Abstract
Interaction of the cytoplasmic adaptor molecule β-arrestin2 with the activated parathyroid hormone (PTH)/PTHrP receptor inhibits G protein mediated signaling and triggers MAPKs signaling. In turn, the effects of both intermittent (i.) and continuous (c.) PTH on bone are altered in β-arrestin2-deficient (Arrb2−/−) mice. To elucidate the expression profile of bone genes responsive to PTH and targeted for regulation by β-arrestin2, we performed microarray analysis using total RNA from primary osteoblastic cells isolated from wild-type (WT) and Arrb2−/− mice. By comparing gene expression profiles in cells exposed to i.PTH, c.PTH or vehicle (Veh) for 2 weeks, we found that i.PTH specifically up-regulated 215 sequences (including β-arrestin2) and down-regulated 200 sequences in WT cells, about two thirds of them being under the control of β-arrestin2. In addition, β-arrestin2 appeared necessary to the down-regulation of a genomic cluster coding for small leucin-rich proteins (SLRPs) including osteoglycin, osteomodulin and asporin. Pathway analyses identified a main gene network centered on p38 MAPK and NFκB that requires β-arrestin2 for up- or down-regulation by i.PTH, and a smaller network of PTH-regulated genes centered on TGFB1, that is normally repressed by β-arrestin2. In contrast the expression of some known PTH gene targets regulated by the cAMP/PKA pathway was not affected by presence or absence of β-arrestin2 in osteoblasts. These results indicate that β-arrestin2 targets prominently p38 MAPK- and NFκB-dependent expression in osteoblasts exposed to i.PTH, and delineates new molecular mechanisms to explain the anabolic and catabolic effects of PTH on bone.
Keywords: PTH, bone, arrestin, osteoblast, gene expression
Introduction
β-arrestins (β-arrestin1 and β-arrestin2) are cytoplasmic multifunctional molecules that play a central role in the regulation of intracellular signaling by a variety of transmembrane receptors, primarily G protein-coupled receptors (GPCRs) [1]. β-arrestins are involved in several physiological and pharmacologic processes, including the β-adrenergic response [2], pain/opioid tolerance [3], allergic asthma, [4] and skeletal response to parathyroid hormone (PTH) [5–7]. Activation of the PTH/PTHrP receptor by its agonists recruits β-arrestins which bind to the phosphorylated C-terminal and third intracellular loop of the PTH/PTHrP receptor and uncouple the receptor from G proteins [8–10]. This results in desensitization of cAMP and IP3 signaling and in the activation of new intracellular signals such as MAPK ERKs [11, 12].
PTH fulfills at least two critically different physiological functions, the first being serum calcium homeostasis, the second being bone formation and mineralization [13]. Intermittent PTH (i.PTH), i.e. daily administration, increases bone mass, reduces fracture risk and is an approved treatment for osteoporosis [4, 14]. On the opposite, continuous PTH (c.PTH) exposure, i.e. hyperparathyroidism or PTH infusion, has predominantly catabolic effects, especially on cortical bone [15, 16]. Indeed, PTH directly stimulates osteoblast-mediated bone formation but also, through osteoblast-osteoclast coupling, bone resorption [17]. This PTH paradox is partly explained by PTH effects on the expression of receptor activator of nuclear factor kappa b (NFκB) ligand (RANKL), a non-redundant activator of osteoclast differentiation and activity, and its antagonist, osteoprotegerin (OPG), by osteoblasts [18–21].
In β-arrestin2-deficient (Arrb2−/−) mice, the response to i.PTH is complex, suggesting an increase of periosteal bone formation, but a predominant resorption at trabecular and endocortical bone surfaces [5–7]. In most circumstances, the catabolic response to PTH in Arrb2−/− mice is explained by an increased RANKL/OPG ratio compared to WT [6, 7], which may however be attenuated in estrogen replete-female mice [5].
These observations led us to hypothesize that β-arrestin2 regulates several genes critical for bone anabolism and catabolism in response to PTH. In this work, we used microarray analysis of primary osteoblast cultures from wild-type (WT) and Arrb2−/− mice, exposed intermittently or continuously to PTH to identify bone genes targeted for regulation by β-arrestin2. Our results identify new gene expression networks that are regulated by β-arrestin2 in response to intermittent PTH.
Materials and methods
Cell culture and PTH treatments
To assess the effects of PTH treatments on Arrb2−/− osteoblast differentiation, primary osteoblast cultures isolated from newborn calvaria were chronically exposed to PTH (10nM) either intermittently or continuously in a medium permissive to mineralization. For this purpose, cells were harvested by sequential collagenase type II (3mg/ml, Sigma-Aldrich) digestions of calvaria from 2–3 day-old WT or Arrb2−/− mice, half issued from male and half from female pups. Cells from the third to fifth digestions were pooled [22] and cultured in αMEM (Gibco), supplemented with 10% fetal calf serum (Amimed), antibiotics (penicillin 100U/ml, streptomycin 100µg/ml, Gibco), glutamine (200mM, Gibco), amino acids (Amimed) and amphotericin B (0.25µg/ml, Amimed). At confluence cells were split and plated in at 6000 cells/cm2. After 3 days, cells were incubated in fresh medium supplemented by β-glycerophosphate (10µM, Sigma) and treated with bovine PTH (1–34) (10nM, Sigma) intermittently (i.e incubation with PTH during 6 hours, wash out with medium and incubation for the next 42 hours in medium without PTH) or continuously (i.e incubation in medium with PTH during 48 hours) or vehicle. These different treatment cycles of 48 hours were repeated in fresh medium supplemented by β-glycerophosphate during 2 weeks.
Alizarin Red staining
Calcium deposition was revealed by Alizarin Red staining. Cells were washed twice with cold PBS (Gibco), fixed for 10 minutes in 70% ethanol before staining with 0.5% Alizarin Red S (Sigma) in water (pH 4) for 30 seconds. Following staining, the cells were washed three times with water and once with 70% ethanol.
RNA isolation from cells
Total RNA from WT and Arrb2−/− primary osteoblasts was extracted after 15 days of PTH treatments, 6 hours after the last exposure to PTH using peqGold Trifast (peQLab Biotechnologie GmbH). Total RNA was finally purified on mini-columns (RNeasy Mini kit, Qiagen) in combination with a deoxyribonuclease treatment (RNase-free DNase Set, Qiagen) to avoid DNA contamination. The quality of each sample was verified with a 2100 bioanalyzer (Agilent Technologies AG, Basel).
Microarray analysis
Microarray analysis was performed to identify the genes regulated by β-arrestin2 in primary i.PTH-differentiated osteoblasts, as compared to poorly differentiated cells treated with vehicle and c.PTH. Gene expression profiles (n=3/condition) were carried out with Affymetrix Mouse Genome 430A 2.0 arrays (approximately 14000 genes/chip). Technically, double-stranded cDNA was synthesized from 5µg of total RNA from each sample according to the GeneChip Expression Analysis Technical Manual (Affymetrix), using the SuperScript Choice system (Invitrogen). Following a phenol/chloroform extraction and ethanol precipitation cDNA was used as template in an in vitro transcription reaction to synthesize a biotin-labeled cRNA with the BioArray HighYield RNA Transcript Labeling Kit (Enzo Biochem). After purification (RNeasy Mini Kit, Qiagen) and quantification, 10–15 µg of cRNA were fragmented. Each fragmented cRNA (6.5µg) was then hybridized to the GeneChip. Hybridization, washing and scanning were performed according to the Affymetrix manual. Arrays were visualized on an Agilent 2500 GeneArray scanner, and image files were processed using Affymetrix GeneChip® Operating Software (GCOS). The percentage of the probe sets called “Present” relative to the total of 22690 probe sets ranged from 57.6% to 62.9% with an average of 60.3%. Differences in gene expression levels were evaluated by Affymetrix GCOS and GeneSpring GX 7.3 software (Silicon Genetics, Agilent Technologies AG, Basel) was used to screen for differentially expressed genes. Each of the experimental samples in WT or Arrb2−/− (i.PTH or c.PTH) was compared with each of the reference samples (vehicle), resulting in 9 pairwise comparisons. This approach based on the Mann-Whitney pairwise comparison test allows the ranking of results by concordance as well as the calculation of significance (p value) of each identified change in gene expression [23, 24]. Genes for which the concordance in the pairwise comparisons exceeded a threshold (i.e. 60%) were considered to be statistically significant. This conservative analytical approach was used to limit the number of false-positives. A 77% cut off in consistency of change (at least 7 of 9 comparisons were either increased or decreased) was then applied to identify PTH-regulated genes. PTH-regulated genes/sequences were defined as follows: i) gene expression was detectable (called “present”) in all 3 PTH- and/or 3 vehicle-treated samples, ii) the average level of gene expression in PTH-treated samples was at least 1.5-fold higher or lower than in vehicle-treated samples, iii) gene expression levels differed by ≥1.5 fold between PTH and vehicle-treated samples in at least 7 out of 9 comparisons.
