Abstract
Kallikrein-4 (KLK4) is a serine protease expressed during enamel maturation, and proteolytic processing of the enamel matrix by KLK4 is critical for proper enamel formation. KLK4 is secreted as an inactive zymogen (pro-KLK4), and identification of its activator remains elusive. Dipeptidyl peptidase I (DPPI) is a cysteine aminopeptidase that can activate several serine proteases. In this study, we sought to examine DPPI expression in mouse enamel organ and determine if DPPI could activate KLK4. Real-time PCR showed DPPI expression throughout amelogenesis, with highest expression at maturation, and immunohistochemical staining of mouse incisors confirmed DPPI expression by ameloblasts. We demonstrate in vitro that DPPI activates pro-KLK4 to cleave a fluorogenic peptide containing a KLK4 cleavage site. Examination of mature enamel from DPPI null mice by FTIR showed no significant accumulation of protein; however, microhardness testing revealed that loss of DPPI expression significantly reduced enamel hardness.
Keywords: enamel, dipeptidyl peptidase I, kallikrein-4, amelogenesis, mineralization
Introduction
Dental enamel is formed in three major stages. During the secretory stage, the ameloblast cells secrete large amounts of proteins (amelogenin, ameloblastin, enamelin) and matrix metalloproteinase-20 (MMP20), which cleaves these enamel proteins. The ameloblasts then go through transition and maturation phases where they secrete the serine protease kallikrein-4 (KLK4) (Hu et al., 2000, 2002). KLK4 further degrades the enamel proteins, which are ultimately removed from the enamel matrix, resulting in virtually protein-free mature enamel. Proteolytic processing of the enamel matrix by KLK4 is critical for proper enamel formation, and homozygous mutation of KLK4 causes Amelogenesis Imperfecta, characterized by enamel that is hypomineralized and protein-rich (Hart et al., 2004; Wright et al., 2006).
KLK4, like other kallikreins, is secreted as an inactive zymogen. These serine proteases are synthesized as prepro-enzymes that are proteolytically processed to secreted pro-forms via removal of their signal peptide. The inactive pro-KLKs are then activated extracellularly by specific release of their amino-terminal propeptide. Activation of kallikrein family pro-forms has been well-studied (Yoon et al., 2007; Emami and Diamandis, 2008); however, identification of the KLK4 activator remains elusive. The tooth-specific protease MMP20 has been shown to activate KLK4 in vitro (Ryu et al., 2002); however, KLK4 and MMP20 expression overlap only briefly during enamel development (Hu et al., 2002). KLK4 is secreted throughout maturation and is not autocatalytic, which suggests that another protease activates KLK4 in vivo. KLK4 is also expressed in prostate and ovaries and is overexpressed in prostate and ovarian cancer (Dong et al., 2001; Klokk et al., 2007). Normal MMP20 expression within these tissues has not been demonstrated, indicating the strong possibility of additional activator(s) of KLK4.
Dipeptidyl peptidase I (DPPI), also known as cathepsin C, is a ubiquitously expressed cysteine aminopeptidase that sequentially removes 2 N-terminal amino acid residues from proteins. In addition to its lysosomal activity, DPPI can be secreted (Wolters et al., 1998) and activates pro-enzymes of chymotrypsin-like serine proteases. DPPI activates granzymes A and B in vivo (Pham and Ley, 1999) as well as cathepsin G, elastase (Adkison et al., 2002), and chymases (Wolters et al., 2001).
In this study, we sought to characterize DPPI expression within mouse enamel organ and determine if DPPI could activate pro-KLK4. We also examined enamel from DPPI-null incisors to determine if DPPI is essential for normal enamel development.
