them. Each peptide contained a single cysteine at the second amino acid position (C2). We modified this cysteine with mBBr (Fig. 1B), a small fluorophore with a cysteine-reactive methyl bromide linker consisting of only a single methylene group (17). The peptides also contained a pair of histidines along one face of the helix (Fig. 1B). Pairs of histidines have been shown to bind transition metal ions with high affinity (18, 19). Coordination of the metal with a dihistidine motif offers several advantages: (i) it binds the metal at a well-defined location very close to the peptide backbone, eliminating the long linker normally associated with dyes used to label proteins; (ii) it can be engineered into existing structural elements (α-helices and β-sheets) with minimal perturbation of the protein backbone; and (iii) the site can bind different transition metal ions tightly, yet reversibly, affording the possibility of using different acceptor dyes in the course of the same experiment.
To test the distance dependence of transition metal ion FRET, we performed fluorescence quenching experiments on the above peptides in solution. We added increasing concentrations of metal to solutions containing mBBr-modified peptides and monitored the decrease in the donor's emission. The fraction of donor quenching (1 − Fmetal/F) is a direct measurement of FRET efficiency. In a peptide with a metal binding site adjacent to the fluorophore (C2H3H6), we observed substantial quenching with increasing concentrations of both Ni2+ (Fig. 1C) and Cu2+ (Fig. 1D). There was no change in the peak or the shape of the emission spectrum consistent with quenching being the result of FRET. The quenching with Cu2+ is greater than the quenching with Ni2+, as expected from its larger R0.
We determined the distance dependence of the FRET by measuring the quenching of different model peptides that contain metal binding sites at increasing distances from the mBBr-labeled cysteine. For each of the peptides, the average remaining fluorescence (Fmetal/F) was plotted as a function of metal ion concentration for both Ni2+ and Cu2+ (Fig. 1 E and F). The C2 control peptide (no histidines) was quenched only at very high concentrations of metal (>1 mM). This effect was likely the result of quenching by metal ions in solution (10). The other peptides exhibited a second, higher-affinity component, which was the result of metal binding to the introduced site. To quantify the FRET efficiency in each peptide (1 − Fmetal/F), we fit curves with a two-site binding model. This analysis allowed us to separate the high-affinity quenching caused by metal ions bound specifically to the engineered dihistidine motifs from the low-affinity solution quenching observed in the control peptides. The fraction of the donor's fluorescence quenched in the high-affinity component represents the FRET efficiency (E). FRET to Cu2+ and Ni2+ was greatest when the binding site was located closest to the C2 position (C2H3H6: Ni, 55 ± 1%, n = 4; Cu, 90 ± 3%, n = 4). The C2H6H10 peptide showed on average less FRET (Ni2+, 44 ± 3%, n = 4; Cu2+, 92 ± 3%, n = 4), and the peptide with the binding site farthest from the C2 position showed the smallest amount of FRET (Ni2+, 9 ± 3%, n = 4; Cu2+, 30 ± 2%, n = 4).
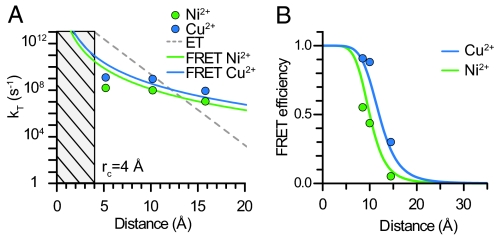
Fluorophores can transfer energy to acceptors through processes other than FRET (4, 20–22). These processes include Dexter exchange and electron transfer and occur predominantly at very close distances (<5 Å), because they require the direct overlap of the electronic orbitals of the fluorophore and quencher (4, 20–22). For electron transfer, the rate of energy transfer falls off with an exponential dependence on distance, whereas the rate of FRET falls off with an inverse sixth power dependence on distance (4, 22). To determine whether the quenching that we observed was caused by FRET and not another form of energy transfer, we determined the distance dependence of the rate of energy transfer by using the distances predicted from the β-carbon positions of an α-helix. Fig. 2A shows that our data do not fit to an exponential decay and conform better to an inverse sixth power of distance dependence. Moreover, the measured transfer rate closely approximated the theoretical predictions of the Förster model for both Ni2+ and Cu2+.