According to the MIAME guidelines [25], the complete microarray dataset was deposited in the public data repository of the European Bioinformatics Institute (ArrayExpress) with accession number E-TABM-624.
Comparison of PTH-regulated genes in WT and Arrb2−/− cells
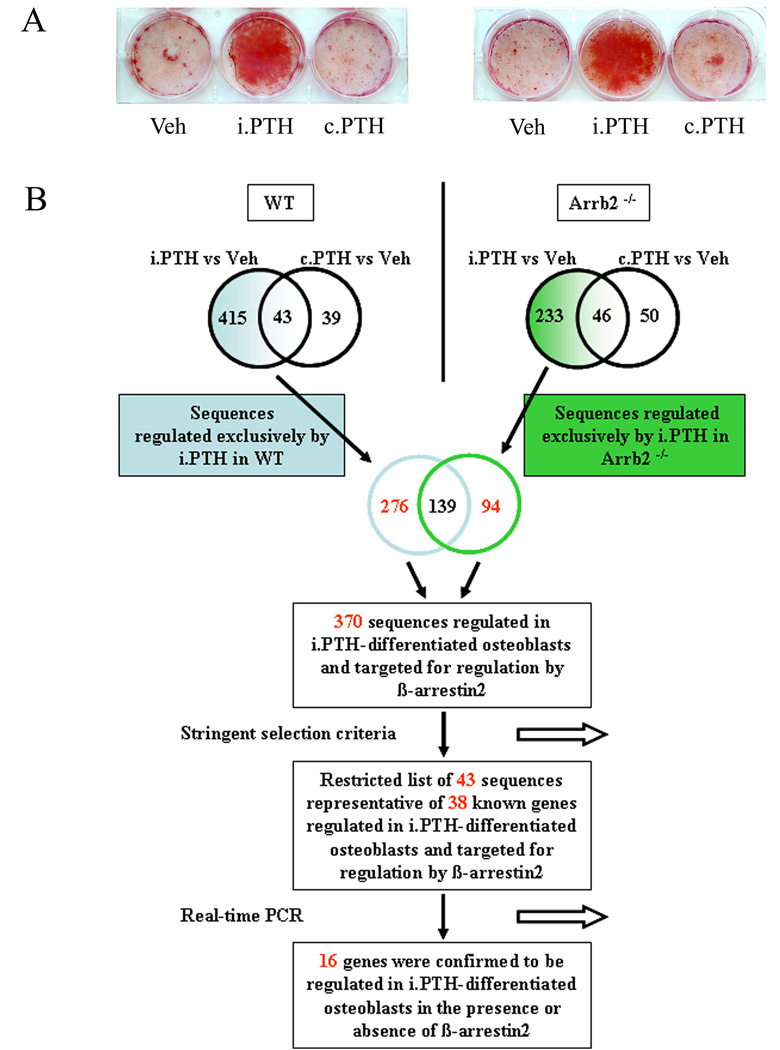
To identify sequences regulated uniquely by i.PTH, i.PTH- and c.PTH-regulated gene expression profiles were compared in WT and Arrb2−/− osteoblasts, respectively (accordingly to the previously defined rule). Thus, sequences regulated solely by c.PTH or regulated by both i.PTH and c.PTH were excluded. Then, sequences regulated uniquely by i.PTH in WT and Arrb2−/− cells were compared to identify those targeted by β-arrestin2. Hence, the sequences regulated specifically by i.PTH in both WT and Arrb2−/− were excluded (accordingly to the previously defined rule). In this case, a sequence/gene which expression increased or decreased ≥1.5 fold in i.PTH-treated WT cells, but <1.5 fold in Arrb2−/− cells, would be called to be targeted for regulation by β-arrestin2.
Additional selection criteria
One of the limitations of GCOS analysis which could lead to false positive results is the fact that a gene up-regulated (respectively down-regulated) by i.PTH in 8/9 comparisons in one group (i.e. WT or Arrb2−/−) and 6/9 comparisons in the other group (i.e. Arrb2−/− or WT) would be called to be differentially regulated. To refine our analysis, we set up the following additional criteria to establish that a gene was definitely differentially regulated by i.PTH in WT and Arrb2−/− osteoblasts: i) a ≥1.5 fold increase of gene expression by i.PTH in WT osteoblasts (or Arrb2−/−, respectively) when i.PTH-induced changes in Arrb2−/− cells (or WT, respectively) are ≤1.2 fold or the sequence is down-regulated (i.e. ≥-1.5 fold decrease of gene expression), ii) alternatively, a ≥-1.5 fold decrease of gene expression by i.PTH in WT osteoblasts (or Arrb2−/−, respectively) when the decrease in Arrb2−/− cells (or WT, respectively) is ≤-1.2 fold or the sequence is up-regulated (i.e. ≥1.5 fold increase of gene expression).
Quantitative real-time PCR
Single-stranded cDNA templates were carried out using SuperScript II Reverse Transcriptase (Invitrogen AG, Basel) following the manufacturer’s instructions. Quantitative real-time PCR were performed using pre-designed TaqMan® Gene Expression Assays (for references see Supplemental Table 1) made of two unlabeled primers and a FAM™ dye-labeled TaqMan® MGB probe, and the correspondent buffer TaqMan® Universal PCR Master Mix (Applied Biosystems, Rotkreuz, Switzerland). A Biomek 2000 robot (Beckman Coulter, Nyon, Switzerland) was used for liquid handling (10µl) in 384-well plates with 3 replicates per sample. The cDNA was PCR amplified in a 7900HT SDS System and raw threshold-cycle (Ct) values were obtained from SDS 2.0 software (Applied Biosystems, Rotkreuz, Switzerland). Relative quantities (RQ) were calculated with the formula RQ=E-Ct using an efficiency (E) of 2 by default. For each gene the highest quantity was arbitrarily designed as a value of 1.0. The mean quantity was calculated from triplicates for each sample and this quantity was normalized (as a ratio) to the similarly calculated mean quantity of the β2-microglobulin normalization gene (B2m). Finally, fold changes (treated versus vehicle samples) were calculated using normalized quantities, as for the results of microarray experiment (see Microarray analysis).
Results
Gene expression profile of continuous versus intermittent PTH in WT osteoblasts
Primary osteoblasts isolated from WT mouse calvariae and exposed to i.PTH for two weeks mineralized in vitro, as judged by Alizarin Red staining, whereas vehicle- and c.PTH-treated cells did not (Fig. 1A). Accordingly gene expression profiling revealed that 458 sequences were regulated in i.PTH-treated WT osteoblasts compared to only 82 sequences in c.PTH-treated WT cells, resulting in 415 sequences uniquely regulated in i.PTH-differentiated osteoblasts; 43 sequences regulated in both i.PTH- and c.PTH-treated cells; and 39 sequences regulated solely in osteoblasts exposed to c.PTH (Fig. 1B, Supplemental table 2 for detailed gene lists). The sequence corresponding to the Arrb2 gene it-self was up-regulated +2.4 fold in WT osteoblasts exposed to i.PTH (Table 2) and the maximal level of changes in gene expression by i.PTH were of +25 and −3.9 fold for Ramp3 and Cys1, respectively. Classification into Gene Ongology (GO) biological process categories revealed that the sequences regulated uniquely by i.PTH were significantly associated to bone-related terms (skeletal development, ossification and bone remodeling) and cell growth, whereas it was not the case for the sequences regulated solely by c.PTH or commonly regulated by both treatments (Table 1).