Materials & Methods
All animals used in this study were housed in facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
Real-time PCR Analysis of Gene Expression in Mouse Enamel Organ
Mouse molars were harvested at 3, 5, 7, 9, and 11 days post-natally and subjected to qPCR analysis by iQ SYBR green (Bio-Rad, Hercules, CA, USA). RNA was extracted and reverse-transcribed as previously described (Turk et al., 2006). Gene-specific primers were designed with DNAStar software (DNASTAR, Inc., Madison, WI, USA). Primers for DPPI expression were: forward (5′-GGTTGTATCTTGCAGCCCCTATG-3′); and reverse (5′-TTCTTCCAC CACCCCAAAATCTT-3′). The PCR temperature profile was 3 min at 95°C, 20 sec at 95°C, 30 sec at 64°C for 45 cycles, and 30 sec at 95°C for 1 cycle, followed by stepwise increases to generate the melt curve. Results were normalized to EF1α1 RNA expression as previously described (Pfaffl, 2001) and are presented as fold-increase in expression over day 3 levels. Data are representative of 6 individual mice per time-point, with measurements for each mouse repeated twice. Statistical significance of each time-point compared with day 3 (secretory stage) was analyzed by Bonferroni’s Multiple-comparison Test.
Immunohistochemistry of Mouse Incisors
Immunohistochemical analysis of demineralized, paraffin-embedded, and sectioned mouse mandibular incisor was performed. Sections were incubated in 10 mM sodium citrate buffer, pH 6.0, for 10 min at 92˚C for antigen retrieval and in blocking agent for 20 min, followed by overnight incubation in 10 µg/mL DPPI antibody (R&D Systems, Minneapolis, MN, USA). Staining was visualized by incubation in peroxidase-conjugated antibody [Vectastain Elite ABC Kit (Goat IgG), Vector Laboratories, Burlingame, CA, USA] and Sigma Fast 3,3′-diaminobenzidine substrate (Sigma, St. Louis, MO, USA). Sections were counterstained with 0.1% Fast Green and examined by light microscopy. A negative control section was not treated with the DPPI-specific antisera.
In vitro Activation of Recombinant KLK4
Inactive, recombinant human pro-KLK4 (0.1 µg/µL; R&D Systems) was incubated with active recombinant mouse DPPI (0.01 µg/µL; R&D Systems) in 50 mM MES, 50 mM NaCl, pH 5.5, at 37ºC for 4 hrs. As a positive control, the bacterial protease thermolysin was used to activate pro-KLK4. Inactive pro-KLK4 (0.1 µg/µL) was incubated with thermolysin (1 µg/mL; Sigma) in 50 mM Tris, 10 mM CaCl2, 150 mM NaCl, pH 7.5, for 2 hrs at 37°C.
To determine if KLK4 had become activated, we measured activity by incubation of 100 ng of DPPI- or thermolysin-activated KLK4 with 100 µM Boc-Val-Pro-Arg-AMC (R&D Systems) in 100 µL of 50 mM Tris, pH 9.0, at room temperature in a 96-well plate with agitation. We measured hydrolysis of the substrate by monitoring fluorescence every 1 min for 2 hrs with 380-nm excitation and 460-nm emission. Inactive pro-KLK4 (100 ng), active DPPI (100 ng), and thermolysin (10 ng) were also incubated with 100 µM fluorogenic peptide, and fluorescence was monitored.
DPPI Null Mice
DPPI−/− mice were generated in 129/SvJ, DBA/1J, and C57BL/6 strains, as previously described (Pham and Ley, 1999; Hu and Pham, 2005; Pagano et al., 2007).