Fig. 2.
Mechanism of energy transfer. (A) Comparison of the predicted rate of energy transfer for electron transfer (gray line) or FRET with Ni2+ (green line) and Cu2+ (blue line) with values generated from experimental data for Ni2+ (green points) and Cu2+ (blue points) for all three sites. (B) Comparison of modeled distances with experimental FRET values in all peptides with Ni2+ (green) and Cu2+ (blue). Theoretical distance dependence of FRET is shown as solid lines.
To further show that our data fit a FRET mechanism, we modeled the position of the donor fluorophore in space by using experimental FRET values (Fig. 2B). Assuming an α-helical peptide structure and a modeled position of the metal binding site, the FRET efficiencies obtained for both Ni2+ and Cu2+ at all three metal ion binding sites are fit well by the Förster equation with the theoretical R0 values of 10 Å for Ni2+ and 12 Å for Cu2+. These data indicate that the small fluorophore mBBr can be used as an ultrashort distance-dependent FRET donor for transition metal ions, and that FRET between mBBr and metal ions can be used to measure very short distances in peptides.
Optimal Spacing of Histidines for Metal Coordination in an α-Helix.
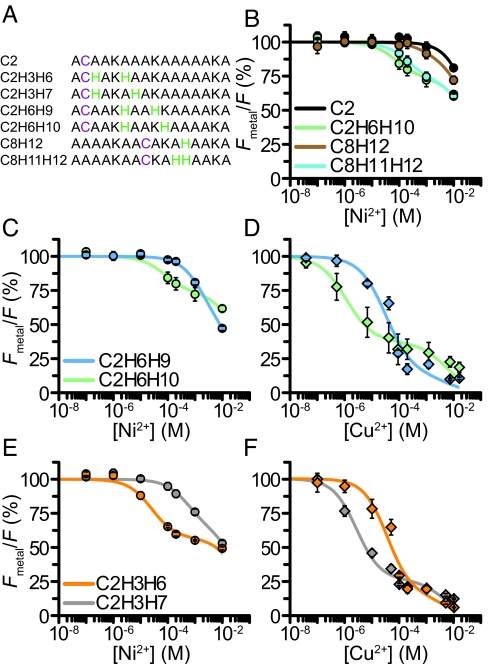
Binding sites for transition metal ions can be engineered into existing secondary elements by introducing as few as two histidines into the protein (18). For example, two histidines separated by one turn of an α-helix can produce a binding site with micromolar affinity (18). The strength of this interaction has been suggested to depend on the exact spacing of the histidines along the helix (18). To determine the optimal spacing of histidines in our system, we measured the metal binding affinity for a series of peptides by transition metal ion FRET (Fig. 3A). The peptides were conjugated with fluorescein-5-maleimide (F5M) and studied with increasing concentrations of Ni2+ or Cu2+. As seen for mBBr, a peptide with no histidines (C2) showed quenching only at high concentrations of Ni2+ (>1 mM) (Fig. 3B). Similarly, a peptide with a single histidine located close to the labeled cysteine (C8H12) exhibited quenching only at relatively large concentrations of Ni2+. These data indicate that a single histidine does not provide a strong binding site. However, a peptide with two adjacent histidines positioned near the cysteine (C8H11H12) showed robust quenching with a higher affinity (KD, 318 ± 100 μM) than the control or single histidine peptides. A peptide with the histidines spaced one turn away on the α-helix (C2H6H10) showed the highest affinity (125 ± 40 μM). From these data, we conclude that two histidines bind Ni2+ with high affinity and that spacing the histidines one turn away on an α-helix provides the most robust binding site.
Fig. 3.
Optimal histidine spacing for metal binding. (A) Sequences of peptides. (B) Average Ni2+ quenching for the F5M-reacted peptides C2F5M, C8F5MH12, C8F5MH11H12, and C2F5MH6H10. Data were fit as in Fig. 1. (C) Average Ni2+ quenching for the F5M-reacted peptides C2F5MH6H9 and C2F5MH6H10. (D) Average Cu2+ quenching for the peptides in C. (E) Average Ni2+ quenching for the F5M-reacted peptides C2F5MH3H6 and C2F5MH3H7. (F) Average Cu2+ quenching values for the peptides in E.
