Abstract
In E. coli, a four gene operon, sbm-ygfD-ygfG-ygfH, has been shown to encode a putative cobalamin-dependent pathway with the ability to produce propionate from succinate in vitro (Haller et al., Biochemistry 39; 4622–4629, 2000). However, the operon was thought to be silent in vivo, illustrated by the eponym describing its first gene, “sleeping beauty mutase” (methylmalonyl-CoA mutase, MCM). Of the four genes described, only ygfD could not be assigned a function. In this study, we have evaluated the functional integrity of YgfD and Sbm and show that, indeed, both proteins are expressed in E. coli and that YgfD has GTPase activity. We show that YgfD and Sbm can be co-immunoprecipitated from E. coli extracts using antibody to either protein, demonstrating in vivo interaction, a result confirmed using a strain deleted for ygfD. We show further that, in vitro, purified His-tagged YgfD and Sbm behave as a monomer and dimer, respectively, and that they form a multi-subunit complex that is dependent on pre-incubation of YgfD with non-hydrolysable GTP, an outcome that was not affected by the state of Sbm, as holo- or apoenzyme. These studies reinforce a role for the in vivo interaction of YgfD and Sbm.
Keywords: YgfD, Sbm, methylmalonyl-CoA mutase, cobalamin, vitamin B12
Introduction
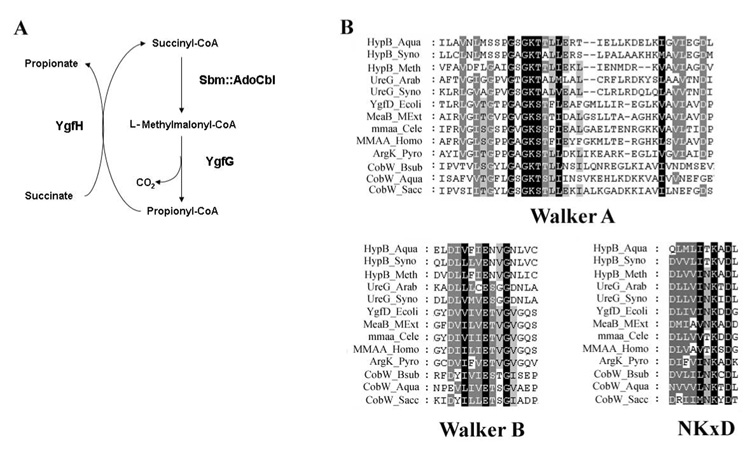
E. coli has the ability to synthesize methionine from homocysteine using either a vitamin B12-(cobalamin)-dependent (MetH) or a cobalamin-independent (MetE) methionine synthase (Foster et al., 1961; Foster et al., 1964; Guest et al., 1964). Therefore, E. coli are not obligate cobalamin users, but utilize it with availability. Not surprisingly, they do not have all the machinery required for cobalamin synthesis, but have retained all components required for its uptake and modification to active cofactor forms. Recently, a previously unrecognized cobalamin-dependent pathway was proposed for E. coli that catalyzes the conversion of succinate to propionate (Haller et al., 2000). The genes associated with this pathway are members of a four-gene operon: sbm-ygfD-ygfG-ygfH. Three of the encoded proteins were assigned functions based on enzymatic analysis of the expressed proteins in vitro: Sbm, shown to be methylmalonyl-CoA mutase, catalyzes the rearrangement of succinyl-CoA to L-methylmalonyl-CoA; YgfG, as methylmalonyl-CoA decarboxylase, catalyzes the decarboxylation of methylmalonyl-CoA to form propionyl- CoA; YgfH, as propionyl-CoA:succinate-CoA transferase, transfers the CoA of the propionyl-CoA product to an available succinate, thus priming another round of succinate to propionate decarboxylation (Fig. 1A) (Haller et al., 2000). The fourth protein, YgfD, however, could not be assigned a function, although the authors speculated that it might act as a possible kinase/phosphatase that is involved in the regulation of the other enzymes (Haller et al. 2000). This operon, while shown to be functional in vitro, was not thought to function in vivo due to the lack of a functioning promoter, a notion reflected in the name of the first gene, “sleeping beauty mutase” (Haller et al., 2000; Roy and Leadlay, 1992)
Figure 1.