Fourier Transform Infrared Spectroscopy (FTIR)
Mandibular incisors were obtained from 3 DPPI null mice and 3 wild-type mice (C57BL/6 strain). The heads from mice were skinned after death and fixed in 4% neutral formalin/saline for 24 hrs, then rinsed and incubated in phosphate-buffered saline at 4ºC. Erupted portions of mandibular incisors from wild-type and DPPI null mice were embedded horizontally (to expose both enamel and dentin) in a hard-formulation epoxy embedding medium (EpoFix, EMS, Hatfield, PA, USA). Samples were polished to 0.25 µm with a diamond suspension (EMS). Enamel and dentin from the erupted portions of incisors were studied in reflectance mode on a Multiscope FTIR microscope coupled with a Spectrum One FTIR spectrometer (Perkin Elmer, Boston, MA, USA). Spectra were collected from 4000 to 550 cm−1 at a resolution of 4 cm−1with 64 accumulations per run. Background spectra were collected from a gold mirror. Reflectance spectra were processed with the Kramers-Konig transformation in Spectra 5.0 to obtain an absorbance-like spectrum. Corrected maximum heights of protein Amide I absorption peaks in the 1600 to 1700 cm−1 region and v3PO4 absorption peaks in the 1000 to 1100 cm−1 region were measured. The Amide I/v3PO4 ratio was used to assess protein to mineral content in wild-type and DPPI null enamel. Statistical significance was determined by the t test.
Vickers Microhardness Testing
The polished samples of 3 wild-type and 3 DPPI null incisors (described above) were tested for enamel microhardness on a LECO M 400 HI testing machine (LECO, St. Joseph, MI, USA). Mandibular incisors from 2 additional strains of DPP−/− mice (129SvJ and DBA/1J) were also examined. Incisors from 2 mice of each strain were embedded and polished as described above.
Microhardness testing was performed with a load of 25 g for 5 sec with a Vickers tip. Thirty indentations per enamel sample were performed. Statistical significance was determined by t test.
Results
DPPI is Expressed by the Enamel Organand its Ameloblasts
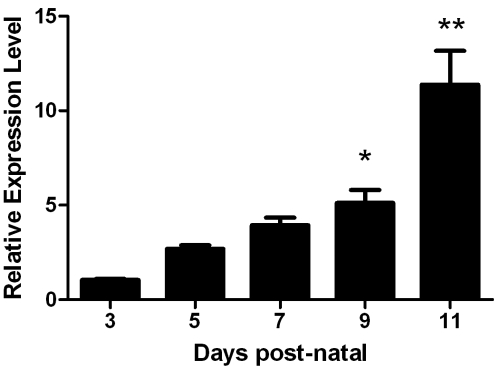
Expression of DPPI with respect to enamel formation was examined by real-time PCR. DPPI expression was quantified at days 3, 5, 7, 9, and 11, where day 3 represents early-secretory-stage and day 11 represents maturation-stage enamel organ. DPPI was expressed throughout amelogenesis, and expression increased dramatically, with an 11-fold increase at the maturation over the secretory stage (Fig. 1).
Figure 1.
DPPI is expressed in enamel organ. Mouse molars were harvested at defined time-points and subjected to real-time PCR analysis with DPPI specific primers. Expression was quantified at days 3, 5, 7, 9, and 11, where day 3 represents early-secretory-stage and day 11 represents maturation-stage enamel organ. Data represent measurements of 6 individual mice per time-point, with each mouse repeated in triplicate (n = 6). Results are presented as fold-increase in expression over day 3 and normalized to EF1α1 mRNA expression. DPPI expression increased dramatically, with an 11.38 ± 1.78-fold increase at the maturation stage over the secretory stage (*p < 0.05; **p < 0.001). Data are presented as mean ± SEM.
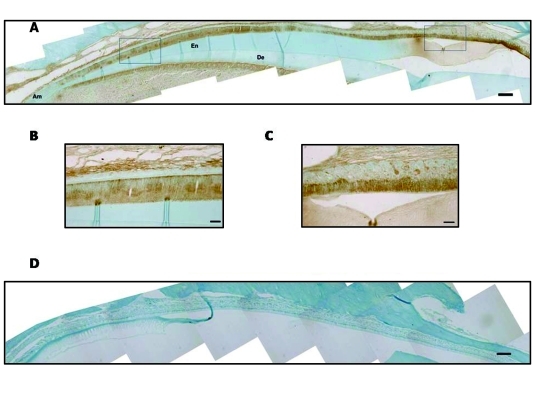
We performed immunohistochemical analysis of mouse mandibular incisors with DPPI antibody to confirm that ameloblasts expressed DPPI (Fig. 2). Consistent with the qPCR results, DPPI was expressed within secretory- and maturation-stage ameloblasts. DPPI expression was also detected within the odontoblast cells and pulp.
