Abstract
Low-density lipoprotein receptor-related protein 1 (LRP1) is a multifunctional endocytic receptor that plays critical roles in the pathogenesis of several human diseases including tumor metastasis and Alzheimer’s disease. However, mechanisms that regulate LRP1 expression under physiological and pathophysiological conditions are not unclear. In human cell lines, we found that miR-205 down-regulates the expression of LRP1 by targeting sequences in the 3′UTR of LRP1 mRNA. This effect was abolished by deleting the miR-205 seed site in the 3′UTR of LRP1. The ectopic expression of miR-205 also significantly mitigated migration of both U87 and SK-LU-1 cells. These results, for the first time, demonstrate that expression of human LRP1 is regulated in part by a specific miRNA, leading to decreased tumor cell migration.
Keywords: microRNA, LRP1, cell migration, tumor
Introduction
The low-density lipoprotein receptor (LDLR)-related protein 1 (LRP1) is a large endocytic receptor that belongs to the LDLR family. LRP1 binds and endocytoses over 30 structurally and functionally distinct ligands, including apolipoprotein, proteinases, proteinase-inhibitor complexes, and extracellular matrix proteins such as matrix-metalloproteinases (MMPs) and urokinase-type plasminogen activator (uPA) [1]. Accumulating evidence suggests that altered expression of LRP1 is closely related to various diseases such as Alzheimer’s disease (AD), cancer, and atherosclerosis. For example, LRP1 plays a critical role in clearing amyloid-β (Aβ) peptide, the toxic species and principal component of senile plaques found in AD brains [2]. In a previous report, we showed that knockdown of LRP1 expression in human smooth muscle cells by siRNA resulted in significantly decreased cell migration [3]. We further showed that LRP1 promotes cancer cell migration and invasion by inducing the expression of matrix metalloproteinase (MMP) 2 and 9 [4]. In addition, mice lacking LRP1 in vascular smooth muscle cells exhibit hyperplasia of the aortic wall, disruption of the elastic layer, and increased susceptibility to developing atherosclerotic lesions [5]. Collectively, these results suggest that altered LRP1 expression could influence the pathogenesis of several human diseases. Indeed, reduced LRP1 expression is found in AD brains [6] and LRP1 overexpression has been described in several human cancers [7,8]. Despite these important observations, the cellular mechanisms regulating LRP1 expression are largely unknown.
MicroRNAs (miRNAs) are small noncoding RNAs that control post-transcriptional gene expression. Recent evidences demonstrate that some miRNAs are differentially expressed in AD brains and in tumor tissues [9,10]. Moreover, miRNAs are known to regulate key cellular events related to tumorigenesis such as metastasis, invasion, cellular proliferation, and apoptosis. Tavazoie et al. reported that expression of miR-126 and miR-335 are lost in metastatic breast cancer cells. Furthermore, overexpression of miR-126 and miR-335 in cancer cells decreases metastasis into lung and bone in vivo [11]. Conversely, miR-373 and miR-520c enhance tumor cell invasion and metastasis both in vitro and in vivo breast cancer models [12], which suggests that these miRNAs may target genes required to suppress cellular growth and metastasis.
In this study, we hypothesized that LRP1 expression is regulated by specific microRNAs, which act to suppress cellular migration and tumor progression. Herein, we present evidence that miR-205 down-regulates LRP1 level, resulting in decreased tumor cell migration.
Materials and Methods
Materials and microRNAs
Human α2-macroglobulin (α2M) was purified from human plasma and activated with methylamine (α2M*) as previously described [13]. Isolation of rabbit polyclonal anti-LRP1 antibody has been described previously [14]. Peroxidase-labeled anti-rabbit antibody and ECL system were from GE Healthcare. Carrier-free Na125I was purchased from Perkin Elmer Lifescience. pMIR-REPORT vector and pre-miR precursor molecules were obtained from Ambion. Pre-miR precursor molecules 205, 338-5p, and 545 (For convenience, the miR-xxx precursor molecule is termed miR-xxx throughout this article.) are small, chemically modified, double-stranded RNA molecules designed to mimic endogenous mature miRNAs in transfected cells. Scrambled oligonucleotides that do not produce identifiable effects on known miRNA function were used as negative control.