Fig.1. Osteoblast differentiation and gene expression in response to PTH.
(A) Effects of intermittent PTH (i.PTH), continuous PTH (c.PTH) or vehicle (Veh) on mineralization of primary osteoblasts isolated from WT and Arrb2−/− mice. Mineralization was revealed by Alizarin Red staining. (B) Scheme representing the step-by-step approach used to delineate the sequences/genes regulated by i.PTH and targeted for regulation by β-arrestin2. See Materials and methods section for details.
Table 2.
Sequences and corresponding genes regulated by i.PTH and targeted for regulation by β-arrestin2.
| Probe Set | Gene Symbol | Description | Fold change i.PTH vs Vehicle | |
|---|---|---|---|---|
| WT | Arrb2−/− | |||
| 1451987_at | Arrb2* | arrestin, beta 2 | +2.4 (+1.9) | - |
| 1436905_x_at | Laptm5 | lysosomal-associated protein transmembrane 5 | + 1.9 | +1.2 |
| 1423768_at | Unc93b1* | unc-93 homolog B1 (C. elegans) | + 1.8 (+1.8) | +1.1 (+1.2) |
| 1420361_at | Slc11a1* | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | + 1.7 (+1.8) | +1.0 (+1.1) |
| 1448883_at | Lgmn* | Legumain | + 1.7 (+1.5) | +1.1 (1.0) |
| 1448417_at | Ninj1* | ninjurin 1 | + 1.7 | +1.1 |
| 1425423_at | Glis1* | GLIS family zinc finger 1 | + 1.7 | +1.1 |
| 1417856_at | Relb* | avian reticuloendotheliosis viral (v-rel) oncogene related B | + 1.6 | +1.1 |
| 1423233_at | Cebpd* | CCAAT/enhancer binding protein (C/EBP), delta | + 1.6 (+1.4) | +1.1 (−1.4) |
| 1450355_a_at | Capg | capping protein (actin filament), gelsolin-like | + 1.6 | +1.2 |
| 1430700_a_at | Pla2g7* | phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) | + 1.6 (+1.7) | +1.2 (+1.1) |
| 1452414_s_at | Ccdc86 (D19ERTD678E) | Coiled-coil domain containing 86 | + 1.6 | +1.2 |
| 1423871_at | Tmem63a (BC014795) | Transmembrane protein 63a | + 1.6 | +1.2 |
| 1418841_s_at | Cdc2l1* | cell division cycle 2-like 1 | + 1.6 | +1.2 |
| 1416468_at | Aldh1a1 | aldehyde dehydrogenase family 1, subfamily A1 | − 1.5 | −1.2 |
| 1460258_at | Lect1 | leukocyte cell derived chemotaxin 1 | − 1.5 | −1.2 |
| 1426945_at | Ranbp5* | RAN binding protein 5 | − 1.5 | −1.1 |
| 1419666_x_at | Nupr1* | nuclear protein 1 | − 1.5 | −1.1 |
| 1454610_at | Sept7* | septin 7 | − 1.6 (-1.5) | +1.0 (+1.3) |
| 1422243_at | Fgf7* | fibroblast growth factor 7 | − 1.6 | +1.1 |
| 1419663_at | Ogn* | Osteoglycin | − 1.7 (−1.8) | +1.2 (+1.9) |
| 1450647_at | Hps3* | Hermansky-Pudlak syndrome 3 homolog (human) | − 1.7 | −1.1 |
| 1460224_at | Snx2 | sorting nexin 2 | − 1.7 | −1.1 |
| 1416652_at | Aspn* | Asporin | − 1.7 (−1.6) | +1.0 (+1.3) |
| 1416483_at | Ttc3* | tetratricopeptide repeat domain 3 | −1.7 (−1.5) | −1.0 (1.0) |
| 1425491_at | Bmpr1a* | bone morphogenetic protein receptor, type 1A | −1.7 (−1.5) | −1.1 (−1.2) |
| 1425829_a_at | Steap4 (Tnfaip9)* | STEAP family member 4 | − 1.7 (−1.5) | −1.1 (+1.3) |
| 1423919_at | BC023882 | cDNA sequence BC023882 | − 1.8 | +1.3 |
| 1450889_at | Hltf (Smarca3)* | helicase-like transcription factor | − 1.8 (−1.7) | −1.0 (+1.1) |
| 1425493_at | Bmpr1a* | bone morphogenetic protein receptor, type 1A | − 1.8 (−1.5) | −1.2 (−1.2) |
| 1450739_at | Tbl1xr1* | transducin (beta)-like 1X-linked receptor 1 | − 1.9 | −1.2 |
| 1418745_at | Omd* | Osteomodulin | − 1.9 (−1.7) | +1.1 (1.0) |
| 1421871_at | Sh3bgr*l | SH3-binding domain glutamic acid-rich protein like | − 1.9 (−1.5) | +1.1 (+1.5) |
| 1419662_at | Ogn* | Osteoglycin | − 1.9 (−1.8) | −1.0 (+1.9) |
| 1450537_at | Mid2* | midline 2 | − 2.3 | −1.2 |
| 1416318_at | Serpinb1a* | serine (or cysteine) proteinase inhibitor, clade B, member 1a | − 1.0 (1.0) | +1.8 (+1.8) |
| 1450846_at | Bzw1* | basic leucine zipper and W2 domains 1 | + 1.0 | + 1.8 |
| 1423040_at | Bzw1* | basic leucine zipper and W2 domains 1 | + 1.1 | + 1.7 |
| 1450845_a_at | Bzw1* | basic leucine zipper and W2 domains 1 | + 1.1 | + 1.7 |
| 1452058_a_at | Rnf11* | ring finger protein 11 | − 1.2 | − 1.5 |
| 1455812_x_at | Vasn (Slitl2) | vasorin | − 1.2 | − 1.6 |
| 1449370_at | Sox4* | SRY-box containing gene 4 | − 1.2 | − 1.7 |
| 1417654_at | Sdc4* | syndecan 4 | + 1.2 | − 2.2 |
This table shows a limited number of genes that were differentially regulated in WT and Arrb2−/− osteoblasts exposed to i.PTH, according to most stringent criteria for comparison. Genes with an asterisk were further investigated by quantitative real-time PCR. Fold changes in bold represent significant differences in gene expression upon i.PTH treatment by microarray analysis, with in parenthesis fold changes confirmed by real-time PCR. (for details, see Materials and methods section).
Table 1.
Gene Ontology (GO) sub-categories significantly influenced by PTH according to the mode of exposure.
| Biological process (GO:8150) | ||||
|---|---|---|---|---|
| Treatment | Development (GO:7275) | Physiological process (GO:7582) | Growth (GO:40007) | Response to stimulus (GO:50896) |
| i.PTH | -regulation of cell size c | -immune response c | -cell growth c | -response to chemical stimuli c |
| -vascular development c | -ossification a | -defense response c | ||
| -skeletal development b | -bone remodeling a | -response to external stimuli c | ||
| i.PTH & c.PTH | -glial cell differentiation b | -copper ion transport c | -defense response a | |
| -segmentation a | -death a | |||
| -thyroid hormone catabolism a | ||||
| c.PTH | -copper ion transport b | -response to wounding a | ||
| -inflammatory response a | -response to external stimuli a | |||
| -cytosolic calciumion homeostasis a | -chemotaxis a | |||
The list of sequences regulated uniquely by i.PTH, c.PTH or both in WT osteoblast cultures was subjected to Gene Ontology analysis. The mentioned categories are the most significant and relevant with regard to bone biology.
p<0.05
p<0.001
p<0.0001.
The list of 415 and 39 sequences regulated uniquely by i.PTH and c.PTH, respectively were also subjected to Ingenuity Pathway Analysis (IPA) bioinformatic approach. This analysis resulted in a large number of canonical pathways possibly associated with i.PTH-regulated genes in osteoblasts, including actin cytoskeleton signaling, VDR/RXR activation and integrin signaling (Fig. 2A). In contrast, the genes uniquely regulated by c.PTH lead to only one significant association with a chemokine signaling pathway (Fig. 2B).