Figure 2.
DPPI is expressed by ameloblasts. Immunohistochemical analysis of demineralized, paraffin-embedded sections of a mouse mandibular incisor counterstained with 0.1% Fast Green. Ameloblasts (Am), enamel space (En), and dentin (De) are depicted. (A) Staining for DPPI in an adult mouse incisor (20X magnification). (B) Enlargement of the indicated secretory-stage ameloblasts from panel A (40X magnification). (C) Enlargement of the indicated maturation-stage ameloblasts from panel A (40X magnification). (D) Negative control section treated with the secondary, but not primary, antisera (20X magnification). Scale bar represents 140 mM for panels A and D. Scale bar in panels B and C represents 50 µm.
DPPI Activates KLK4 in vitro
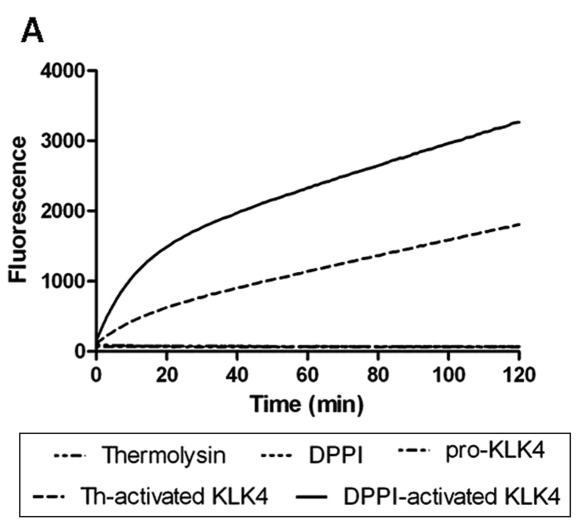
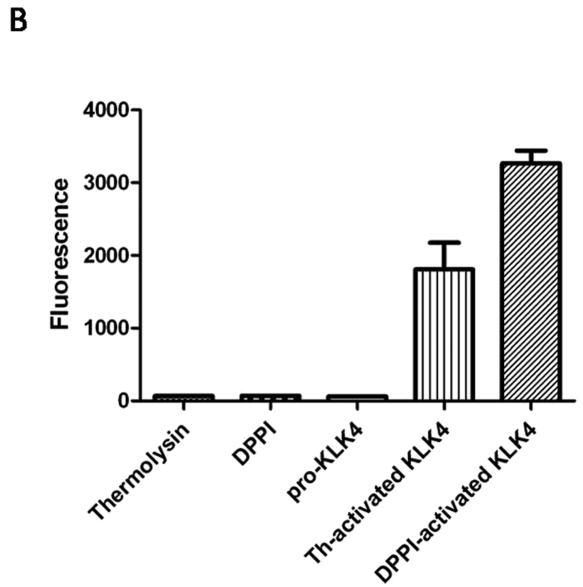
Activation of pro-KLK4 by DPPI was demonstrated by the use of a quenched fluorescent peptide. When the peptide is cleaved by active KLK4, it fluoresces, and this fluorescence can be quantified. Specificity of KLK4 for the peptide was demonstrated by incubation of the quenched peptide with thermolysin and DPPI. Neither enzyme was capable of cleaving the peptide (Fig. 3). Pro-KLK4 was also shown to be inactive and unable to cleave the peptide. After pro-KLK4 was incubated with DPPI, the peptide substrate was cleaved, as indicated by the increase in fluorescence. Thermolysin, which was previously demonstrated to activate KLK4 in vitro, was used as a positive control.