A. Pathway for the AdoCbl-dependent conversion of succinate to propionate in E. coli by Sbm, YgfG and YgfH. B. Conserved domains in the G3E family of GTPases including members of the YgfD, CobW, HypB and UreG protein families. The alignment shows the three most highly conserved regions, the Walker A and the surrounding strands (block 1), the Walker B and surrounding area (block 2), and the area including the NKxD motif (block 3). Species names are abbreviated as follows: Aqua, Aquifex aeolicus; Arab, Arabidopsis thaliana; Bsub, Bacillus subtilis; Cele, Caenorhabditis elegans; Ecoli, Escherichia coli; Homo, Homo sapians; Meth, Methanobacterium thermoautotrophicum; MExt, Methylobacterium extorquens; Pyro, Pyrococcus abyssi; Sacc, Saccharomyces cerevisiae; and Syno, Synechococcus PCC8801. Based on studies by Leipe (Leipe et al., 2002).
While the function of YgfD remains unknown, a few clues to its possible role exist. Homologs of YgfD are often found associated with MCM in bacterial operons (Dobson et al., 2002; Bobik and Rasche, 2001), suggesting a role in the production of AdoCbl, the cofactor required for MCM function, or a role in the protection or facilitation of MCM function. As well, structurally, YgfD belongs to the G3E family of P-loop GTPases, a family defined by the glutamate residue in the Walker B motif and an intact NKxD, members of which include: UreG, HypB, CobW, and MeaB (Leipe et al., 2002) (Fig. 1B). Of these, MeaB is the closest homolog of YgfD, and MeaB from Methylobacterium extorquens has been shown to form a complex with M. extorquens MCM in vitro and that binding of MeaB to MCM was altered depending on the state of MCM, holo- vs. apoenzyme (Korotkova and Lidstrom, 2004; Padovani et al., 2006). Based on these data, we were interested to discover whether YgfD had a similar role as its homolog MeaB, and whether the interaction with methylmalonyl-CoA mutase would occur in vivo, as well as in vitro.
In the work described here, we report that contrary to its name, Sbm is indeed endogenously expressed in E. coli cells and is therefore not “sleeping”. We also demonstrate that YgfD and Sbm physically interact both in vitro and in vivo. Finally, we demonstrate that like its P-loop orthologs, YgfD binds and cleaves GTP, and this binding is integral to its interaction with Sbm. These studies reveal a functional role for YgfD, as the fourth member of the Sbm operon, and extend earlier work by establishing the in vivo interaction of YgfD and Sbm.
Materials and Methods
Materials
AdoCbl, OHCbl, GTP, guanosine 5′-[β,γ-imido]triphosphate (GMPPNP), GDP, ATP and methylmalonic acid were purchased from Sigma-Aldrich (Oakville, ON). All other chemicals were reagent grade. The plasmid vectors containing pET16-b:sbm and pET16- b:ygfD, each containing N-terminal His tags, have been previously described (Haller et al., 2000).
YgfD knockout
A mutant of E. coli deleted for ygfD was created using a method for allelic replacement by homologous recombination (White et al., 1999; White et al., 2007). Briefly, PCR was used to clone 5’ and 3’ segments of the ygfD gene (overlapped into the adjacent genes), which were combined and subcloned into pHSG415 (Hashimoto-Gotoh et al., 1981) so as to effectively delete amino acid residues 8–283 of the 331 residue protein. The primers used were ygfD-A (GAATTCTGCGCAAAGCTATCATCAGTCTGAG [an EcoRI site is underlined]) and ygfD-B (CTGCAGCAGCGTGGCTTCATTAATCATGCTG [a PstI site is underlined]) for the first fragment and ygfD-C (CTGCAGAAGAAGTACTGAATCACCTGTTCGC [a PstI site is underlined]) and ygfD-D (AAGCTTACCTTCCACCATCGAAATGATCGGT [a HindIII site is underlined]) for the second fragment. The two resulting PCR fragments were digested with PstI, ligated, cloned into pGEM and sequenced to verify the correct insert. The fragment was released using EcoRI and HindIII and ligated into pHSG415. Recombinant pHSG415∷Δygfd was used to generate a Δygfd mutant strain following established procedures (White et al., 1999; White et al., 2007). Presence of the chromosomal ygfD deletion was detected by PCR and DNA sequencing using the primers ygfDKo1 (TGGTGCCAAGCCAGTGTGTT) and ygfDKo2 (CCGCCACTTTGTTGATAGTGAC).