Right-handed α-helices found in proteins consist of 3.6 residues for each turn of the helix. Thus, histidines at the i and i + 3 and i and i + 4 position of the helix could both, in principal, form metal coordination sites. We next asked whether the i + 3 or i + 4 spacing created a better metal ion binding site in our model α-helix. At a position one helical turn from the dye-modified cysteine, both Ni2+ and Cu2+ bound with a higher affinity to the i + 4 configuration (C2H6H10) (Fig. 3 C and D). However, when the first histidine was located directly adjacent to the dye-modified cysteine, Ni2+ preferred the i + 3 spacing (C2H3H6), whereas Cu2+ preferred the i + 4 spacing (C2H3H7) (Fig. 3 E and F). We conclude that the i + 4 spacing generally creates a better binding site; however, other local factors, including neighboring residues, backbone stability, and solvent exposure, may influence the ion specificity or affinity of the dihistidine site. These factors should be considered and tested when applying transition metal ion FRET to the study of protein structure.
Labeling Peptides with Cysteine-Reactive Metal-Binding Molecules.
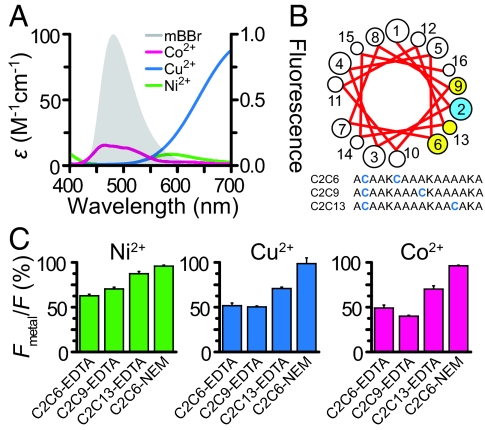
Although dihistidine sites offer a number of advantages for binding metal ions, under some circumstances, their use may be problematic: (i) when the protein contains a nearby native metal binding site of equal or higher affinity, (ii) when the site is uncoiled or its structure is unknown, and (iii) when metal coordination by two histidines causes a change in structure or constrains a conformational rearrangement. For these situations, we used an alternative method for introducing metal quenchers: using small cysteine-reactive metal binding groups to coordinate the metals (23). Synthetic metal binding molecules such as nitrilotriacetic acid (NTA), EDTA, and iminodiacetic acid (IDA), can bind transition metal ions such as Ni2+, Cu2+, and Co2+, with affinities in the picomolar range, higher than the affinity of most native sites in proteins. Similar chelators have been used to coordinate lanthanide ions for use in luminescence resonance energy transfer studies (24). Metals bound to EDTA have similar absorption spectra to metals bound to dihistidine peptides (Fig. 4A). The R0 between bimane and EDTA-chelated metals was calculated to be 9.2 Å for Ni2+, 10.3 Å for Cu2+, and 11.7 Å for Co2+. Also, the metal binding groups can be introduced at a cysteine residue without any previous knowledge of the structure of the protein and with minimal perturbation to the backbone. Thus, the use of synthetic metal binding molecules might mitigate some of the issues associated with endogenous metal sites or structural constraints and extends the usefulness of the transition metal ion FRET method.
Fig. 4.
FRET between mBBr and transition metal ions bound to MTS-EDTA in helical peptides. (A) Spectral overlap between the emission of mBBr reacted with glutathione and the absorbance of Co2+, Cu2+, and Ni2+ bound to EDTA. (B) Sequences of model peptides. A helical wheel projection of cysteine residues in the model peptide (yellow and blue). (C) Average quenching by 100 μM Ni2+, 10 μM Cu2+, and 100 μM Co2+ in double-labeled peptides.