Figure 3.
DPPI activates pro-KLK4 in vitro. (A) We measured hydrolysis of the KLK4 specific substrate (Boc-Val-Pro-Arg-AMC) by monitoring fluorescence every 1 min over 2 hrs with 380-nm excitation and 460-nm emission wavelengths. The fluorescent substrate was incubated with thermolysin, DPPI, pro-KLK4, thermolysin-activated KLK4 (th-activated KLK4), and DPPI-activated KLK4. (B) Fluorescence measurement with the same enzymes after two-hour incubation of the Boc-Val-Pro-Arg-AMC substrate (n = 4). Incubation with thermolysin, DPPI, or pro-KLK4 did not result in substrate cleavage. Thermolysin-activated KLK4 (th-activated KLK4) and DPPI-activated KLK4 both cleaved the substrate, as indicated by the fluorescence. Data are presented as mean ± SEM.
DPPI Null Mice Have Decreased Enamel Hardness
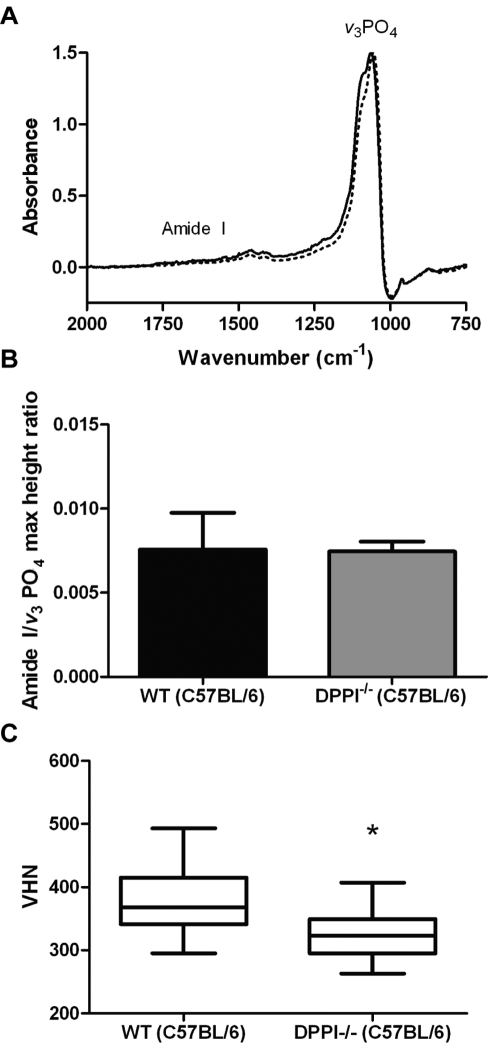
We performed FTIR analysis to assess protein and mineral content. Mandibular incisors from wild-type and DPPI null C57BL/6 mice were examined by FTIR. We used the ratio between maximal intensities of protein Amide I and v3PO4 absorption bonds to assess the matrix-to-mineral ratio of the enamel. The DPPI null mice showed results comparable with those of wild-type mice, with no significant difference in protein/mineral ratio (Figs. 4A, 4B). The Amide I/v3PO4 maximum height ratio from the enamel of the DPPI null samples (0.0075 ± 0.0006) was not significantly different from that of the wild-type controls (0.0076 ± 0.0022). The dentin was also analyzed, and no significant difference in the calculated Amide I/v3PO4 maximum height ratio was observed between wild-type (0.155 ± 0.006) and DPPI null mice (0.151 ± 0.003).
Figure 4.