Purification of expressed proteins
YgfD and Sbm His-tagged proteins were expressed in E. coli (BL21) transformed with pET16-b:ygfD or pET16-b:sbm, respectively, and purified as described by Haller et al. (Haller et al., 2000), with some exceptions. Cells were grown in LB and lysed by passing through a French Pressure Cell (SLM Instruments; Urbana IL) two times at 12,000 psi and centrifuged at 27,000 × g for 30 min. The soluble fraction was incubated with a 5 mL slurry of Ni-NTA beads (Qiagen; Mississauga, ON), rinsed in binding buffer (5 mM imidazole, 0.5 M NaCl and 20 mM Tris-HCl, pH 8.0), and allowed to mix by end-over-end rotation overnight at 4°C. After washing with binding buffer and wash buffer (60 mM imidazole, 0.5 M NaCl, and 20 mM Tris-HCl, pH 8.0), the purified protein was eluted in elution buffer (100 mM imidazole, 0.5 M NaCl, and 20 mM Tris-HCl, pH 8, 20% glycerol). Purified Sbm protein was concentrated and stored at −80°C. Purified YgfD was concentrated, exchanged into storage buffer (50 mM HEPES, pH 8.0, containing 300 mM KCl, 2.5 mM MgCl2 and 5% glycerol), and stored at −80°C. Protein concentrations were determined using the Bradford protein assay (Bio-Rad; Mississauga, ON) with bovine serum albumin as a standard according to the manufacturer’s directions.
Antibody production
The full-length, N-terminal His-tagged Sbm and YgfD were used for the generation of polyclonal antibodies made in rabbit (SACRI Hybridoma Facility, University of Calgary). An enzyme-linked immunosorbant assay (ELISA) was performed using procedures described by Garkavtsev (Garkavtsev et al., 1997) to ensure antibody reactivity to the antigen. Immune and pre-immune sera were tested by Western blot and visualized using the ECL detection system (Amersham/GE Healthcare; Baie d’Urfe, QC). Antibodies were affinity purified using purified proteins bound to HiTrap NHS-activated HP columns, following manufacturer’s protocols (Amersham/GE Healthcare).
Immunoprecipitation
Immunoprecipitation experiments were performed using affinity purified antibodies that were immobilized using the Profound Co-immunoprecipitation kit (Pierce; Rockford, IL), according to manufacturer’s instructions. Briefly, antibodies were immobilized to aldehyde-activated agarose beads by cross-linking in the presence of 5 M cyanoborohydride. Immobilized antibodies were incubated over-night with 300 µl E. coli total cell lysate at 4°C with rotation. This complex was washed with 400 µl PBS and centrifuged for 1 min at 5000 × g three times. Protein was eluted using 100 µl low pH buffer (pH 2.8) and pH neutralized by addition of 10 µl 1 M Tris, pH 9.5. Protein was then mixed with 30 µl 2× SDS buffer and the solution was heated for 2 min at 95°C. Proteins in the supernatant were resolved by SDS-PAGE on a 10% polyacrylamide gel and visualized by staining with Coomassie brilliant blue R-250 (EMD; Gibbstown, NJ) or used for immunoblotting.
Immunobloting procedures
Proteins from SDS-PAGE were transferred to nitrocellulose membrane by capillary action. The membranes were blocked with 5% skim milk in PBS overnight at 4°C with shaking. Membranes were then incubated with primary antibody (anti-YgfD at 1:2000, anti-Sbm at 1:2000 in Tris-buffered saline plus 1% Tween-20, TBST) for 1 h, washed 3 × with TBST and incubated with secondary antibody (1:2500 Goat anti-Rabbit IgG HRP, Biorad, in TBST) for 45 min. Immune complexes were detected using ECL Reagent (Amersham/GE Healthcare) according to manufacturer’s recommendations and exposed to Hyperfilm (Amersham/GE Healthcare).
Size exclusion chromatography
Gel filtration was accomplished by fast protein liquid chromatography (FPLC) using a Superdex 200 HR 10/30 column (AKTA Purifier, Amersham/GE Healthcare) monitored by following the change in OD280. A 500 µg solution of YgfD or Sbm in sample buffer (10 mM Tris-HCl, pH 8.0, 10 mM KCl, 50 mM MgCl2, 3 mM GTP and 5 mM AdoCbl) was used in each experiment. GTP and AdoCbl were used in the sample buffer in an effort to stabilize YgfD. For the experiment where YgfD and Sbm were run together, they were first pre-incubated in the dark for 15 min. The column was calibrated using proteins of known molecular weight, including bovine milk α-lactalbumin (14.2 kDa), bovine erythrocytes carbonic anhydrase (29 kDa), chick egg albumin (45 kDa), BSA (66 kDa – monomer, 132 kDa – dimer), and Jack bean urease (272 kDa – trimer, 545 kDa – hexamer) (Sigma) as standards. These proteins were also used as a protein milieu for the gel filtration of YgfD and Sbm mixtures.