To test whether cysteine-reactive chelators could be used to coordinate metals for transition metal ion FRET, we created a set of short peptides that contained two incrementally spaced cysteines (Fig. 4B). The peptides were partially labeled with mBBr and then completely labeled with an excess of MTS-EDTA, a cysteine-reactive metal chelator. This procedure ensured that virtually every fluorescent peptide that had a donor (mBBr) also had an acceptor (MTS-EDTA). Reversed-phase liquid chromatography showed that the fluorescent peptides were nearly equally divided between the two orientations of donor and acceptor, and virtually no peptide contained two donor fluorophores. Using saturating concentrations of metal (10–100 μM), we tested whether EDTA-bound metals could FRET with mBBr. Peptides showed position- and metal-specific quenching (Fig. 4C). Control peptides modified with N-ethylmaleimide (NEM), a molecule that does not bind metal, showed no appreciable quenching (Fig. 4C). These data indicate that metal binding molecules such as EDTA can be used to coordinate metals at defined locations within proteins for use as FRET acceptors. Similar data were acquired by using F5M for the donor and metal bound to a cysteine-reactive NTA (MNTA) as the acceptor (Fig. S2).
Linker Effects on Transition Metal Ion FRET.
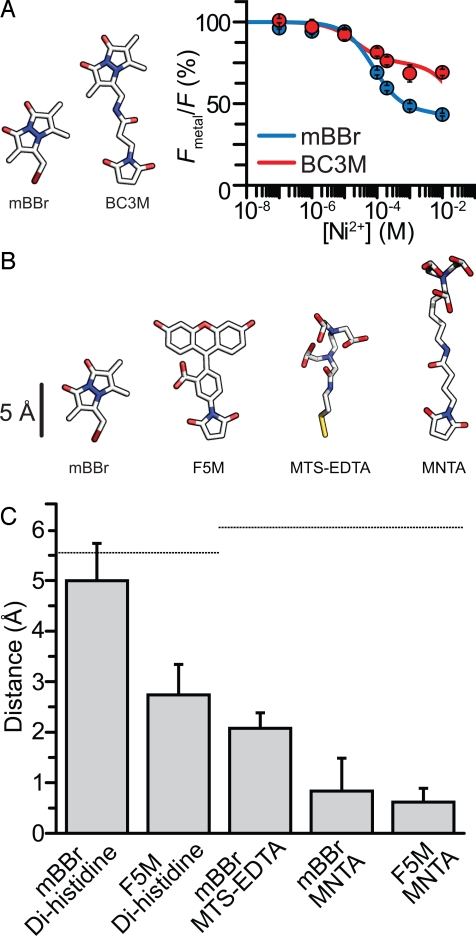
Many fluorophores used to label cysteines in biological molecules are attached to reactive groups through long, flexible linkers. Long linkers can increase the diffusional area occupied by a fluorophore or an acceptor molecule and can skew FRET measurements (4). For example, movement of the fluorophore during the excited-state lifetime can bias FRET measurements toward shorter distances. This bias occurs because of the nonlinear dependence of distance on energy transfer (4, 8, 25). Even a distribution of static distances can skew FRET measurements (7). Specifically, distances measured by FRET appear closer to the R0 distance when there is heterogeneity of fluorophore positions (4, 7). To explore the effect of linker length on transition metal ion FRET, we compared FRET between dihistidine-bound Ni2+ and bimane with essentially no linker (mBBr) to bimane attached by a long, flexible linker (bimane-C3-maleimide, BC3M) (Fig. 5A). Fig. 5A shows a comparison of Ni2+ quenching in the two differently labeled peptides. In C2H3H6, the BC3M-labeled peptide had less FRET (31 ± 5%) than mBBr-labeled peptide (55 ± 5%). Thus, the long linker of BC3M likely positioned the dye farther on average from the metal binding site. These data indicate that close-range distance measurements of backbone structure can be affected by the length of linkers connecting probes to the backbone. Clearly, for direct measurements of backbone distances with FRET, it is important to use short linkers.
Fig. 5.
Effect of fluorophore size and linker length on distance measurements. (A) Diagram of mBBr and BC3M. Average Ni2+ quenching of mBBr and BC3M attached to C2H3H6. Data were fit as in Fig. 1. (B) Diagram of mBBr, F5M, MTS-EDTA, and MNTA. (C) Distance between adjacent metal binding sites in peptides determined from FRET. Plot compares mBBr and F5M and metal ions coordinated by different chemical groups. Dotted lines indicate the predicted distances between sites along the peptide backbone.