Characterization of enamel from DPPI null mice. (A) Representative normalized FTIR absorption spectra of enamel from wild-type (solid line) and DPPI null C57BL/6 mice (dashed line) collected in reflectance mode. Amide I protein band and v3PO4 band are shown. (B) Amide I/v3PO4 maximum height ratio from the enamel of DPPI null mice compared with that of wild-type C57BL/6 mice (n = 3). No difference in protein-to-mineral ratio is observed in null mice. Data are presented as mean ± SEM. (C) Box chart representing Vickers microhardness data for enamel from DPPI null and control incisors with 30 indentations per incisor (n = 3). The Y-axis shows the Vickers hardness number (VHN). Whiskers represent minimum and maximum. Student’s t test showed that the means were significantly different (*p < 0.05).
Microhardness of C57BL/6 DPPI null [324.2 VHN, SEM ± 16.70] enamel was significantly decreased (p < 0.05) as compared with that of control [403.7 VHN, SEM ± 29.86] (Fig. 4C). The decrease in enamel hardness does not appear to be strain-specific, since 129SvJ [297.0 VHN, SEM ± 16.93] and DBA/1J [342.9 VHN, SEM ± 28.97] strain DPPI null mice also show decreased enamel hardness.
Discussion
The activator of KLK4 in vivo remains uncertain. We therefore asked if DPPI, a widely expressed protease known to activate numerous serine proteases, could also activate pro-KLK4. Herein we examined the expression of DPPI with respect to enamel formation and determined if DPPI null mice have defective dental enamel.
DPPI is expressed throughout enamel formation, with expression increasing at transition stage and with even greater increases seen at maturation. This increase in expression coincides with the expression of KLK4, which begins to appear at transition and peaks at maturation.
Human prepro-KLK4 contains 254 amino acids, the inactive zymogen has 228 amino acids, and the active protein has 224 amino acids. The amino acid sequence of the propeptide is not highly conserved across species; however, the propeptides of humans, the mouse, rat, pig, dog, and cow contain amino acids that DPPI is able to cleave. DPPI cannot cleave dipeptides if there is an N-terminal Lys or Arg, or a P1 Pro residue (McDonald et al., 1969; McGuire et al., 1992). It is of note that active KLK4 from all these species begins with Ile-Ile. The active mature form of the chymases, cathepsin G and granzymes A and B, all of which are activated by DPPI, also begins with Ile-Ile. Using synthetic peptides, investigators found that DPPI is unable to cleave Ile-Ile-AMC (Tran et al., 2002). This supports DPPI as a likely activator of KLK4, and our in vitro activity assay demonstrated that DPPI is capable of activating human pro-KLK4.
If DPPI is the sole activator of KLK4 in vivo, we would expect DPPI−/− mice to have inactive KLK4 and the phenotype to mimic KLK4 null mice. The enamel from KLK4 null mice has yet to be described; however, the enamel from individuals with Amelogenesis Imperfecta and with a mutation in the KLK4 gene does have a hypomaturation phenotype. Radiographically, the teeth appear morphologically normal in shape, with only a slight increase in x-ray opacity compared with dentin, indicative of decreased enamel mineral content (Hart et al., 2004). Closer examination showed the enamel to be of normal thickness and prismatic (Wright et al., 2006).
We examined the mandibular incisors from DPPI null mice, and while no decrease in mineral content was evident based on our FTIR analysis of the protein-to-mineral ratio, the hardness studies showed that enamel quality was affected. The cleavage of enamel proteins by proteases is critical to crystal growth, and any alteration to this cleavage will likely affect the overall structural properties of the enamel crystals. We propose that the loss of DPPI resulted in reduced activation of KLK4. While it appears that the overall protein-to-mineral ratio is unaltered, the timing of protein removal, and therefore crystal formation, is likely to have been affected. The result is the weakened or softer enamel seen by microhardness testing.