PAGE analysis of complex formation
To analyze complex formation between YgfD and Sbm, 10 µg of apo-Sbm or holo-Sbm (prepared by incubating Sbm with 50 µM AdoCbl and 0.05 mM methylmalonyl-CoA at 20°C in storage buffer) was incubated with 15 µg of YgfD, either alone or pre-incubated with (i) 5 mM GTP, (ii) 5 mM GDP or (iii) 5 mM GMPPNP at 20°C for 10 min. The mixtures were then separated by PAGE under non-denaturing conditions in a 4–15% gradient gel (Bio-Rad) for 12 h at room temperature.
GTPase Assay
GTPase activity was determined by using the ATPase/GTPase ELIPA Biochem Kit (Cytoskeleton; Denver, CO), according to the manufacturer’s instructions. For reactions, 15 µl of YgfD (750 ng) in storage buffer was added to 135 µl reaction mix (25 nmol MESG, 0.135 units PNP, 12 mM PIPES and 5 mM MgCl2) containing various concentrations of GTP (0–1000 µM). Reactions were carried out in 96 well plates (Corning; Corning, NY) in duplicate, read by a PowerWave × (Bio-Tek Instruments; Winooski, VT) at 360 nm and plotted using KaleidaGraph (V3.6, Synergy Software; Reading, PA).
Results
YgfD and Sbm characterization
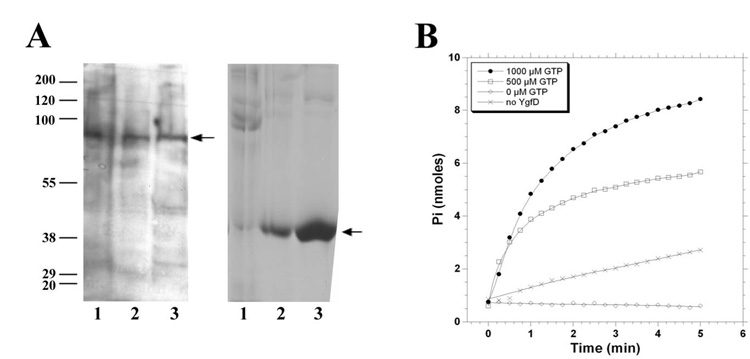
Although Sbm was shown to express a functional methylmalonyl CoA mutase in vitro, it was not thought to be expressed in vivo because of its placement in a presumed promoterless operon (Haller et al., 2000). We evaluated the endogenous expression of Sbm and YgfD in E. coli using polyclonal antibodies generated in rabbit against recombinant YgfD and Sbm. These antibodies were used to probe cell lysates from two wild type strains of E. coli, DH5α and MGC1876, as well as the ygfD knock-out strain MGC1876ΔygfD (Fig. 2A). In both wild-type strains, proteins of the appropriate size, ~78 kDa for Sbm and ~37 kDa for YgfD, were detected on immunoblots (Fig. 2A). For MGC1876ΔygfD, the appropriately sized-band was detected for Sbm but not YgfD, as expected (Fig. 2A). Therefore, we conclude that the Sbm operon is functional in E. coli.
Figure 2.
A. Western blot analysis of whole cell lysates from E. coli MGC1876ΔygfD (lane 1), DH5α (lane 2), and MGC1876 (lane 3), probed with anti-Sbm (left panel) and anti-YgfD (right panel). B. GTPase activity of YgfD as measured by the production of inorganic phosphate (Pi) from GTP, monitored at A360.
Given its identification as a G3E P-loop GTPase, we expected that YgfD could also function as a GTPase. Assay of GTPase activity confirmed GTP hydrolysis (Fig. 2B), however, the protein proved to be extremely unstable during isolation and purification and was observed to precipitate in pure and semi-pure extracts. Therefore, we were able to demonstrate hydrolysis but could not obtain reliable kinetic data.