Reporter vectors and DNA constructs
Minireceptor of LRP1 (mLRP4) was described in previous report [15]. The 3′UTR region of LRP1 was subcloned into the end of the ORF of the Luciferase reporter vector and mLRP4. Vectors containing microRNA specific target sites were generated by site-directed mutagenesis. We utilized established methods [16] to clone these synthetic versions of putative miRNA target sites into a luciferase reporter gene (pMIR-REPORT; Ambion).
Cell culture and transfection
Human small cell lung cancer SK-LU-1, Human embryonic kidney 293 (HEK293), and glioblastoma U87 cells were cultured in Dulbecco’s minimum essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, and 1 mM sodium pyruvate. For transfection, cells were grown to 80% confluence and transfected with miRNAs using Lipofectamine2000 (Invitrogen) according to the manufacturer’s specifications. Forty-eight hours after transfection, cells were collected for migration assay, real-time PCR, and Western blotting.
Quantitative real-time PCR
Quantitative RT-PCR was carried out using SYBR Green reporter. Total RNA isolated using the RNeasy Mini Kit (Qiagen) was subsequently reverse transcribed to cDNA with the SuperScript First-strand Synthesis System (Invitrogen). The reaction mix was subjected to quantitative real-time PCR to detect levels of corresponding GAPDH and LRP1. GAPDH was used as an internal control for each specific gene. The relative levels of expression were quantified and analyzed using Bio-Rad iCycler iQ software. Three independent experiments were performed to analyze relative gene expressions and each sample was tested in triplicate. The real-time value for each sample was averaged and compared using the CT method. The amount of target RNA (2−ΔΔCT) was normalized to the endogenous GAPDH reference (ΔCT) and to the amount of target gene in the control sample, which was set as the calibrator at 1.0.
Western blotting
Cells were lysed in lysis buffer (phosphate-buffered saline (PBS) containing 1% Triton X-100, protease inhibitor cocktail, and 1 mM phenylmethylsulfonyl fluoride (PMSF)) at 4°C for 30 min. Equal quantities of protein were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and followed by transfer to Immobilon-P transfer membrane. Successive incubations with anti-LRP1 antibody or anti-actin antibody and horseradish peroxidase-conjugated secondary antibody were carried out to detect the immunoreactive proteins using the ECL system. Kodak Digital Science1D image analysis software was used for quantification.
Cell migration assay
Cell migration activities were examined by three-dimensional Boyden chamber assay and two-dimensional wound healing assay. Boyden chamber assay was carried out in 6.5-mm diameter transwell chambers with pore size of 8.0 m. Twenty-four hours after transfection with miRNAs, cells were resuspended in the migration medium of DMEM containing 0.1% BSA and 2 mM L-glutamine, and placed in the upper compartment of Transwell chambers coated with collagen I on the lower surface (5×104 cells in 100 μl). The lower compartment was filled with 600 μl of the same medium. After incubation for 6 h at 37°C, cells on the lower surface of the filter were fixed and stained, and five random fields/filter were counted at ×200 magnifications.
pMIR luciferase assay
Cells were co-transfected with the appropriate cDNAs: pMIR or pMIR-3′UTR of LRP1, and microRNAs or control microRNA. β-gal reporter construct was also co-transfected for normalizing transfection efficiency. Twenty-four hours after transfection, the luciferase activity and β-gal activity were measured by the Luciferase Assay System and β-gal Assay System, respectively, following the manufacturer’s instructions (Promega).
Statistical Analysis
All quantified data represent an average of at least triplicate samples. Error bars represent standard deviation of the mean. Statistical significance was determined by Student’s t-test and p < 0.05 was considered significant.
Results
LRP1 3′UTR suppresses reporter gene expression
To test whether the 3′UTR of LRP1 impacts gene expression, we appended the full-length 3′UTR of LRP1 to the end of coding sequence for a previously defined functional LRP1 minireceptor mLRP4 [15], and investigated whether this 3′UTR alone would affect the protein expression level of mLRP4. As shown in Fig. 1A, the protein expression of mLRP4 (mLRP4+3′UTR) was significantly decreased by >30% compared to mLRP4 control without this 3′UTR, in both HEK293 and SK-LU-1 lung cancer cells. Figure 1B represents densitometric analyses from three independent Western blots. To further confirm that the 3′UTR of LRP1 plays a role in regulating gene expression, the 3′UTR was cloned into the 3′ end of the luciferase ORF in pMIR-Luciferase Reporter vector. We observed a significant decrease in luciferase activity, ~30% and ~50% in HEK293 and SK-LU-1 cells, respectively, when transfected with pMIR+3′UTR vector (Fig. 1C). These results indicate that the expression level of LRP1 is regulated post-transcriptionally by its 3′UTR.