Fig. 2. Relevant canonical pathways associated with the 415 and the 39 sequences regulated uniquely by i.PTH (A) and c.PTH (B) in WT osteoblasts.
The threshold line across the bars represents the point where for a given pathway the significance value for the observation is of 0.05. The ratio represents the number of molecule of interest (in a specific list) divided by the total number of molecules in the pathway. Ref: IPA 7.0; Ingenuity Systems, Mountain View, CA, web site: http://www.ingenuity.com/products/pathways_analysis.html.
Profiling of β-arrestin2-regulated genes in osteoblasts
Primary osteoblasts isolated from Arrb2−/− calvariae exposed to i.PTH for two weeks mineralized in vitro, whereas cells exposed to c.PTH or vehicle treatments did not, as observed in WT osteoblasts (Fig.1A). In differentiated osteoblasts from Arrb2−/− mice, 233 sequences were identified to be exclusively regulated by i.PTH versus c.PTH, i.e. −44% less than observed in WT cells. By comparing the sequences uniquely regulated by i.PTH in WT and Arrb2−/− cells, we identified 139 common sequences, i.e. regulated independently of β-arrestin2; 276 sequences regulated solely in the presence of β-arrestin2, i.e. requiring β-arrestin2 for up- or down-regulation by PTH; and 94 sequences regulated solely in absence of β-arrestin2, i.e. which expression was normally repressed by β-arrestin2 (Fig. 1B). In total, 370 sequences regulated by i.PTH were targeted for regulation by β-arrestin2, 189 positively and 181 negatively. Moreover, upon the 189 sequences positively regulated, 83% of the sequences were up-regulated in presence of β-arrestin2, while only 17% of the sequences were up-regulated in its absence, indicating that β-arrestin2-mediated signaling events exert a predominantly positive control on i.PTH-induced gene expression.
Clustering and gene ontology (GO) analyses
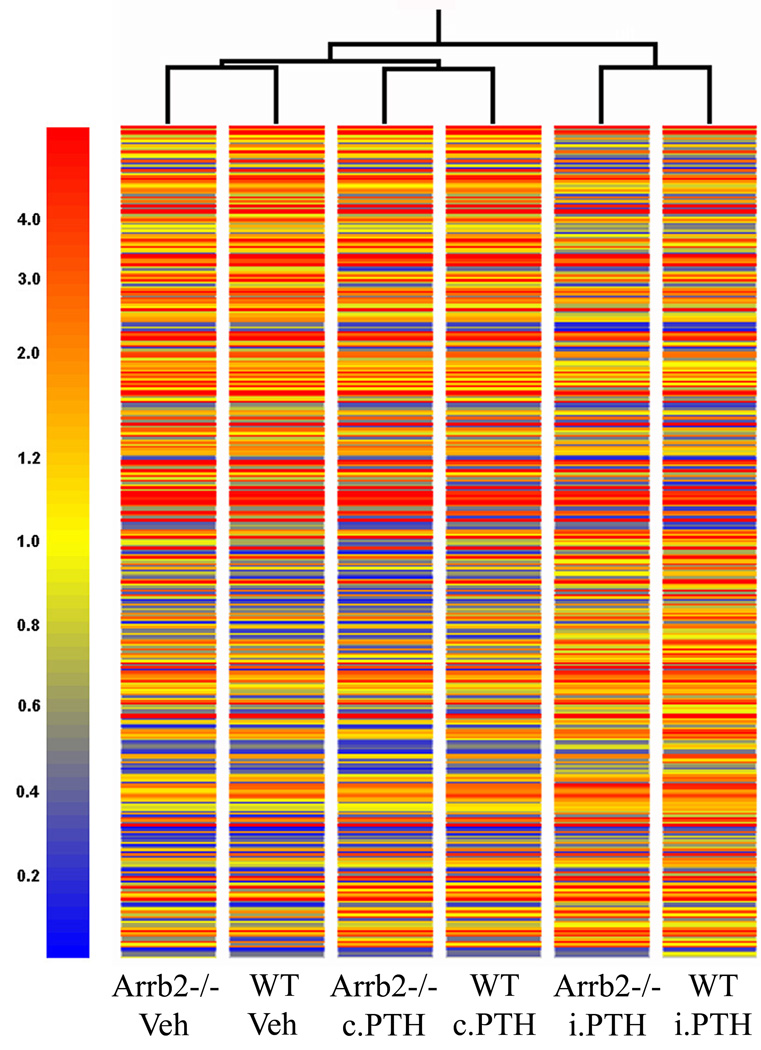
To further investigate the osteoblastic genes targeted for regulation by β-arrestin2, we performed in silico clustering analysis of the 370 β-arrestin2-targeted sequences up- or down-regulated by i.PTH. This analysis first indicated that the differential display of these 370 sequences is greater between i.PTH- and c.PTH- treated cells (or vehicle-treated cells) than between WT and Arrb2−/− osteoblasts (Fig. 3). Moreover, a number of genes known to respond to i.PTH in osteoblasts were similarly up-regulated (Tnfsf11 (RANKL), Igf1, Vdr, Alpl (ALP)) or down-regulated (Tnfrsf11b (OPG), Pth1r) in presence or absence of β-arrestin2 (data not shown). Taken together with a similar pattern of mineralization in WT and Arrb2−/− cultures treated with i.PTH (Fig. 1A), these data suggested that β-arrestin2 does not primarily target genes involved in the terminal differentiation of osteoblasts in vitro, whereas the mode of exposure to the hormone remains the primary determinant of its effects on osteoblast differentiation.
Fig.3. Gene tree clustering analysis of 370 β-arrestin2-regulated sequences in response to i.PTH.
This analysis grades the gene expression profiles according to their resemblance, with short branches indicating more similar profiles and long branches less similar profiles. The colorscale is according to the scaled signal of Affymetrix microarray experiment (mean genes expression value equal 1.0 for each GeneChip, in yellow). Higher than mean gene expression tends to the red color, whereas lower than mean gene expression tends to the blue color. This gene tree cluster was generated with the GeneSpring GX 7.3 software (Silicon Genetics, Agilent Technologies AG, Basel).
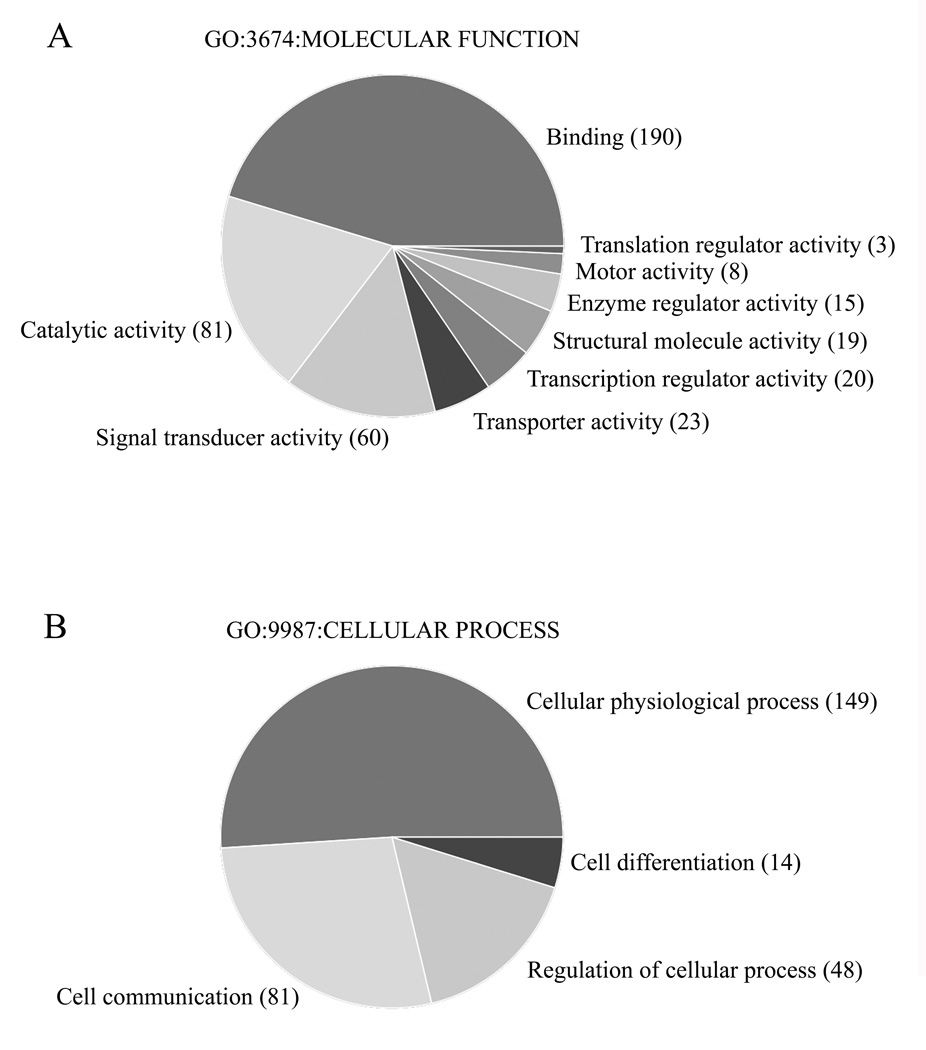
The 370 sequences were then classified into GO molecular function categories. As shown in figure 4A, the sequences were principally involved in binding (i.e. mostly protein, nucleic acid and nucleotide binding), catalytic activity (i.e. mostly transferase, hydrolase and oxydoreductase activity) and signal transducer activity (i.e mostly receptor activity and binding), possibly reflecting induction of physiological process and activation of cell-cell communication (Fig. 4B). (For detailed gene distribution in GO categories, see the supplemental Tables 3A and 3B).