YgfD and Sbm physically interact in vivo
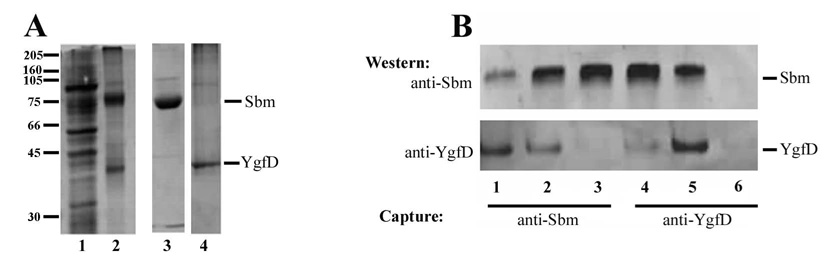
Based on the observation that MeaB physically interacts with MCM in Methylobacterium extorquens in vitro (Korotkova and Lidstrom, 2004), we evaluated whether YgfD and Sbm might interact in vivo. For this experiment, we made use of immobilized anti-YgfD to capture protein from a lysate of wild type E. coli grown in LB (Fig. 3A). Western blot analysis of the protein released from the immobilized antibody revealed the presence of both YgfD and Sbm (Fig. 3A, lanes 3, 4). A more elaborate experiment in which both anti-YgfD and anti-Sbm were used as the immobilized antibody followed by Western blot analysis using antibodies against the two proteins to identify the immunoprecipitated species is shown in Fig. 3B. Using wild type strains BL21 and MGC1876, immunocapture by either anti-YgfD or anti-Sbm recovered both YgfD and Sbm proteins (Fig. 3B, lanes 1,2,4,5). In contrast, in MGC1876ΔygfD, anti-Sbm but not anti-YgfD identified Sbm in the knockout strain lacking YgfD (Fig. 3B, lanes 3,6). These experiments confirm that YgfD and Sbm physically interact in vivo and that detection of Sbm following immunocapture using anti-YgfD is dependent on the presence of YgfD protein. They also imply that YgfD is not essential for either the viability of these cells or the expression of Sbm, as neither was affected by knockout of the YgfD protein.
Figure 3. Interaction of YgfD and Sbm in vivo.
A. Proteins from MGC1876 crude cell lysate (lane 1) or immunoprecipitated from crude cell lysate with anti-YgfD (lanes 2–4) were analyzed by Coomassie blue staining (lanes 1 and 2) or Western blot analysis, probed with anti-Sbm (lane 3) or anti-YgfD (lane 4). Note anti-YgfD captures a complex of YgfD and Sbm (lane 2). B. Immunocapture of YgfD and Sbm using immobilized anti-Sbm (lanes 1–3) or anti-YgfD antibodies (lanes 4–6) followed by SDS-PAGE and Western blot analysis using anti-Sbm (upper) or anti-YgfD (lower). Lanes 1, 4, strain BL21; lanes 2, 5, MG1876; lanes 3, 6, MGC1876ΔygfD.
Interaction of YgfD and Sbm in vitro
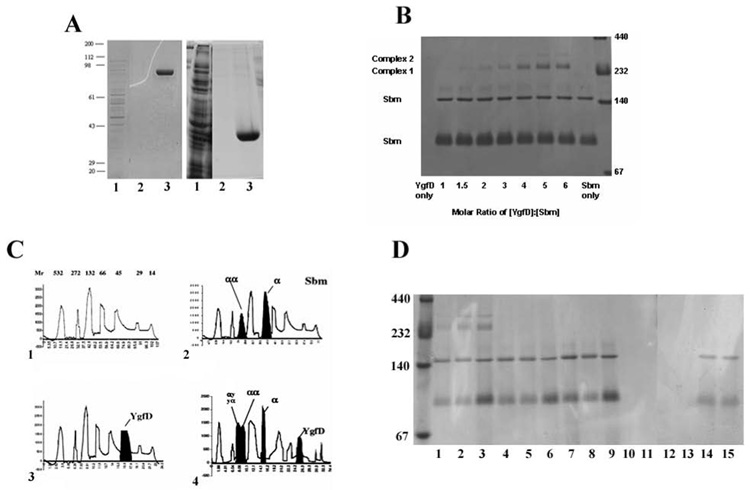
To investigate the nature of the interaction between YgfD and Sbm, we examined cofactor requirements for interaction in vitro. Recombinant YgfD and Sbm containing N-terminal His tags were expressed in E. coli and purified by nickel affinity chromatography, as described previously (Haller et al., 2000). Both proteins were obtained in high yield (10–15 mg/L) and migrated with molecular masses of ~40 kDa for YgfD and ~80 kDa for Sbm, as monitored by SDS-PAGE (Fig. 4A). When apo-Sbm was separated on a native gel, it migrated both as a monomer and as a dimer (Fig. 4B). This result was supported by size-exclusion chromatography, which also showed peaks corresponding to an Sbm monomer and dimer (Fig. 4C panel 2). This was not surprising as most methylmalonyl-CoA mutases form dimers, whether known to be encoded by two genes (heterodimeric) (Korotkova and Lidstrom, 2004; Marsh et al., 1989) or a single gene (homodimeric) (Miyamoto et al., 2003). In contrast, while clearly visible as a band on an SDS gel (Fig. 4A), YgfD by itself could not be visualized on a native gel (Fig. 4B), a behavior that was also observed with MeaB (Padovani et al., 2006). However, native YgfD could be detected by size exclusion chromatography and eluted as a monomer (Fig. 4C panel 3), although it was very unstable and had to be run in the presence of molecular weight standards to minimize precipitation. Therefore, all subsequent gel exclusion chromatography experiments were run in the presence of molecular