Fig. 1.
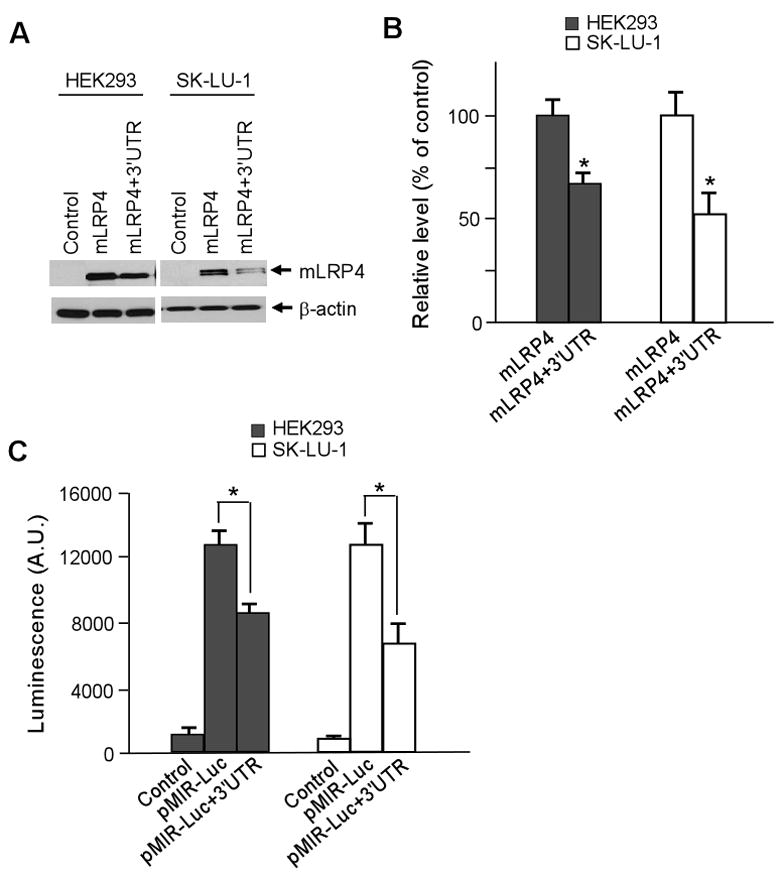
LRP1 3′UTR regulates target gene expression. A. The expression levels of mLRP4 without or with LRP1 3′UTR were analyzed by Western blot. Cells were co-transfected with HA-tagged mLRP4 or mLRP4 containing the 3′UTR of LRP1 and RAP. Cells were harvested 48 h later and analyzed for mLRP4 expression levels using HA antibody. B. Densitometric analysis of three independent triplicate Western blots. C. Luciferase reporter assays in cells expressing pMIR-Luc vector without or with LRP1 3′UTR. Cells were harvested 24 h later and analyzed for luciferase activity. β-gal expression vector was co-transfected for normalization of transfection efficiency. Values are the average of triple determinations with the S.D. indicated by error bars. *p < 0.05.
MiR-205 down-regulates endogenous LRP1 expression
To investigate whether microRNAs regulate LRP1 levels, we scanned 3′UTR of LRP1 for miRNA targeting sites using target miRNA prediction program, mirDB, and found that it contains seeding sites for hsa-mir-205, hsa-mir-338-5p, and hsa-mir-545, which were chosen due to having larger free energy among several candidates. The seed regions for these miRNAs to LRP1 3′UTR are shown in Fig. 2A. To test whether these miRNAs indeed regulate LRP1 expression, we used U87 glioblastoma and SK-LU-1 cells because they express relatively high levels of endogenous LRP1. MiRNAs and negative miRNA were transfected at a final concentration of 40nmol/L. In a previous study, we also found that the metastatic properties of U87 and SK-LU-1 cell lines are significantly influenced by LRP1 expression levels [4]. As shown in Fig. 2B, the endogenous levels of LRP1 were decreased compared to negative miRNA controls by >40% and >25% in mir-205-transfected U87 and SK-LU-1 cells, respectively. Our quantitative real-time PCR results showed that the mRNA levels of LRP1 were not altered by miR-205 transfection in U87 and SK-LU-1 cells, indicating that miR-205 suppresses gene expression by inhibiting translation rather than altering target gene mRNA levels. We observed no significant changes in LRP1 expression in miR338-5p or miR-545 transfected cells.
