Abstract
For cell morphogenesis, the cell must establish distinct spatial domains at specified locations at the cell surface. Here, we review the molecular mechanisms of cell polarity in the fission yeast Schizosaccharomyces pombe. These are simple rod-shaped cells that form cortical domains at cell tips for cell growth and at the cell middle for cytokinesis. In both cases, microtubule-based systems help to shape the cell by breaking symmetry, providing endogenous spatial cues to position these sites. The plus ends of dynamic microtubules deliver polarity factors to the cell tips, leading to local activation of the GTPase cdc42p and the actin assembly machinery. Microtubule bundles contribute to positioning the division plane through the nucleus and the cytokinesis factor mid1p. Recent advances illustrate how the spatial and temporal regulation of cell polarization integrates many elements, including historical landmarks, positive and negative controls, and competition between pathways.
The morphogenesis of fission yeast cells is regulated by microtubule-based mechanisms that position sites of actin assembly at cell tips and the cell division plane.
One of the ultimate goals in cell biology is to understand how cells are assembled. As in the development of multicellular organisms, single cells need to form distinct spatial domains with specific form, structure, and functions. How do cells organize themselves in space to form a specific shape and size?
The fission yeast Schizosaccharomyces pombe is an attractive, simple unicellular model organism for studying cell morphogenesis. These are nonmotile cells with highly invariant shape 8–14 µm long and 3 µm in diameter. The relative simplicity of the cells and the powers of genetic approaches and live cell imaging facilitate rigorous and quantitative studies.
Here, we review the current understanding of spatial regulation in fission yeast. The cell defines distinct cortical domains at each of the cell tips, along the sides of cells, and at the cell division plane. Each cortical domain is characterized by different sets of molecules, which impart distinct functions. In particular, as it proceeds through its cell cycle, the cell delineates distinct actin-rich cortical regions at cell tips for polarized cell growth and at the middle for cell division. In both cases, a self-organizing network of microtubules directly or indirectly contributes to the proper localization of these markers. In cell polarity, microtubule ends transport polarity factors to the plasma membrane, where they function to recruit protein complexes involved in actin assembly. In cytokinesis, a medial cortical site is marked by an interacting system of microtubules, the nucleus, and cell tip factors, and functions to organize actin filaments into a cytokinetic ring. This reliance on microtubules contrasts with polarity mechanisms in budding yeast in which spatial cues are dependent on septins and actin, but not microtubules. As many of these processes involve conserved proteins, this work in fission yeast contributes toward understanding the more complex microtubule-based regulation of cell migration, cytokinesis, and cell shape regulation in animal cells. This work in fission yeast thus provides a paradigm for how a self-organizing system can shape a cell.
A FISSION YEAST PRIMER
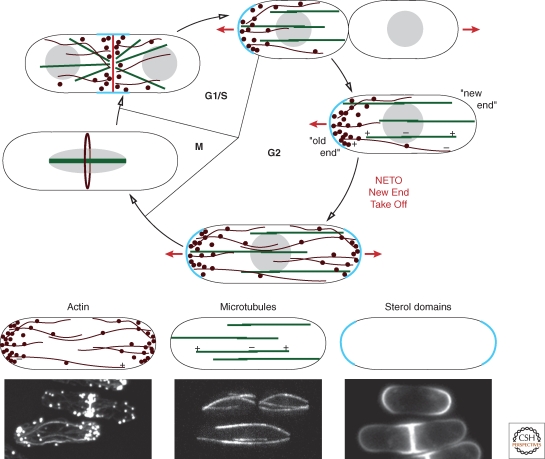
Fission yeast cells grow by tip extension and then divide medially (Fig. 1). Growth is regulated both temporally and spatially, with several important cell–cycle-regulated transitions. Each of these transitions in cell polarization is predicted to be controlled by cell cycle and polarity regulators (Mitchison and Nurse 1985). Immediately after division, cells initially grow from only one end, the “old end” (the end that was the cell end in the previous cell cycle). Then, at a certain point in G2 phase, cells begin growing also from the new end, in a process termed “new end take off” (NETO). In mitosis, cells cease growth, and at the end of mitosis, the growth machinery is directed toward the division site for septation. On cell division, growth is once again redirected to the old cell tip, for “old end take off.” Studies on these transitions during the cell cycle reveal how polarization is established, turned off, or remembered. Thus, in the course of the normal cell cycle, sites of growth may be already marked by spatial cues located at sites where cells have either divided or grown before.
Figure 1.
Cytoskeleton organization during the cell cycle. The cell growth cycle of fission yeast is represented, with sites of active growth labeled by the red lines and arrows. The microtubules (green), actin cytoskeleton (dark red), and sterol-rich membrane domains (light blue) are depicted at representative cell cycle stages. Individual images of these structures are shown at the bottom, where the actin cytoskeleton is stained with phalloidin, microtubules are shown with GFP-atb2, and sterol-rich domains are labeled with filipin.
However, fission yeast cells can also establish the rod shape from seemingly symmetric situations. For instance, on sporulation, the small round spore germinates and immediately begins to grow into a rod shape. Thus, it is clear that in these spheres, internal cues direct the formation of the rod shape. Do spores retain a “memory” of marked polarization sites? More intriguing perhaps, are cellular components capable of self organizing a zone of polarization with the proper dimensions and orientation de novo from a symmetric situation?
Tip growth in fungal cells is a complex process involving the cell wall, membranes, and the cytoskeleton. High turgor pressure within the cell may be the primary force generator (Harold 1990; Minc et al. 2009). As turgor pressure itself exerts an outward force in all directions, cells must carefully modulate cell wall elasticity at specific sites, by cell wall remodeling enzymes, for controlled extension at the cell tips. Thus, some of the ultimate downstream events in cell growth in these fungal cells may be at the level of the cell wall and turgor pressure.