weight standards. When YgfD and Sbm were mixed and incubated in the absence of cofactor and nucleotide substrate, novel high molecular weight species indicative of complex formation could not be detected on a native gel. However, when YgfD was pre-incubated with the non-hydrolysable GTP analog (GMPPNP) and then incubated with Sbm, two new migrating species were observed with molecular masses greater than the 232 kDa molecular weight marker (Fig. 4B, Complex 1 and Complex 2). Maximal complex formation occurred when the YgfD:GMPPNP mix was at a molar ratio of 4:1 with Sbm. Such a high ratio may have been required to drive complex formation owing to the instability of YgfD. Complex formation was not affected by whether or not B12 was bound to Sbm, and the complex was not observed when YgfD was pre-incubated with GTP or GDP as the nucleotide (Fig. 4D). By size exclusion chromatography, the YgfD:Sbm complex could be detected in the presence of GTP and AdoCbl, revealing that under these milder conditions, complex formation between the two proteins could be observed in the absence of GMPPNP. Under these conditions, the complex migrated with a size corresponding to ~258 kDa, suggestive of a stoichiometry of 2 YgfD + 2 Sbm (Fig. 4C panel 4). However, the accuracy of molecular weight determinations in this range is limited, so our estimate is only approximate.
Figure 4. Interaction of YgfD and Sbm in vitro.
A. Expression and purification of His-tagged Sbm (left panel) and YgfD (right panel) run on 12% SDS-PAGE and stained by Coomassie blue. For both: lane 1, total cell lysate; lane 2, final wash fraction before elution; lane 3, eluate containing purified protein. B. Native 4–20% polyacrylamide gel containing Sbm apoenzyme incubated with increasing molar equivalents of YgfD pre-incubated with GMPPNP. Sbm ran at ~80 kDa monomer and ~160 kDa dimer (Sbm only lane), while YgfD (~40 kDa) was run off this gel but not seen previously (YgfD only lane). Note appearance of species (Complex 1 and Complex 2) with masses in excess of the 232 kDa MW standard at increasing YgfD ratio (maximal band shift obtained at 4:1 YgfD:Sbm incubation mix). C. Purified Sbm, YgfD, or an equal mixture of the two, were analyzed by gel filtration. Proteins were run in the presence of MW standards (bovine milk α-lactalbumin (14.2 kDa), bovine erythrocytes carbonic anhydrase (29 kDa), chick egg albumin (45 kDa), BSA (66 kDa – monomer, 132 kDa – dimer), and Jack Bean urease (272 kDa – trimer, 545 kDa – hexamer)) to prevent precipitation and loss of proteins during sieving. Panel 1, MW standards with molecular masses in kDa indicated above each peak; Panel 2, Sbm plus MW standards, with Sbm monomer (“α”) and dimer (“αα”) peaks filled in black; Panel 3, YgfD plus MW standards with YgfD peak in black; Panel 4, YgfD plus Sbm with each protein and putative complex (“ααyy”) filled in black. Peaks in black were determined by comparison with peaks in Panel 1. The x- and y-axis refer to time (min) and elution profile at OD280, respectively, for all charts. D. Native 4–20% polyacrylamide gel containing different combinations of YgfD and Sbm with or without prior incubation to cofactor/coenzyme demonstrating that only YgfD pre-incubated with GMPPNP results in complex formation with Sbm. Lane 1, YgfD:GMPPNP & Sbm:AdoCbl; Lane 2, YgfD:GMPPNP & Sbm:HOCbl; Lane 3, YgfD:GMPPNP & Sbm; Lane 4, YgfD & Sbm:AdoCbl; Lane 5, YgfD & Sbm:HOCbl; Lane 6, YgfD & Sbm; Lane 7, Sbm:AdoCbl; Lane 8, Sbm:HOCbl; Lane 9, Sbm; Lane 10, YgfD:GMPPNP; Lane 11, YgfD; Lane 12, YgfD:GTP; Lane 13, YgfD:GDP; Lane 14, YgfD:GTP & Sbm; Lane 15, YgfD:GDP & Sbm.