Fig. 2.
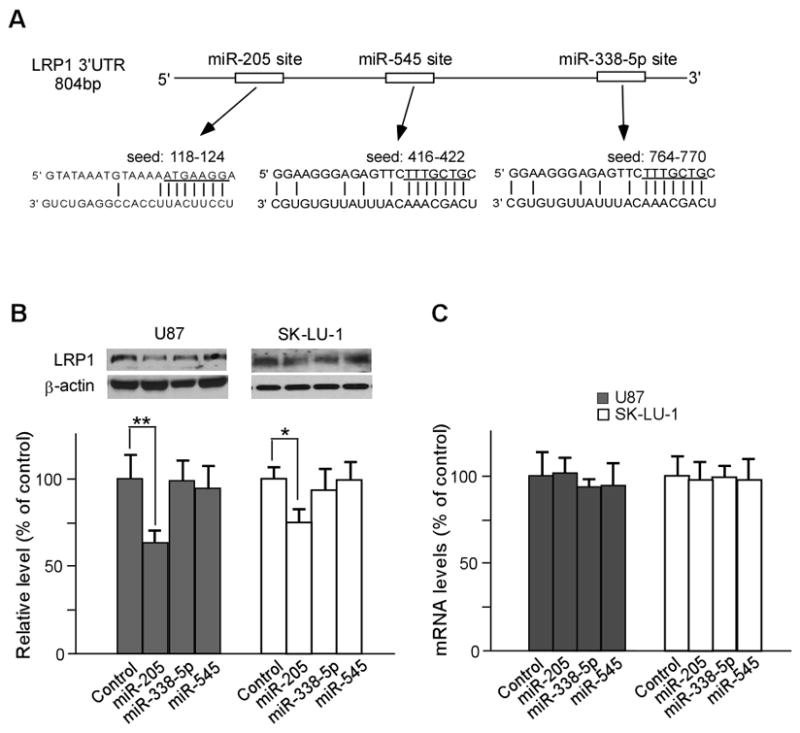
Mir-205 down-regulates the expression of endogenous LRP1 post-transcriptionally. A. miR-205, miR-338-5p, and miR-545 seed sites within LRP1 3′UTR are shown. B. The effects of ectopic expression of miRNAs on endogenous LRP1 expression. U87 and SK-LU-1 cells were transfected with each miRNA and the endogenous expression level of LRP1 was analyzed by Western blot. Densitometric analysis of three independent Western blots is shown by bar graph. C. The expression levels of LRP1 mRNA were determined by quantitative real-time PCR following miRNA overexpression. Values are the average of triple determinations with the S.D. indicated by error bars. *p < 0.05, **p < 0.01.
MiR-205 modulates cell migration by regulating LRP1 expression
To examine whether the miR-205-mediated down-regulation of LRP1 expression suppresses LRP1 function, ligand degradation assays were performed using an LRP1-specific ligand, α2M. Degradation of 125I-α2M was reduced by ~30% in cells transfected with miR-205, indicating that LRP1 expression and its endocytic function were significantly reduced in these cells (Fig. 3A).
Fig. 3.