Fig.4. Gene Ontology (GO) categories of 370 β-arrestin2-regulated sequences in response to i.PTH.
(A) GO categories by Molecular functions (ref. GO:3674). (B) GO categories by Cellular processes (ref. GO:9987). Numbers in parenthesis represent the number of sequences found in the respective sub-categories. These pie charts were generated with the GeneSpring GX 7.3 software (Silicon Genetics, Agilent Technologies AG, Basel).
Pathway Analysis
Next, we subjected the list of 370 differentially-regulated sequences in presence or absence of β-arrestin2 to additional selection criteria (see Materials and methods), resulting in a restricted list of 35 sequences (32 known genes) regulated by i.PTH only in presence of β-arrestin2 and 8 sequences (6 genes) regulated in its absence (Table 1). Of note, none of the genes that were up- or down-regulated more than 2.5 fold by i.PTH in WT cells were differentially regulated by β-arrestin2 accordingly to the additional selection criteria, which is consistent with the role of β-arrestin2 to modulate intracellular signaling (and in contrast with the on-off effects of a transcription factor on gene expression, for instance).
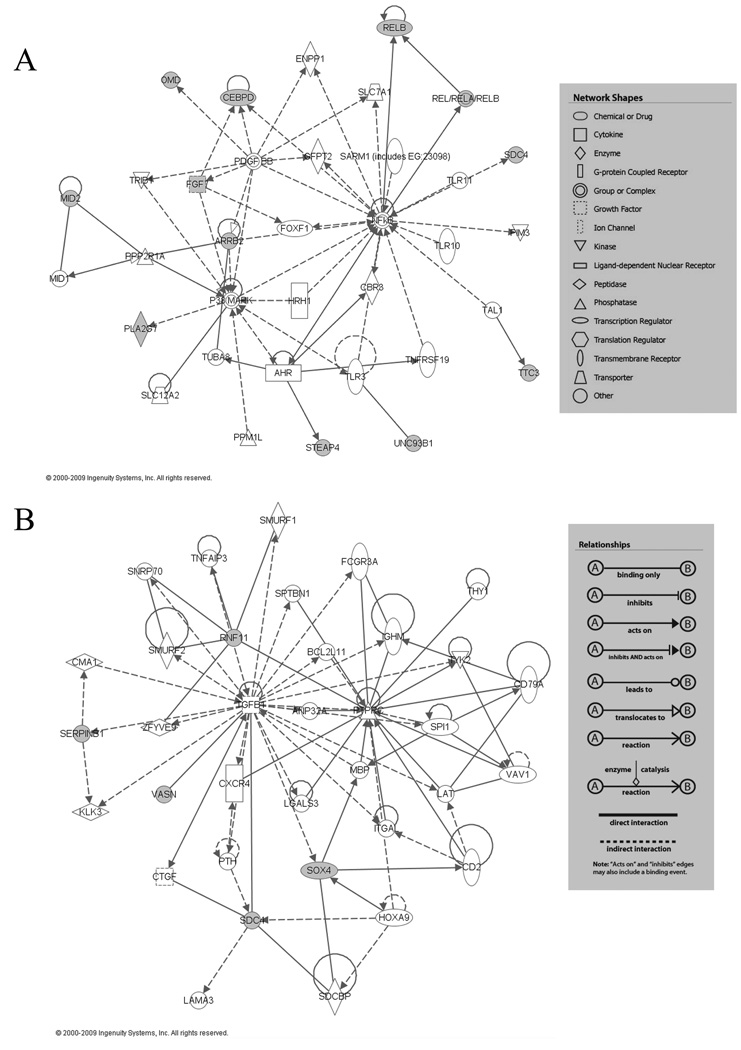
We used the Ingenuity Pathway Analysis (IPA) bioinformatic approach to set up potential functional networks based on these 38 genes (IPA 7.0; Ingenuity Systems, Mountain View, CA). This analysis resulted in a main network containing 12 targeted genes, centered on NFκB and p38 MAPK and including the Arrb2 gene itself (Fig. 5A), which were either up- or down-regulated by i.PTH in presence of β-arrestin2. A separate analysis of the 6 genes regulated in i.PTH-differentiated cells in absence of β-arrestin2, resulted in a network containing 5 of the 6 focused genes, centered on TGFB1 (Fig. 5B).
Fig.5. Principal networks of β-arrestin2-regulated genes in response to i.PTH.
(A) Network based on the 38 genes up- or down-regulated by i.PTH in presence/absence of β-arrestin2, centered on p38 MAPK and NFκB genes. (B) Network based on the 6 genes regulated by i.PTH in absence βarrestin2, centered on TGFB1. Closed symbols represent differentially regulated genes identified in the microarray analysis (A: Arrb2, Cebpd, Fgf7, Mid2, Omd, Pla2g7, Relb, Sdc4, Steap4, Ttc3 and Unc93b1, B: Rnf11, Serpinb1, Sdc4, Sox4 and Vasn). For gene name description see Table 2. Boxes contain the legends for relationship between genes and network shapes. Ref: IPA 7.0; Ingenuity Systems, Mountain View, CA, web site: http://www.ingenuity.com/products/pathways_analysis.html.
Real-time PCR analyses
Based on the microarray analysis, 30 genes of potential biological interest were further investigated by quantitative real-time PCR (see genes with an asterisk in Table 2). Approximately half of them were confirmed to be differentially regulated by i.PTH in WT and Arrb2−/− osteoblasts (see fold changes in parenthesis in Table 2). In addition, out of the 12 genes of interest contained in the NFκB and p38 MAPK network, 9 were confirmed to be regulated by i.PTH, including Arrb2 itself, and targeted for regulation by β-arrestin2, whereas among the 5 target genes involved in the TGFB1 network, only one gene (i.e. Serpinb1a) was confirmed by real-time PCR.