Loss-of-function mutations in the DPPI gene result in early-onset periodontitis and palmoplantar keratosis, characteristic of Papillon-Lefèvre syndrome (PLS) (Hart et al., 1999; Toomes et al., 1999; Pham et al., 2004). Very little information describing the enamel in PLS-affected individuals can be found within the literature, which may be attributed to the lack of dentition in many of these individuals. Typically, individuals with PLS shed all their deciduous teeth by the age of 4, and most secondary teeth are shed by age 14 (Hart and Shapira, 1994). Kerebel et al. (1975) performed an ultrastructural odontological study of a 12-year-old female with PLS and noted structural abnormalities of the enamel, including anomalies of the prism structure. A high degree of caries, which may be indicative of weakened or softened enamel, was also mentioned in the original identification of this syndrome (Papillon and Lefèvre, 1924). Radiographically, the teeth of PLS-affected individuals appear normal. A more complete examination of enamel from PLS-affected individuals would be of great interest; however, the rarity of this syndrome (1-4 in 1,000,000) may make these studies difficult (Hart and Shapira, 1994).
Identification of the in vivo activator of KLK4 is not only important for furthering the understanding of enamel formation, but also has exciting potential as a therapeutic target for both ovarian and prostate cancer. While it remains to be determined if DPPI is the predominant activator of KLK4 in vivo, this study provides the first evidence of a ubiquitously expressed protease that can activate KLK4.
Footnotes
This investigation was supported by Research Grants DE016276 from the National Institute of Dental and Craniofacial Research (J.D.B.) and AI049261 from the National Institute of Allergy and Infectious Diseases (C.T.P.), National Institutes of Health, Bethesda, MD 20892, USA.
References
- Adkison AM, Raptis SZ, Kelley DG, Pham CT. (2002). Dipeptidyl peptidase I activates neutrophil-derived serine proteases and regulates the development of acute experimental arthritis. J Clin Invest 109:363-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Kaushal A, Bui L, Chu S, Fuller PJ, Nicklin J, et al. (2001). Human kallikrein 4 (KLK4) is highly expressed in serous ovarian carcinomas. Clin Cancer Res 7:2363-2371 [PubMed] [Google Scholar]
- Emami N, Diamandis EP. (2008). Human kallikrein-related peptidase 14 (KLK14) is a new activator component of the KLK proteolytic cascade. Possible function in seminal plasma and skin. J Biol Chem 283:3031-3041 [DOI] [PubMed] [Google Scholar]
- Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, et al. (2004). Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet 41:545-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart TC, Shapira L. (1994). Papillon-Lefèvre syndrome. Periodontol 2000 6:88-100 [DOI] [PubMed] [Google Scholar]
- Hart TC, Hart PS, Bowden DW, Michalec MD, Callison SA, Walker SJ, et al. (1999). Mutations of the cathepsin C gene are responsible for Papillon-Lefèvre syndrome. J Med Genet 36:881-887 [PMC free article] [PubMed] [Google Scholar]
- Hu JC, Ryu OH, Chen JJ, Uchida T, Wakida K, Murakami C, et al. (2000). Localization of EMSP1 expression during tooth formation and cloning of mouse cDNA. J Dent Res 79:70-76 [DOI] [PubMed] [Google Scholar]
- Hu JC, Sun X, Zhang C, Liu S, Bartlett JD, Simmer JP. (2002). Enamelysin and kallikrein-4 mRNA expression in developing mouse molars. Eur J Oral Sci 110:307-315 [DOI] [PubMed] [Google Scholar]
- Hu Y, Pham CT. (2005). Dipeptidyl peptidase I regulates the development of collagen-induced arthritis. Arthritis Rheum 52:2553-2558 [DOI] [PubMed] [Google Scholar]