Discussion
We report the expression of Sbm and YgfD, contiguous members of an E. coli operon comprised of sbm-ygfD-ygfG-ygfH. Past attempts to demonstrate enzyme activity associated with Sbm had failed (Haller et al., 2000; Dayem et al., 2002), leading to the gene name “sleeping beauty mutase.” Although Haller et al. (Haller et al., 2000) successfully expressed all four proteins in vitro, they were able to assign functions to only three of them, predicting a cycle for the conversion of succinyl-CoA to propionyl-CoA. The function of YgfD could not be assessed. In this study, we confirmed the expression of native Sbm and YgfD in E. coli and demonstrated that the two proteins form a complex in vitro and in vivo. We showed further that YgfD has GTPase activity and that binding a non-hydrolyzable GTP analog stabilizes the association in vitro.
Identified as a member of the G3E family of P-loop GTPases (Leipe et al., 2002), YgfD shares 48% sequence identity with MeaB, the orthologous protein from M. extorquens, and 46% with MMAA, its counterpart in human cells. While MeaB has previously been shown to interact with MCM in vitro, this was not demonstrated in vivo (Korotkova and Lidstrom, 2004; Padovani et al., 2006). Our finding that YgfD and Sbm physically interact in vivo was made by co-immunoprecipitation of the two proteins using antibodies to either YgfD or Sbm as the capture reagent, a result that was confirmed with the use of a ygfD knockout strain. Further, we showed in vitro that recombinant YgfD and Sbm form a stable complex that can be separated on a non-denaturing gel or by size exclusion chromatography. The interaction depended on pre-incubation of YgfD with non-hydrolyzable GTP for detection by electrophoresis. A YgfD-Sbm complex could also be detected on the sieving column in the presence of GTP and AdoCbl, although the conditions for interaction were not further evaluated in this medium owing to the extreme lability of the proteins. Our results are consistent with a 2YgfD:2Sbm structure by molecular sieving, in contrast to MeaB:MCM which behaves as a heterodimer. While we found that Sbm was detected as a mixture of monomer and dimer, YgfD was only seen as a monomer in contrast to MeaB in which a dimer was observed (Padovani et al., 2006).
The sbm operon does not appear to be essential for cell growth, at least under the culture conditions used in this study. Further, Haller et al. were unable find evidence for an effect of hydroxocobalamin on cell growth under either aerobic or anaerobic conditions with succinate or propionate as the carbon source (Haller et al., 2000). Likewise, when the ygfD gene was disrupted, we did not observe any effect on cell viability.
While we have characterized the interaction between Sbm and YgfD, the significance of this interaction remains incompletely understood. The stabilization of the complex using a non-hydrolyzable form of GTP suggests that GTP is required for complex formation in vivo. Yet, co-immunoprecipitation of the complex was obtained from extracts of cells grown in standard LB medium. As well, while YgfD and Sbm are expressed endogenously, previous studies failed to demonstrate a functional pathway (Haller et al., 2000; Dayem et al., 2002). Since recombinant Sbm has been shown to be functional when overexpressed in E. coli in medium containing vitamin B12 (Dayem et al., 2002), one can only speculate that under standard growth conditions, expression of Sbm, while detectable, must be too low to produce a functional pathway.
Acknowledgments
These studies were supported by a grant from the Canadian Institute for Health Research (CIHR). Scholarship support was provided from the CIHR Training Grant in Genetics, Child Health and Development to D.S.F. and from the Alberta Heritage Foundation for Medical Research for C.M.D. and A.P.W.