To determine the influence of probe size and linker length on distance calculation, we measured distances for five donor–acceptors pairs of various sizes (Fig. 5B). Fig. 5C shows a plot of the change in apparent distance between two labeled positions within the peptide calculated from FRET changes compared with the change in distance expected from an α-helical model. From the model, the distance between two backbone positions was measured to be ≈5.5 Å. The change in distance observed with the small dye mBBr and dihistidine-coordinated metals was closest to the value predicted from the backbone structure (5 Å) (Fig. 5C). However, when we used a larger fluorophore (F5M), the apparent distance change was reduced to 2.7 Å. Also, using mBBr with cysteine-reactive metal chelators produced an underestimate of the distance change (2 Å), although the shorter MTS-EDTA was better than the longer MNTA chelator (0.8 Å). When we used both a larger dye and long linkers on the metal chelator (F5M and MNTA), we observed the smallest difference in apparent distance between the probes (0.6 Å). From this analysis, we conclude that the use of large dyes and long linkers can result in a dramatic underestimate of backbone distance changes in proteins. This problem was largely alleviated by using the smallest dye (mBBr) in combination with the shortest linker (dihistidine coordination of transition metals).
Discussion
On the scale of most proteins, dyes typically used for FRET are large. In transition metal ion FRET, the probes used are small. Here, we used bimane, a fluorophore that emits at visible wavelengths with virtually no linker to position the fluorophore as close as possible to the peptide backbone while still maintaining conformational flexibility of the probe. Also, we have used metal ion acceptors that were coordinated very near to the peptide backbone and minimally perturb the backbone structure. Thus, distances obtained with transition metal ion FRET more accurately represent the backbone architecture of the protein than standard FRET. Also, distance changes of the protein backbone are better recapitulated in changes in FRET when using this system. Minimizing the effects of probe size and linker length should extend the fidelity and sensitivity of FRET for measuring backbone distances and conformational motions of the backbone.
FRET depends on the relative orientation between the probes (9). Because transition metal ions have multiple absorption dipoles, these orientation effects should be mitigated (12). Again, this advantage of transition metal ion FRET has the potential to improve the accuracy and reliability of distance measurements within biological molecules.
Last, with transition metal ion FRET, multiple acceptors can be used in a single experiment. Dihistidine binding sites and metal chelators accommodate ions with a range of absorption spectra and extinction coefficients. This feature of transition metal ion FRET provides the opportunity to obtain multiple separate measurements of distance. Indeed, in our measurements, both Ni2+ and Cu2+ predicted distances that were within 1 Å of one another. Because FRET estimates of distance are most accurate when the distance is near R0, the acceptor used can be tuned to match the range of distances in question.
Several important considerations should be made when applying transition metal ion FRET to the study of protein structures. FRET efficiency is influenced by the quantum yield of the donor (2). Changes in quantum yields have been observed in proteins undergoing conformation changes. Because of this issue, controls should be performed to ensure that apparent changes in FRET are not caused by changes in the fluorophore itself (26). Previous methods to address this issue include exchanging the position of the donor and acceptor site and measuring distances from multiple positions (10, 26–28). Also, as we have shown, regional differences in the binding affinity of the dihistidine site may occur in peptides and proteins. Indeed, dihistidine sites in proteins have been shown to have a range of binding affinities (10, 18, 19). These differences in affinity might be caused by surrounding residues, the stability of the helix, the local pKa of the histidines, or the solvent exposure of the site. We recommend that a full concentration-response curve be performed to ensure that each metal binding site is saturated. Third, transition metals might bind to native metal binding sites on the protein and produce FRET (10). The contribution of native metal binding sites to the FRET measurements should be assessed in the absence of engineered sites. Last, transition metal ions can affect the activity and behavior of enzymes. These effects might occur either through the oxidation of cysteine residues or the direct binding of the ion to native sites. In this regard, the activity of the protein should be tested in the absence and presence of fluorophore and metals.
When large fluorophores with long linkers are used, determining the location of the fluorophore in space is nontrivial (29). Several methods have been developed to account for this uncertainty. These methods include Bayesian statistical methods and molecular dynamics modeling to calculate a region that the fluorophore attached to the protein will likely occupy during the excited-state lifetime (30, 31). Although these computational methods improve the ability to estimate the location of the fluorophore in space, an experimental method that precludes the use of computation to find backbone positions would be advantageous. In transition metal ion FRET, because both probes are small and attached near the peptide backbone, estimating the likely area the dye occupies is much simpler. Also, because of the close proximity between the dye and attachment site, transition metal ion FRET better reflects the structure of the backbone.