Other genes were confirmed to require β-arrestin2 for up-regulation in i.PTH-differentiated osteoblasts: Slc11a1, also called Nramp (for natural resistance-associated macrophage protein) that belongs to a family of proton-coupled transporters facilitating the cellular absorption of divalent ions; Unc93b1, a multi-transmembrane-domain-containing protein having an essential role in signaling by the nucleotide-sensing Toll-like receptors in the immune response; Pla2g7, an enzyme involved in lipoprotein metabolism and inflammatory pathways; legumain (Lgmn), a cysteine endopeptidase known to be involved in the regulation of osteoclast activity; and Cebpd, a transcription factor critical for normal cellular differentiation and metabolic functions in a variety of tissues. In contrast, the up-regulation of Serpinb1a gene by i.PTH was repressed in presence of β-arrestin2. Other genes were confirmed to require β-arrestin2 for down-regulation in i.PTH-differentiated osteoblasts: Sept7, a GTP-binding protein; Ttc3, a gene of unknown function mapped to the Down syndrome critical region; Steap4, a metalloreductase playing a role in cellular import of iron and copper involved in inflammation and insulin resistance; Bmpr1a, a receptor for the bone morphogenetic proteins well expressed in bone; Sh3bgrl, that encodes for a small protein of unknown function; Hltf, that belongs to the SWI/SNF protein family of transcription factor. In addition, among the genes that require β-arrestin2 to be down-regulated by i.PTH, 3 genes belong to the Small Leucine Rich Proteoglycans (SLRPs) family, namely osteoglycin/mimecan (Ogn), osteomodulin/osteoadherin (Omd) and asporin (Aspn), and are located in a gene cluster on human chromosome 9 and on the syntenic mouse chromosome 13. This family of molecules is known to be important for collagen fibrillogenesis, cellular growth, differentiation and migration.
Discussion
Exposure of primary osteoblastic cultures to i.PTH, i.e. 6 hours every 48 hour, or c.PTH, results in profound differences of osteoblast differentiation, as previously shown by Ishizuya et al. and confirmed in our study [26]. Hence this model provides a valuable tool to analyze the gene expression profile associated with osteoblast differentiation in response to PTH. The array of genes regulated in WT osteoblasts in response to i.PTH versus c.PTH, revealed that 83.5% of the sequences were specifically regulated by i.PTH, whereas only 7.8% of them were regulated uniquely by c.PTH. By comparing these results with array of genes regulated by i.PTH versus c.PTH in WT and Arrb2−/− osteoblasts, we made three major findings. First, about two thirds of i.PTH-regulated genes is targeted for regulation by β-arrestin2, either positively or negatively. However, β-arrestin2 has little/no influence on osteoblast mineralization in vitro, nor on the PTH- and cAMP/PKA-dependent expression of some well known gene targets related to osteoblast differentiation. Hence none of the genes which expression was prominently altered by i.PTH (i.e. more than 2.5 fold increased of decreased) was confirmed to significantly differ by real-time PCR, consistently with the modulating function of β-arrestin2 on intracellular signaling (contrasting with transcription factors). Second, gene network analyses suggest a key role for β-arrestin2 in the regulation of i.PTH responsive genes centered on p38 MAPK and NFκB genes. These pathway analyses also suggest that β-arrestin2 may act as a repressor of i.PTH-stimulated TGFB1-related gene expression. Third, β-arrestin2 is crucial for the down-regulation of specific genes in response to i.PTH, including a cluster of 3 SLRPs matrix proteins genes (Ogn, Omd and Aspn).
Two previously published in vivo experiments studied the gene expression profiles induced by i.PTH and c.PTH in trabecular bone of female rats [27, 28]. More than 50% of the c.PTH regulated-genes were identical in both studies [27, 28]. However, Onyia et al. found that only 9% of the genes were solely regulated by i.PTH [28] whereas Li et al. reported that 35% of the genes were regulated specifically by i.PTH [27]. The small number of genes regulated by i.PTH in one study [28] may be explained by the longer delay between the last PTH injection and RNA extraction, as a majority of genes could be back to their basal expression levels 24 hours post-treatment. In our in vitro study where cells were harvested 6 hours after the last exposure to PTH, about 80% of PTH-induced genes were specifically regulated by i.PTH, whereas only 8% of the genes were regulated uniquely by c.PTH. Thus our findings are consistent with a broader activation of bone-related gene expression shortly after exposure to PTH. Interstingly, two canonical pathways, i.e. integrin signaling and PDGF signaling, were potentially associated with i.PTH in both our in vitro study and in vivo experiment [27].
We previously demonstrated that absence of β-arrestin2 was sufficient to increase PTH-stimulated cAMP accumulation in osteoblasts, consistent with the role of β-arrestin2 in uncoupling the PTH/PTHrP receptor from G proteins [6]. It is currently admitted that cAMP/PKA is the main PTH signaling pathway for stimulation of osteoblast differentiation. This was confirmed by the bone anabolic effects of PTH/PTHrP receptor partial agonists that specifically activate the cAMP/PKA pathway, respectively by the absence of anabolic effects of partial agonists of the PLC/IP3 cascade [29–31]. Here we found that absence of β-arrestin2 neither affected in vitro osteoblast mineralization, nor the expression of some genes associated with osteoblast differentiation by i.PTH. Interestingly, these genes including Tnfsf11 (RANKL), Igf1, Vdr, Alpl (ALP), Tnfrsf11b (OPG), and Pth1r are all known to be regulated by PTH in a cAMP/PKA-dependent manner [32–36]. This suggests that inhibition of cAMP production is not a major mechanism for β-arrestin2-mediated regulation of gene expression in osteoblasts. Alternatively, sufficient regulation of cAMP signaling by β-arrestin1 in these cells could alleviate the effects of β-arrestin2-deficiency [9].
Downstream signaling by the PTH/PTHrP receptor also involves mitogen-activated protein kinases (MAPKs) ERK1/2, p38 and JNKs in osteoblasts [36–38]. Moreover, PTH can activate two independent pathways leading to ERK1/2 activation: an early G protein-dependent pathway mediated through PKA/PKC, and a G protein-independent pathway mediated by β-arrestin2 [36, 39]. Furthermore, a PTH-biased ligand that selectively stimulates the β-arrestin2-dependent, G proteinin-dependent, ERK1/2 signaling pathway, has the ability to induce bone formation in WT, but not in Arrb2−/− mice [40]. Consistent with the latter observations, we found that β-arrestin2 regulates a MAPK gene network in response to PTH. These results also suggest a role for the β-arrestin2-MAPK signaling cascade in bone anabolism.
In addition to p38 MAPK, pathway analyses also implicated NFκB in the expression network of β-arrestin2-regulated genes. β-arrestin2 has previously been described as an inhibitor of signal-induced IκBα degradation, leading to subsequent activation of NFκB [41]. NFκB is a key transcription factor involved in bone remodeling and osteoclastogenesis in response to RANKL [42]. We previously showed that RANKL production is increased in primary osteoblasts isolated from Arrb2−/− mice [7]. NFκB might therefore contribute to PTH-induced bone resorption. Our results confirm and extend these observations, by delineating an array of NFκB-related genes targeted for regulation by β-arrestin2 in osteoblasts. Of note, we used primary osteoblasts isolated from a 50:50 ratio of male and female newborn mice. Hence the in vivo regulation of RANKL (and others) gene expression by β-arrestin2 may be further influenced by the level of gonadal steroids [43].
This microarray analysis further indicates a role for β-arrestin2 in the regulation of genes coding for matrix proteins. Hence, the expression of a SLRPs gene cluster containing Ogn, Omd and Aspn was down-regulated in presence of β-arrestin2 in i.PTH-differentiated osteoblasts. Consistent with this coordinated regulation, Tasheva et al. previously suggested that this gene cluster might be regulated through a common cis-regulatory homeodomain and runt domain [44]. Thus this SLRPs gene cluster might be repressed through the action of RUNX2, a key transcription factor in osteoblast differentiation already shown to play a key role in the regulation of proteogylcans [45].
Although the gene expression of only one among the five genes of interest included in the TGFB1 network was confirmed by real-time PCR to be repressed in presence of β-arrestin2, some insights from the literature strongly support a repressive role of β-arrestin2 on TGFB1 signaling. Thus, TGFB1 is known to promote early stages of differentiation with the synthesis of extracellular matrix proteins [46] and inhibition of Arrb2 expression in vivo was shown to enhance TGFB1 signaling in primary keratinocytes [47]. Our results now suggest that this phenomenon also occurs in osteoblasts exposed to i.PTH. In addition to its positive role on osteoblast chemotaxis, proliferation and early stages of differentiation, TGFB1 is implicated in pre-osteoclast recruitment and differentiation to mature osteoclasts, therefore enhancing bone remodeling [46]. Taken together, our results suggest that enhanced TGFB1 signaling could contribute to the low bone mass phenotype and i.PTH-induced alterations of bone remodeling observed in Arrb2−/− compared to WT mice [5, 6].