Reference List
- Bobik TA, Rasche ME. Identification of the human methylmalonyl-CoA racemase gene based on the analysis of prokaryotic gene arrangements. Implications for decoding the human genome. J Biol Chem. 2001;276:37194–37198. doi: 10.1074/jbc.M107232200. [DOI] [PubMed] [Google Scholar]
- Dayem LC, Carney JR, Santi DV, Pfeifer BA, Khosla C, Kealey JT. Metabolic engineering of a methylmalonyl-CoA mutase-epimerase pathway for complex polyketide biosynthesis in Escherichia coli. Biochemistry. 2002;41:5193–5201. doi: 10.1021/bi015593k. [DOI] [PubMed] [Google Scholar]
- Dobson CM, Wai T, Leclerc D, Wilson A, Wu X, Dore C, Hudson T, Rosenblatt DS, Gravel RA. Identification of the gene responsible for the cblA complementation group of vitamin B12-responsive methylmalonic acidemia based on analysis of prokaryotic gene arrangements. Proc Natl Acad Sci U S A. 2002;99:15554–15559. doi: 10.1073/pnas.242614799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster MA, Jones KM, Woods DD. The purification and properties of a factor containing vitamin B12 concerned in the synthesis of methionine by Escherichia coli. Biochem J. 1961;80:519–531. doi: 10.1042/bj0800519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster MA, Tejerina G, Guest JR, Woods DD. Two enzymic mechanisms for the methylation of homocysteine by extracts of Escherichia coli. Biochem J. 1964;92:476–488. doi: 10.1042/bj0920476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garkavtsev I, Boland D, Mai J, Wilson H, Veillette C, Riabowol K. Specific monoclonal antibody raised against the p33ING1 tumor suppressor. Hybridoma. 1997;16:537–540. doi: 10.1089/hyb.1997.16.537. [DOI] [PubMed] [Google Scholar]
- Guest JR, Friedman S, Foster MA, Tejerina G, Woods DD. Transfer of the methyl group from N5-methyltetrahydrofolates to homocysteine in Escherichia coli. Biochem J. 1964;92:497–504. doi: 10.1042/bj0920497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller T, Buckel T, Retey J, Gerlt JA. Discovering new enzymes and metabolic pathways: conversion of succinate to propionate by Escherichia coli. Biochemistry. 2000;39:4622–4629. doi: 10.1021/bi992888d. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Gotoh T, Franklin FC, Nordheim A, Timmis KN. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene. 1981;16:227–235. doi: 10.1016/0378-1119(81)90079-2. [DOI] [PubMed] [Google Scholar]
- Korotkova N, Lidstrom ME. MeaB Is a Component of the Methylmalonyl-CoA Mutase Complex Required for Protection of the Enzyme from Inactivation. J Biol Chem. 2004;279:13652–13658. doi: 10.1074/jbc.M312852200. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- Marsh EN, Harding SE, Leadlay PF. Subunit interactions in Propionibacterium shermanii methylmalonyl-CoA mutase studied by analytical ultracentrifugation. Biochem J. 1989;260:353–358. doi: 10.1042/bj2600353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto E, Watanabe F, Charles TC, Yamaji R, Inui H, Nakano Y. Purification and characterization of homodimeric methylmalonyl-CoA mutase from Sinorhizobium meliloti. Arch Microbiol. 2003;180:151–154. doi: 10.1007/s00203-003-0570-3. [DOI] [PubMed] [Google Scholar]
- Padovani D, Labunska T, Banerjee R. Energetics of interaction between the G-protein chaperone, MeaB, and B12-dependent methylmalonyl-CoA mutase. J Biol Chem. 2006;281:17838–17844. doi: 10.1074/jbc.M600047200. [DOI] [PubMed] [Google Scholar]
- Roy I, Leadlay PF. Physical map location of the new Escherichia coli gene sbm. J Bacteriol. 1992;174:5763–5764. doi: 10.1128/jb.174.17.5763-5764.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AP, Collinson SK, Burian J, Clouthier SC, Banser PA, Kay WW. High efficiency gene replacement in Salmonella enteritidis: chimeric fimbrins containing a T-cell epitope from Leishmania major. Vaccine. 1999;17:2150–2161. doi: 10.1016/s0264-410x(98)00491-5. [DOI] [PubMed] [Google Scholar]
- White AP, len-Vercoe E, Jones BW, DeVinney R, Kay WW, Surette MG. An efficient system for markerless gene replacement applicable in a wide variety of enterobacterial species. Can J Microbiol. 2007;53:56–62. doi: 10.1139/w06-102. [DOI] [PubMed] [Google Scholar]