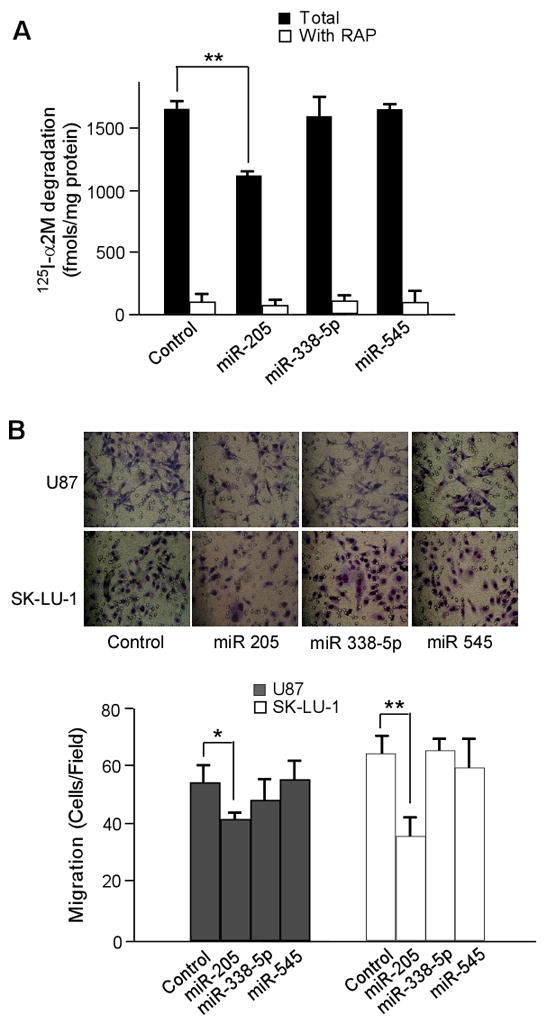
Ectopic expression of mir-205 decreases tumor cell migration. A. 125I-α2M (1 nM) uptake and degradation assays were performed at 37°C for 4 h in the absence or presence of RAP (500 nM). The degradation of 125I-α2M was analyzed as described in the Materials and Methods section. B. Twenty-four hours after overexpression of control or miRNA, three-dimensional migration assays were performed using Boyden chambers. Representative photos of migration are shown taken at x200 magnification under an inverted microscope. The migrated cells were counted from five randomly chosen fields. Values are the average of triple determinations with the S.D. indicated by error bars. *p < 0.05
To further investigate whether miR-205 affects U87 and SK-LU-1 cell migration through down-regulation of LRP1, we conducted three-dimensional cell migration assays using Matrigel pre-coated Transwell chambers. We found that miR-205 transfection into U87 and SK-LU-1 cells decreases cell migration approximately 25% in both cell types, compared to controls (Fig. 3B), indicating that miR-205 is a potential suppressor of tumor metastasis in glioblastoma and lung cancer cells.
Deletion of the miR-205 seed site in LRP1 3′UTR rescues target gene expression
To further confirm that the effect of miR-205 on LRP1 expression is specific, the miR-205 seed site in the 3′UTR of the mLRP4+3′UTR and pMIR+3′UTR constructs was deleted, and the expression of these target genes was tested. Deletion of the miR-205 seed site significantly restored the level of mLRP4 expression in both U87 and SK-LU-1 cells (Fig. 4A and B). In addition, luciferase activities of miR-205 deleted constructs were also restored in U87 and SK-LU-1 cells. As a control, we also deleted the seed site of miR-545 but did not observe any alteration in target gene expression (data not shown). These results support that LRP1 is specifically regulated by miR-205 (Fig. 4C).
Fig. 4.
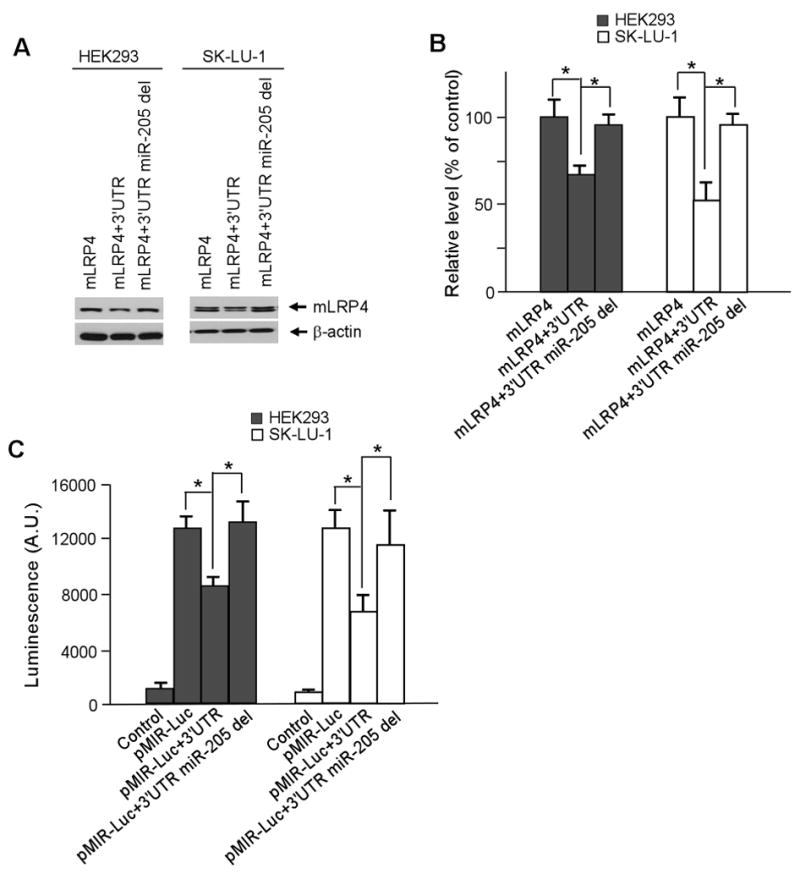
Deletion of MiR-205 seed site in LRP1 3′UTR restores target gene expression. A. Cells were transfected with mLRP4, mLRP4 containing LRP1 3′UTR, or mLRP4 containing LRP1 3′UTR with the miR-205 seed site deleted. Cells were harvested 48 h later and analyzed for expression levels by Western blot. B. Densitometric analysis of three independent Western blots. C. Luciferase assays were performed using cells expressing pMIR-Luc vector containing full-length or miR-205 seed site deleted sequences of LRP1 3′UTR. Cells were harvested 24 h later and analyzed for luciferase expression. β-gal expression vector was co-transfected for normalization of transfection efficiency. Values are the average of triple determinations with the S.D. indicated by error bars. *p < 0.05.
Discussion
In the present study, we provide evidence that miR-205, a specific microRNA directed towards the 3′UTR of LRP1 mRNA, down-regulates LRP1 expression, leading to a significant decrease in glioblastoma and lung cancer cell migration.