- Kerebel B, Clergeau-Guerithault S, Brion M. (1975). [Ultrastructural odontological study of a case of Papillon-Lefevre disease]. Ann Anat Pathol (Paris) 20:283-292 [article in French]. [PubMed] [Google Scholar]
- Klokk TI, Kilander A, Xi Z, Waehre H, Risberg B, Danielsen HE, et al. (2007). Kallikrein 4 is a proliferative factor that is overexpressed in prostate cancer. Cancer Res 67:5221-5230 [DOI] [PubMed] [Google Scholar]
- McDonald JK, Zeitman BB, Reilly TJ, Ellis S. (1969). New observations on the substrate specificity of cathepsin C (dipeptidyl aminopeptidase I). Including the degradation of beta-corticotropin and other peptide hormones. J Biol Chem 244:2693-2709 [PubMed] [Google Scholar]
- McGuire MJ, Lipsky PE, Thiele DL. (1992). Purification and characterization of dipeptidyl peptidase I from human spleen. Arch Biochem Biophys 295:280-288 [DOI] [PubMed] [Google Scholar]
- Pagano MB, Bartoli MA, Ennis TL, Mao D, Simmons PM, Thompson RW, et al. (2007). Critical role of dipeptidyl peptidase I in neutrophil recruitment during the development of experimental abdominal aortic aneurysms. Proc Natl Acad Sci USA 104:2855-2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papillon MM, Lefèvre P. (1924). Deux cas de Keratodermie palmaireet plantaire symétrique familiale (Maladie de Meléda) chez le frère et la soeur. Coexistence dans les deux cas d’altérations dentaires graves. Bull Soc Franç Derm Syph 31:82-87 [Google Scholar]
- Pfaffl MW. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham CT, Ley TJ. (1999). Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proc Natl Acad Sci USA 96:8627-8632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham CT, Ivanovich JL, Raptis SZ, Zehnbauer B, Ley TJ. (2004). Papillon-Lefèvre syndrome: correlating the molecular, cellular, and clinical consequences of cathepsin C/dipeptidyl peptidase I deficiency in humans. J Immunol 173:7277-7281 [DOI] [PubMed] [Google Scholar]
- Ryu O, Hu JC, Yamakoshi Y, Villemain JL, Cao X, Zhang C, et al. (2002). Porcine kallikrein-4 activation, glycosylation, activity, and expression in prokaryotic and eukaryotic hosts. Eur J Oral Sci 110:358-365 [DOI] [PubMed] [Google Scholar]
- Toomes C, James J, Wood AJ, Wu CL, McCormick D, Lench N, et al. (1999). Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat Genet 23:421-424 [DOI] [PubMed] [Google Scholar]
- Tran TV, Ellis KA, Kam CM, Hudig D, Powers JC. (2002). Dipeptidyl peptidase I: importance of progranzyme activation sequences, other dipeptide sequences, and the N-terminal amino group of synthetic substrates for enzyme activity. Arch Biochem Biophys 403:160-170 [DOI] [PubMed] [Google Scholar]
- Turk BE, Lee DH, Yamakoshi Y, Klingenhoff A, Reichenberger E, Wright JT, et al. (2006). MMP-20 is predominately a tooth-specific enzyme with a deep catalytic pocket that hydrolyzes type V collagen. Biochemistry 45:3863-3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters PJ, Raymond WW, Blount JL, Caughey GH. (1998). Regulated expression, processing, and secretion of dog mast cell dipeptidyl peptidase I. J Biol Chem 273:15514-15520 [DOI] [PubMed] [Google Scholar]
- Wolters PJ, Pham CT, Muilenburg DJ, Ley TJ, Caughey GH. (2001). Dipeptidyl peptidase I is essential for activation of mast cell chymases, but not tryptases, in mice. J Biol Chem 276:18551-18556 [DOI] [PubMed] [Google Scholar]
- Wright JT, Daly B, Simmons D, Hong S, Hart SP, Hart TC, et al. (2006). Human enamel phenotype associated with amelogenesis imperfecta and a kallikrein-4 (g.2142G>A) proteinase mutation. Eur J Oral Sci 114(Suppl 1):13-17 [DOI] [PubMed] [Google Scholar]
- Yoon H, Laxmikanthan G, Lee J, Blaber SI, Rodriguez A, Kogot JM, et al. (2007). Activation profiles and regulatory cascades of the human kallikrein-related peptidases. J Biol Chem 282:31852-31864 [DOI] [PubMed] [Google Scholar]