In summary, these findings confirm and extend the critical role of β-arrestin2 in the regulation of osteoblastic gene expression mediated by p38 MAPK and NFκB and provide new gene targets to elucidate the molecular signals of PTH anabolic and catabolic effects on bone. Further work is needed to directly investigate the role of these specific genes on bone metabolism.
Supplementary Material
Acknowledgements
We thank Ms Fanny Cavat and Ms Madeleine Lachize for their technical assistance with mice and cell cultures and Dr Dominique Pierroz for proofreading the manuscript. Real-time PCR, microarray experiments and data analyses were performed with the support from the Genomics Platform, NCCR program "Frontiers in Genetics", University of Geneva, CH-1211 Geneva 4. (web site: http://www.frontiers-in-genetics.org/genomics.htm).
This work was supported by the Swiss National Foundation, grant number 3100AO-116633/1 and the NIH, grant number NIH-RO1 AR049265-04.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lefkowitz RJ. G protein-coupled receptors. III. New roles for receptor kinases and beta-arrestins in receptor signaling and desensitization. J Biol Chem. 1998;273:18677–18680. doi: 10.1074/jbc.273.30.18677. [DOI] [PubMed] [Google Scholar]
- 2.Lefkowitz RJ. Historical review: a brief history and personal retrospective of seven-transmembrane receptors. Trends Pharmacol Sci. 2004;25:413–422. doi: 10.1016/j.tips.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Bohn LM, Gainetdinov RR, Lin FT, Lefkowitz RJ, Caron MG. Mu-opioid receptor desensitization by beta-arrestin-2 determines morphine tolerance but not dependence. Nature. 2000;408:720–723. doi: 10.1038/35047086. [DOI] [PubMed] [Google Scholar]
- 4.Walker JK, Fong AM, Lawson BL, Savov JD, Patel DD, Schwartz DA, Lefkowitz RJ. Beta-arrestin-2 regulates the development of allergic asthma. J Clin Invest. 2003;112:566–574. doi: 10.1172/JCI17265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouxsein ML, Pierroz DD, Glatt V, Goddard DS, Cavat F, Rizzoli R, Ferrari SL. beta-Arrestin2 regulates the differential response of cortical and trabecular bone to intermittent PTH in female mice. J Bone Miner Res. 2005;20:635–643. doi: 10.1359/JBMR.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrari SL, Pierroz DD, Glatt V, Goddard DS, Bianchi EN, Lin FT, Manen D, Bouxsein ML. Bone response to intermittent parathyroid hormone is altered in mice null for {beta}-Arrestin2. Endocrinology. 2005;146:1854–1862. doi: 10.1210/en.2004-1282. [DOI] [PubMed] [Google Scholar]
- 7.Pierroz DD, Rufo A, Bianchi EN, Glatt V, Capulli M, Rucci N, Cavat F, Rizzoli R, Teti A, Bouxsein ML, Ferrari SL. beta-Arrestin2 Regulates RANKL and Ephrins Gene Expression in Response to Bone Remodeling in Mice. J Bone Miner Res. 2008 doi: 10.1359/JBMR.081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bisello A, Chorev M, Rosenblatt M, Monticelli L, Mierke DF, Ferrari SL. Selective ligand-induced stabilization of active and desensitized parathyroid hormone type 1 receptor conformations. J Biol Chem. 2002;277:38524–38530. doi: 10.1074/jbc.M202544200. [DOI] [PubMed] [Google Scholar]
- 9.Ferrari SL, Behar V, Chorev M, Rosenblatt M, Bisello A. Endocytosis of ligand-human parathyroid hormone receptor 1 complexes is protein kinase C-dependent and involves beta-arrestin2. Real-time monitoring by fluorescence microscopy. J Biol Chem. 1999;274:29968–29975. doi: 10.1074/jbc.274.42.29968. [DOI] [PubMed] [Google Scholar]
- 10.Mannstadt M, Juppner H, Gardella TJ. Receptors for PTH and PTHrP: their biological importance and functional properties. Am J Physiol. 1999;277:F665–F675. doi: 10.1152/ajprenal.1999.277.5.F665. [DOI] [PubMed] [Google Scholar]
- 11.Rey A, Manen D, Rizzoli R, Caverzasio J, Ferrari SL. Proline-rich motifs in the parathyroid hormone (PTH)/PTH-related protein receptor C terminus mediate scaffolding of c-Src with beta-arrestin2 for ERK1/2 activation. J Biol Chem. 2006;281:38181–38188. doi: 10.1074/jbc.M606762200. [DOI] [PubMed] [Google Scholar]
- 12.Sneddon WB, Friedman PA. Beta-arrestin-dependent parathyroid hormone-stimulated extracellular signal-regulated kinase activation and parathyroid hormone type 1 receptor internalization. Endocrinology. 2007;148:4073–4079. doi: 10.1210/en.2007-0343. [DOI] [PubMed] [Google Scholar]
- 13.Karaplis AC, Goltzman D. PTH and PTHrP effects on the skeleton. Rev Endocr Metab Disord. 2000;1:331–341. doi: 10.1023/a:1026526703898. [DOI] [PubMed] [Google Scholar]
- 14.Neer RM, Arnaud CD, Zanchetta JR, Prince R, Gaich GA, Reginster JY, Hodsman AB, Eriksen EF, Ish-Shalom S, Genant HK, Wang O, Mitlak BH. Effect of parathyroid hormone (1–34) on fractures and bone mineral density in postmenopausal women with osteoporosis. N Engl J Med. 2001;344:1434–1441. doi: 10.1056/NEJM200105103441904. [DOI] [PubMed] [Google Scholar]
- 15.Lotinun S, Evans GL, Bronk JT, Bolander ME, Wronski TJ, Ritman EL, Turner RT. Continuous parathyroid hormone induces cortical porosity in the rat: effects on bone turnover and mechanical properties. J Bone Miner Res. 2004;19:1165–1171. doi: 10.1359/JBMR.040404. [DOI] [PubMed] [Google Scholar]
- 16.Silverberg SJ, Shane E, de la Cruz L, Dempster DW, Feldman F, Seldin D, Jacobs TP, Siris ES, Cafferty M, Parisien MV, et al. Skeletal disease in primary hyperparathyroidism. J Bone Miner Res. 1989;4:283–291. doi: 10.1002/jbmr.5650040302. [DOI] [PubMed] [Google Scholar]
- 17.Qin L, Raggatt LJ, Partridge NC. Parathyroid hormone: a double-edged sword for bone metabolism. Trends Endocrinol Metab. 2004;15:60–65. doi: 10.1016/j.tem.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Goltzman D. Interactions of PTH and PTHrP with the PTH/PTHrP receptor and with downstream signaling pathways: exceptions that provide the rules. J Bone Miner Res. 1999;14:173–177. doi: 10.1359/jbmr.1999.14.2.173. [DOI] [PubMed] [Google Scholar]
- 19.Iida-Klein A, Zhou H, Lu SS, Levine LR, Ducayen-Knowles M, Dempster DW, Nieves J, Lindsay R. Anabolic action of parathyroid hormone is skeletal site specific at the tissue and cellular levels in mice. J Bone Miner Res. 2002;17:808–816. doi: 10.1359/jbmr.2002.17.5.808. [DOI] [PubMed] [Google Scholar]
- 20.Khosla S. Minireview: the OPG/RANKL/RANK system. Endocrinology. 2001;142:5050–5055. doi: 10.1210/endo.142.12.8536. [DOI] [PubMed] [Google Scholar]
- 21.Martin TJ, Sims NA. Osteoclast-derived activity in the coupling of bone formation to resorption. Trends Mol Med. 2005;11:76–81. doi: 10.1016/j.molmed.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Luben RA, Wong GL, Cohn DV. Biochemical characterization with parathormone and calcitonin of isolated bone cells: provisional identification of osteoclasts and osteoblasts. Endocrinology. 1976;99:526–534. doi: 10.1210/endo-99-2-526. [DOI] [PubMed] [Google Scholar]