Although X-ray crystallography has provided high-resolution static images of many proteins, the conformational architecture and dynamics of most proteins in solution are largely unknown. Techniques to map the structure and dynamics of proteins in their native environment are required for a complete understanding of protein function. We anticipate that transition metal ion FRET will help to fill this gap in understanding by providing Å-scale distance mapping of proteins in solution or in membranes. This information should help reveal how proteins of known structure transition between various states. It also has the potential to aid in the high-resolution modeling of unknown protein structures. Indeed, using only a few distance constraints can substantially improve the ability of current computational methods to predict the structure of a protein (32). Future applications of transition metal ion FRET to single-molecule fluorescence and live cell imaging have the added potential to reveal the complex nature of protein mechanics in individual molecules in a cellular environment.
Methods
Peptides.
Peptides were synthesized with N-terminal acetylation and C-terminal amidation (Sigma or American Peptide) and diluted to a concentration of 20 μM in either Mops buffer (260 mM NaCl/60 mM Mops, pH 7.2) or Hepes buffer (520 mM NaCl/12 mM Hepes, pH 7.2). Single cysteine containing peptides were labeled with 100 μM cysteine-reactive fluorophore (mBBr, BC3M, or F5M; Invitrogen), for 2 h at room temperature. For peptides with two cysteines, 40 μM peptide was reacted with 2 μM fluorescent dye to ensure that no more than one cysteine per molecule was labeled. The remaining cysteines were reacted with an excess of cysteine-reactive metal chelator [400 μM of either N-[5(3′-maleimidopropylamido)-1-carboxypentyl]iminoacetic acid (MNTA) or [S-methanethiosulfonylcysteaminyl] ethylenediamine-N,N,N′,N′-tetraacetic acid (MTS-EDTA); Dojindo or Toronto Research Chemicals]. Labeled peptides were purified as a single peak from unincorporated dye by fluorescence size exclusion chromatography (Shimadzu) using a Superdex Peptide 10/300 column (GE Healthcare).
Spectroscopy and Fluorometry.
Fluorescently-labeled peptide was diluted in 4× fluorescence buffer (520 mM NaCl/12 mM Hepes, pH 7.2). For metal-binding curves, HPLC purified peptide was mixed with an equal volume of metal solutions and twice the volume of 2,2,2-trifluorethanol (TFE) (final concentration 50%) and kept on ice. Steady-state fluorescence emission spectra were acquired with a Fluorolog-3 fluorometer (Horiba). The absorbance of metals bound to unlabeled dihistidine model peptides (ACAAKAAAKHAAAHKA) at ratio 3:1 peptide/metal, or excess EDTA, or NTA were determined with a Beckman DU640 spectrophotometer.
Fluorescence Data Analysis.
Fluorescence values were averaged over a 9-nm window surrounding the peak of the spectrum. All spectra were blank-corrected and normalized to the fluorescence before the addition of metals. Fluorescence was corrected for the inner filter effect (4). Data for the control C2 peptides were fitted to a one-site binding curve model with the following equation to account for nonspecific solution quenching of the fluorophore:
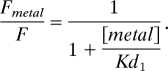 |
Fmetal is the fluorescence of the donor in the presence of metal, F is the fluorescence of the donor without metal, and Kd1 is the equilibrium dissociation constant for solution quenching. Data for the peptides containing engineered metal binding sites were fitted with a two-site binding curve model with the following equation:
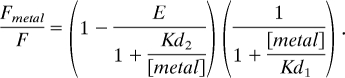 |
E is the FRET efficiency, and Kd2 is the equilibrium dissociation constant for the engineered metal binding sites. Kd1 was fixed at the same value measured for solution quenching in control peptides. The distances between the fluorophore and the metal were calculated by using the Förster equation:
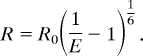 |
R is the distance between the fluorophore and the metal, and R0 is the Förster distance. R0 was calculated as previous described (2, 4, 10). To calculate the theoretical rate of energy transfer for FRET, the following equation was used:
where Kt is the rate of energy transfer and τD is the lifetime of bimane in the absence of acceptor (8 ns) (17). The experimental rate of energy transfer was calculated with the following equation:
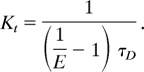 |
To calculate the theoretical rate of energy transfer for electron transfer, the following equation was used (4):
where Kt is the rate of energy transfer, Rc is the van der Waals contact distance (4 Å) between the two probes, and R is the distance between probes. The rate of 1013 s−1 was used as the maximum rate of energy transfer at van der Waals contact distances, and 1.4 Å−1 was used as the exponential coefficient of decay (β) (21).