We utilized two complementary approaches to determine whether LRP1 expression is regulated by miRNA. The 3′UTR of LRP1 was fused to the 3′ end of mLRP4 or a luciferase reporter construct and the translational expression of both fusion constructs were determined by Western blotting or luciferase reporter assay, respectively. Both approaches demonstrated that LRP1 expression is regulated by this 3′UTR region, suggesting that there are specific miRNAs that suppress LRP1 protein expression level (Fig. 1). We observed similar results in both HEK293 and SK-LU-1 lung cancer cells, but found no difference in U87 glioblastoma cells (data not shown).
MiR-205 significantly decreased endogenous LRP1 expression in both U87 and SK-LU-1 cells, whereas miR-338-5p and miR-545 had no effect on LRP1 expression (Fig. 2). In contrast to HEK293 and SK-LU-1 lung cancer cells, the presence of LRP1 3′UTR did not alter target gene expression in U87 cells. However, overexpression of miR-205 significantly decreased LRP1 expression in U87 cells. A possible explanation for these results is that endogenous expression of miR-205 is lost or significantly decreased in U87 glioblastoma cells compared to other cell types. Indeed, miR-205 has been found to be either up-regulated [17] or down-regulated [18] in different tumor types. In addition, several lines of evidence suggest that miR-205 acts as an oncosuppressor in prostate cancer and breast cancer [19,20]. Since SK-LU-1, HEK293, and U87 cells are all transformed cell lines, it will be interesting to investigate the expression levels of miR-205 in human un-transformed cell lines and correlate its expression with metastatic properties as well as LRP1 expression in future study.
Although the role of LRP1 in tumor metastasis has been a subject of debate, we have recently shown that the endogenous levels of LRP1 in lung cancer cells and U87 glioblastoma cells positively correlates with the ability of these cells to invade and migrate in 3D Matrigel assays [4]. In the present study, we observed similar results through ectopic expression of miR-205 in both U87 and SK-LU-1 cells. As demonstrated by α2M degradation assays and cellular migration assays, inhibition of LRP1 expression by miR-205 leads to decreased LRP1 function in ligand uptake and cellular migration.
In conclusion, we have provided strong evidence that further supports a role for LRP1 in tumor cell migration and invasion, making LRP1 a promising therapeutic target to treat tumor metastases. Moreover, we have identified miR-205 as a potential tumor suppressor that modulates LRP1 expression. Future studies should be directly toward examining how miR-205 is regulated in human cancer and other human diseases, and whether both LRP1 and miR-205 are targets for therapy.