- 23.Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- 24.Liu WM, Mei R, Di X, Ryder TB, Hubbell E, Dee S, Webster TA, Harrington CA, Ho MH, Baid J. Smeekens SP. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- 25.Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M, Kemmeren P, Lara GG. Oezcimen A, Rocca-Serra P, Sansone SA. ArrayExpress--a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishizuya T, Yokose S, Hori M, Noda T, Suda T, Yoshiki S, Yamaguchi A. Parathyroid hormone exerts disparate effects on osteoblast differentiation depending on exposure time in rat osteoblastic cells. J Clin Invest. 1997;99:2961–2970. doi: 10.1172/JCI119491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Liu H, Qin L, Tamasi J, Bergenstock M, Shapses S, Feyen JH, Notterman DA, Partridge NC. Determination of dual effects of parathyroid hormone on skeletal gene expression in vivo by microarray and network analysis. J Biol Chem. 2007;282:33086–33097. doi: 10.1074/jbc.M705194200. [DOI] [PubMed] [Google Scholar]
- 28.Onyia JE, Helvering LM, Gelbert L, Wei T, Huang S, Chen P, Dow ER, Maran A, Zhang M, Lotinun S, Lin X, Halladay DL, Miles RR, Kulkarni NH, Ambrose EM, Ma YL, Frolik CA, Sato M, Bryant HU, Turner RT. Molecular profile of catabolic versus anabolic treatment regimens of parathyroid hormone (PTH) in rat bone: an analysis by DNA microarray. J Cell Biochem. 2005;95:403–418. doi: 10.1002/jcb.20438. [DOI] [PubMed] [Google Scholar]
- 29.Armamento-Villareal R, Ziambaras K, Abbasi-Jarhomi SH, Dimarogonas A, Halstead L, Fausto A, Avioli LV, Civitelli R. An intact N terminus is required for the anabolic action of parathyroid hormone on adult female rats. J Bone Miner Res. 1997;12:384–392. doi: 10.1359/jbmr.1997.12.3.384. [DOI] [PubMed] [Google Scholar]
- 30.Hilliker S, Wergedal JE, Gruber HE, Bettica P, Baylink DJ. Truncation of the amino terminus of PTH alters its anabolic activity on bone in vivo. Bone. l1996;19:469–477. doi: 10.1016/s8756-3282(96)00230-x. [DOI] [PubMed] [Google Scholar]
- 31.Rixon RH, Whitfield JF, Gagnon L, Isaacs RJ, Maclean S, Chakravarthy B, Durkin JP, Neugebauer W, Ross V, Sung W, et al. Parathyroid hormone fragments may stimulate bone growth in ovariectomized rats by activating adenylyl cyclase. J Bone Miner Res. l1994;9:1179–1189. doi: 10.1002/jbmr.5650090807. [DOI] [PubMed] [Google Scholar]
- 32.Fu Q, Jilka RL, Manolagas SC, O'Brien CA. Parathyroid hormone stimulates receptor activator of NFkappa B ligand and inhibits osteoprotegerin expression via protein kinase A activation of cAMP-response element-binding protein. J Biol Chem. 2002;277:48868–48875. doi: 10.1074/jbc.M208494200. [DOI] [PubMed] [Google Scholar]
- 33.Huening M, Yehia G, Molina CA, Christakos S. Evidence for a regulatory role of inducible cAMP early repressor in protein kinase a-mediated enhancement of vitamin D receptor expression and modulation of hormone action. Mol Endocrinol. 2002;16:2052–2064. doi: 10.1210/me.2001-0260. [DOI] [PubMed] [Google Scholar]
- 34.Koh AJ, Beecher CA, Rosol TJ, McCauley LK. 3',5'-Cyclic adenosine monophosphate activation in osteoblastic cells: effects on parathyroid hormone-1 receptors and osteoblastic differentiation in vitro. Endocrinology. 1999;140:3154–3162. doi: 10.1210/endo.140.7.6872. [DOI] [PubMed] [Google Scholar]
- 35.McCarthy TL, Centrella M, Canalis E. Cyclic AMP induces insulin-like growth factor I synthesis in osteoblast-enriched cultures. J Biol Chem. 1990;265:15353–15356. [PubMed] [Google Scholar]
- 36.Rey A, Manen D, Rizzoli R, Ferrari SL, Caverzasio J. Evidences for a role of p38 MAP kinase in the stimulation of alkaline phosphatase and matrix mineralization induced by parathyroid hormone in osteoblastic cells. Bone. 2007;41:59–67. doi: 10.1016/j.bone.2007.02.031. [DOI] [PubMed] [Google Scholar]
- 37.Doggett TA, Swarthout JT, Jefcoat SC, Jr, Wilhelm D, Dieckmann A, Angel P, Partridge NC. Parathyroid hormone inhibits c-Jun N-terminal kinase activity in rat osteoblastic cells by a protein kinase A-dependent pathway. Endocrinology. l2002;143:1880–1888. doi: 10.1210/endo.143.5.8759. [DOI] [PubMed] [Google Scholar]
- 38.Swarthout JT, Doggett TA, Lemker JL, Partridge NC. Stimulation of extracellular signal-regulated kinases and proliferation in rat osteoblastic cells by parathyroid hormone is protein kinase C-dependent. J Biol Chem. 2001;276:7586–7592. doi: 10.1074/jbc.M007400200. [DOI] [PubMed] [Google Scholar]
- 39.Gesty-Palmer D, Chen M, Reiter E, Ahn S, Nelson CD, Wang S, Eckhardt AE, Cowan CL, Spurney RF, Luttrell LM, Lefkowitz RJ. Distinct beta-arrestin- and G protein-dependent pathways for parathyroid hormone receptor-stimulated ERK1/2 activation. J Biol Chem. l2006;281:10856–10864. doi: 10.1074/jbc.M513380200. [DOI] [PubMed] [Google Scholar]
- 40.Gesty-Palmer D, Spurney RF, Flannery PJ, Yuan L. The PTH 1 receptor (PTHR1) Stimulates Bone Formation Through a Distinct β-Arrestin Dependent Pathway Independent of G Protein Activation. J Bone Miner Res. l2008;23:S64. [Google Scholar]
- 41.Gao H, Sun Y, Wu Y, Luan B, Wang Y, Qu B, Pei G. Identification of beta-arrestin2 as a G protein-coupled receptor-stimulated regulator of NF-kappaB pathways. Mol Cell. 2004;14:303–317. doi: 10.1016/s1097-2765(04)00216-3. [DOI] [PubMed] [Google Scholar]
- 42.Chen F, Castranova V, Shi X, Demers LM. New insights into the role of nuclear factor-kappaB, a ubiquitous transcription factor in the initiation of diseases. Clin Chem. 1999;45:7–17. [PubMed] [Google Scholar]
- 43.Eghbali-Fatourechi G, Khosla S, Sanyal A, Boyle WJ, Lacey DL, Riggs BL. Role of RANK ligand in mediating increased bone resorption in early postmenopausal women. J Clin Invest. 2003;111:1221–1230. doi: 10.1172/JCI17215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tasheva ES, Klocke B, Conrad GW. Analysis of transcriptional regulation of the small leucine rich proteoglycans. Mol Vis. 2004;10:758–772. [PubMed] [Google Scholar]
- 45.Teplyuk NM, Haupt LM, Ling L, Dombrowski C, Mun FK, Nathan SS, Lian JB, Stein JL, Stein GS, Cool SM, van Wijnen AJ. The osteogenic transcription factor Runx2 regulates components of the fibroblast growth factor/proteoglycan signaling axis in osteoblasts. J Cell Biochem. l2009;107:144–154. doi: 10.1002/jcb.22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janssens K, ten Dijke P, Janssens S, Van Hul W. Transforming growth factor-beta1 to the bone. Endocr Rev. 2005;26:743–774. doi: 10.1210/er.2004-0001. [DOI] [PubMed] [Google Scholar]
- 47.Chen W, Kirkbride KC, How T, Nelson CD, Mo J, Frederick JP, Wang XF, Lefkowitz RJ, Blobe GC. Beta-arrestin 2 mediates endocytosis of type III TGF-beta receptor and down-regulation of its signaling. Science. l2003;301:1394–1397. doi: 10.1126/science.1083195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.