Modeling and Structure Determination.
Models of α-helices, dyes, and metal ion binding sites were built with PYMOL (The PyMOL Molecular Graphics System, DeLano Scientific) and ViewerPro (Accelrys) from ideal α-helical models and a survey of crystal structures of metal-binding proteins from the Protein Data Bank. To model the position of mBBr, distances to each metal binding site were calculated from FRET. A location of the fluorophore was found by using the solver function of Excel (Microsoft) that produced the best fit by a least-squares analysis to all of the experimental data (28). To determine distances of backbone resides, β-carbons of each labeled residue were measured from the ideal helical model.
Supplementary Material
Acknowledgments.
We thank G. Sheridan for technical assistance. This work was supported by the Howard Hughes Medical Institute and National Institutes of Health Grant EY10329 (to W.N.Z.), National Eye Institute National Research Service Award Fellowship 5F32EY018981 (to M.C.P.), a Jane Coffin Childs Foundation fellowship (to J.W.T.), and National Institutes of Health Pathway to Independence Award 1K99NS064213 (to J.W.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905207106/DCSupplemental.
References
- 1.Forster T. Experimentelle und theoretische untersuchung des zwischenmolekularen ubergangs von elektronenanregungsenergie. Z Naturforsch A. 1949;4:321–327. [Google Scholar]
- 2.Selvin PR. Fluorescence resonance energy transfer. Methods Enzymol. 1995;246:300–334. doi: 10.1016/0076-6879(95)46015-2. [DOI] [PubMed] [Google Scholar]
- 3.Stryer L, Haugland RP. Energy transfer: A spectroscopic ruler. Proc Natl Acad Sci USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd Ed. New York: Springer; 2006. p. xxvi.p. 954. [Google Scholar]
- 5.Best RB, et al. Effect of flexibility and cis residues in single-molecule FRET studies of polyproline. Proc Natl Acad Sci USA. 2007;104:18964–18969. doi: 10.1073/pnas.0709567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakowicz JR, et al. Influence of end-to-end diffusion on intramolecular energy transfer as observed by frequency-domain fluorometry. Biophys Chem. 1990;38:99–109. doi: 10.1016/0301-4622(90)80044-8. [DOI] [PubMed] [Google Scholar]
- 7.Wiczk W, et al. Distance distributions recovered from steady-state fluorescence measurements on thirteen donor–acceptor pairs with different Forster distances. J Fluoresc. 1991;1:273–286. doi: 10.1007/BF00865251. [DOI] [PubMed] [Google Scholar]
- 8.Posson DJ, Selvin PR. Extent of voltage sensor movement during gating of shaker K+ channels. Neuron. 2008;59:98–109. doi: 10.1016/j.neuron.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dale RE, Eisinger J, Blumberg WE. The orientational freedom of molecular probes. The orientation factor in intramolecular energy transfer. Biophys J. 1979;26:161–193. doi: 10.1016/S0006-3495(79)85243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taraska JW, et al. Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat Methods. 2009;6:532–537. doi: 10.1038/nmeth.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosower NS, et al. Bimane fluorescent labels: Labeling of normal human red cells under physiological conditions. Proc Natl Acad Sci USA. 1979;76:3382–3386. doi: 10.1073/pnas.76.7.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figgis BN, Hitchman MA. Special Topics in Inorganic Chemistry. New York: Wiley; 2000. Ligand field theory and its applications; pp. xviii–354. [Google Scholar]
- 13.Horrocks WD, Jr, Holmquist B, Vallee BL. Energy transfer between terbium (III) and cobalt (II) in thermolysin: A new class of metal–metal distance probes. Proc Natl Acad Sci USA. 1975;72:4764–4768. doi: 10.1073/pnas.72.12.4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richmond TA, et al. Engineered metal binding sites on green fluorescence protein. Biochem Biophys Res Commun. 2000;268:462–465. doi: 10.1006/bbrc.1999.1244. [DOI] [PubMed] [Google Scholar]