Acknowledgments
We thank Kathleen Baysac for careful reading of this manuscript. This work was supported by grants from the National Institutes of Health R01-AG027924 and R01-AG031784.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Herz J, Strickland DK. LRP1: a multifunctional scavenger and signaling receptor. J Clin Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deane R, Wu Z, Sagare A, Davis J, Du Yan S, Hamm K, Xu F, Parisi M, LaRue B, Hu HW, Spijkers P, Guo H, Song X, Lenting PJ, Van Nostrand WE, Zlokovic BV. LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron. 2004;43:333–344. doi: 10.1016/j.neuron.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Lu W, Bu G. Essential role of the low density lipoprotein receptor-related protein in vascular smooth muscle cell migration. FEBS Lett. 2003;555:346–350. doi: 10.1016/s0014-5793(03)01272-9. [DOI] [PubMed] [Google Scholar]
- 4.Song H, Li Y, Lee J, Schwartz AL, Bu G. Low-density lipoprotein receptor-related protein 1 promotes cancer cell migration and invasion by inducing the expression of matrix metalloproteinases 2 and 9. Cancer Res. 2009;69:879–886. doi: 10.1158/0008-5472.CAN-08-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boucher P, Gotthardt M, Li WP, Anderson RG, Herz J. LRP: role in vascular wall integrity and protection from atherosclerosis. Science. 2003;300:329–332. doi: 10.1126/science.1082095. [DOI] [PubMed] [Google Scholar]
- 6.Kang DE, Pietrzik CU, Baum L, Chevallier N, Merriam DE, Kounnas MZ, Wagner SL, Troncoso JC, Kawas CH, Katzman R, Koo EH. Modulation of amyloid β-protein clearance and Alzheimer’s disease susceptibility by the LDL receptor–related protein pathway Search. J Clin Invest. 2000;106:1159–1166. doi: 10.1172/JCI11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto M, Ikeda K, Oshshima K, Tsugo H, Kimura H, Tomonaga M. Increased expression of low density lipoprotein receptor-related protein/α2-macroglobulin receptor in human malignant astrocytomas. Cancer Res. 1997;57:2799–2805. [PubMed] [Google Scholar]
- 8.Bu G, Maksymovitch EA, Geuze H, Schwartz AL. Subcellular localization and endocytic function of low density lipoprotein receptor-related protein in human glioblastoma cells. J Biol Chem. 1994;269:29874–29882. [PubMed] [Google Scholar]
- 9.Lukiw WJ. Micro-RNA speciation in fetal, adult and alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 10.Kristen MN, Glen JW. MicroRNAs and cancer: past, present, and potential future. Mol Cancer Ther. 2008;7:3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 11.Tavazoie SF, Alarcón C, Oskarsson T, David P, Qiongqing W, Bos PD, Gerald WL, Joan M. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Q, Gumireddy K, Schrier M, Sage CI, Nagel R, Nair S, David AE, Li A, Huang G, Andres JK, Gimotty PA, Katsaros D, Coukos G, Zhang L, Pur E, Agami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 13.Williams SE, Ashcom JD, Argraves WS, Strickland DK. A novel mechanism for controlling the activity of alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein. Multiple regulatory sites for 39-kDa receptor-associated protein. J Biol Chem. 1992;267:9035–9040. [PubMed] [Google Scholar]
- 14.Bu G, Geuze HJ, Strous GJ, Schwartz AL. 39 kDa receptor-associated protein is an ER resident protein and molecular chaperone for LDL receptor-related protein. EMBO J. 1995;14:2269–2280. doi: 10.1002/j.1460-2075.1995.tb07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bu G, Rennke S. Receptor-associated protein is a folding chaperone for low density lipoprotein receptor-related protein. J Biol Chem. 1996;271:22218–22224. doi: 10.1074/jbc.271.36.22218. [DOI] [PubMed] [Google Scholar]
- 16.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human mirnas and indications for an involvement of miRNA in cell growth and apoptosis. Nucl Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iorio MV, Visone R, Di LG, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Mard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 18.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 19.Gandellini P, Folini M, Longoni N, Pennati M, Binda M, Colecchia M, Salvioni R, Supino R, Moretti R, Limonta P, Valdagni R, Daidone MG, Zaffaroni N. miR-205 exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cε. Cancer Res. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 20.Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, Mard S, Croce CM, Tagliabue E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]