- 15.Sandtner W, Bezanilla F, Correa AM. In vivo measurement of intramolecular distances using genetically encoded reporters. Biophys J. 2007;93:L45–L47. doi: 10.1529/biophysj.107.119073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marqusee S, Robbins VH, Baldwin RL. Unusually stable helix formation in short alanine-based peptides. Proc Natl Acad Sci USA. 1989;86:5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansoor SE, McHaourab HS, Farrens DL. Mapping proximity within proteins using fluorescence spectroscopy. A study of T4 lysozyme showing that tryptophan residues quench bimane fluorescence. Biochemistry. 2002;41:2475–2484. doi: 10.1021/bi011198i. [DOI] [PubMed] [Google Scholar]
- 18.Arnold FH, Haymore BL. Engineered metal-binding proteins: Purification to protein folding. Science. 1991;252:1796–1797. doi: 10.1126/science.1648261. [DOI] [PubMed] [Google Scholar]
- 19.Suh SS, Haymore BL, Arnold FH. Characterization of His-X3-His sites in α-helices of synthetic metal-binding bovine somatotropin. Protein Eng. 1991;4:301–305. doi: 10.1093/protein/4.3.301. [DOI] [PubMed] [Google Scholar]
- 20.Dexter DL. A theory of sensitized luminescence in solids. J Chem Phys. 1953;21:14. [Google Scholar]
- 21.Moser CC, et al. Nature of biological electron transfer. Nature. 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 22.Turro NJ. Modern Molecular Photochemistry. Menlo Park, CA: Benjamin/Cummings; 1978. [Google Scholar]
- 23.Dvoretsky A, Gaponenko V, Rosevear PR. Derivation of structural restraints using a thiol-reactive chelator. FEBS Lett. 2002;528:189–192. doi: 10.1016/s0014-5793(02)03297-0. [DOI] [PubMed] [Google Scholar]
- 24.Burmeister Getz E, Cooke R, Selvin PR. Luminescence resonance energy transfer measurements in myosin. Biophys J. 1998;74:2451–2458. doi: 10.1016/s0006-3495(98)77953-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakowicz JR, et al. Site-to-site diffusion in proteins as observed by energy-transfer and frequency-domain fluorometry. Photochem Photobiol. 1994;59:16–29. doi: 10.1111/j.1751-1097.1994.tb04996.x. [DOI] [PubMed] [Google Scholar]
- 26.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 27.Majumdar ZK, et al. Measurements of internal distance changes of the 30S ribosome using FRET with multiple donor-acceptor pairs: Quantitative spectroscopic methods. J Mol Biol. 2005;351:1123–1145. doi: 10.1016/j.jmb.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 28.Taraska JW, Zagotta WN. Structural dynamics in the gating ring of cyclic nucleotide-gated ion channels. Nat Struct Mol Biol. 2007;14:854–860. doi: 10.1038/nsmb1281. [DOI] [PubMed] [Google Scholar]
- 29.Deng HY, Odom OW, Hardesty B. Localization of L11 on the Escherichia coli ribosome by singlet–singlet energy transfer. Eur J Biochem. 1986;156:497–503. doi: 10.1111/j.1432-1033.1986.tb09608.x. [DOI] [PubMed] [Google Scholar]
- 30.Margittai M, et al. Single-molecule fluorescence resonance energy transfer reveals a dynamic equilibrium between closed and open conformations of syntaxin 1. Proc Natl Acad Sci USA. 2003;100:15516–15521. doi: 10.1073/pnas.2331232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muschielok A, et al. A nano-positioning system for macromolecular structural analysis. Nat Methods. 2008;5:965–971. doi: 10.1038/nmeth.1259. [DOI] [PubMed] [Google Scholar]
- 32.Zheng W, Brooks BR. Normal-modes-based prediction of protein conformational changes guided by distance constraints. Biophys J. 2005;88:3109–3117. doi: 10.1529/biophysj.104.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.