Actin is organized during interphase into actin cables and actin patches at growing cell tips (Fig. 1) (Marks et al. 1986). The actin patches are sites of endocytosis (Kaksonen et al. 2003; Huckaba et al. 2004; Gachet and Hyams 2005). Actin cables are long bundles of short actin filaments arranged largely in a parallel configuration (Kamasaki et al. 2005). These cables act as tracks for guiding myosin V-based transport of cargoes, such as vesicles to the growing cell tips (Motegi et al. 2001). During mitosis, actin is reorganized into a medial contractile ring structure responsible for cytokinesis. Although actin filaments are strictly required for polarized growth, actin is probably not directly exerting force for growth. Rather, it may serve several functions in targeting membrane traffic to the cell tips and in endocytic recycling of key membrane components. Key cargoes include membrane proteins such as the cell wall synthases and remodeling enzymes (Cortes et al. 2002; Cortes et al. 2005; Mulvihill et al. 2006).
Microtubules (MTs) are organized in bundles oriented along the long axis of the cells during interphase and in bundles comprising the mitotic spindle in mitosis (Fig. 1) (Marks et al. 1986). The interphase bundles contain MTs overlapping in an antiparallel configuration and attached to the nuclear envelope at multiple sites near the middle of the cell (Drummond and Cross 2000; Tran et al. 2001). The plus ends of MTs constantly probe the cell tips. These MT ends generally grow toward the cell tips, contact the cell tip for about 1–2 minutes, and then shrink. These bundles, which are nucleated from gamma tubulin particles present on the MTs, “self organize” through the action of motors and antiparallel bundlers (Janson et al. 2005; Carazo-Salas and Nurse 2006; Daga et al. 2006a; Sawin and Tran 2006; Janson et al. 2007; Bratman and Chang 2008). The interaction of MTs with the cortex and regulation of MT catastrophe regulate the orientation of these MTs (Brunner and Nurse 2000). As described in the following section, MTs are not essential for polarized cell growth or cytokinesis in fission yeast, but regulate polarity by transporting important regulators to correct locations in the cell.
Membrane domains may also contribute as a platform for polarity. Many polarity factors dock at specific sites on the plasma membrane, even in the absence of the microtubule or actin cytoskeleton. Staining with the sterol-dye filipin, for instance, reveals large (μm2) membrane domains covering sites of cell growth at cell tips, and at the cell division site (Fig. 1) (Wachtler et al. 2003). Although the precise functions of these membrane domains are not yet clear, these may regulate the distribution of membrane proteins and lipids critical for polarization.
FROM FUNNY-SHAPED CELLS TO GENES
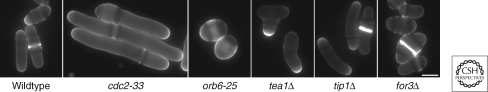
The utility of S. pombe as a genetically tractable model for morphogenesis was established by finding mutants with altered morphology. Wild-type S. pombe cells are very uniform in their rod shape and size, making it easy to spot mutants with altered morphology. In genetic screens for conditional mutants, it quickly became apparent that large numbers of genes regulating cell size and shape could be identified (Verde et al. 1995; Brunner and Nurse 2000; Snaith and Sawin 2003). This approach discovered a veritable “zoo” of morphological mutants, ranging from midgets to monsters (Fig. 2). Initial mutants characterized were those of elongated morphology, which were mutants in the cell division cycle (cdc mutants). Other morphological mutants include those with a bent phenotype, branching phenotypes, and rounded or bottle-shaped phenotypes.
Figure 2.
Shape mutants. Calcofluor staining showing the normal rod shape of wild-type cells and the aberrant elongated, round, T, curved, and dumpy shapes of cdc2-33, orb6-25, tea1Δ, tip1Δ, and for3Δ cells, respectively. Bar, 5 µm.
In general, these cell-shape mutants served as a foundation for subsequent studies on morphology. Mutated genes have been cloned and characterized to varying degree. Reverse genetic approaches have also contributed to the identification of additional genes. With the advent of a genome-wide deletion library, it is likely that almost all of the nonessential cell morphology genes will be described in the near future. These different mutant phenotypes delineate several major processes involved in cell polarity. Mutants with a rounded phenotype represent genes with general functions in cell polarization, including the small GTPases cdc42 and ras genes, and their regulators. Second, bent and branched mutants have phenotypes similar to tubulin mutants or cells treated with MT-destabilizing drugs. These genes represent regulators of interphase MTs, whereas others affect more specifically the MT polarity pathways (such as the tea1 pathway; see the following section). These mutants are also often defective in NETO. Third, mutants in actin and actin regulators often have primary defects in cytokinesis, as they cannot form the actin-based contractile ring. However, mutants with more specific actin defects, for instance in actin cable assembly during interphase or in actin patch-based endocytosis, often form partially depolarized dumpy, fat cells. In general, each of these sets of mutants defines a network of functions. In the following sections, we describe these different modules responsible for cell polarization and some of the important molecular links between these modules.
REGULATORY MODULES CONTROLLING CELL POLARITY IN THE FISSION YEAST
The cell polarization machinery organizes cytoskeletal components and membrane traffic to establish an axis of polarity. The loss of the general polarization machinery can lead to loss of polarity and isotropic growth, producing round cells (or nearly round), instead of rod-shaped cells. In this section, we briefly summarize some conserved signaling modules in the general polarization machinery. In general, many of the basic signaling modules that have been characterized in budding yeast polarity are also likely to be operating in fission yeast, but with of course some interesting and often significant variations.
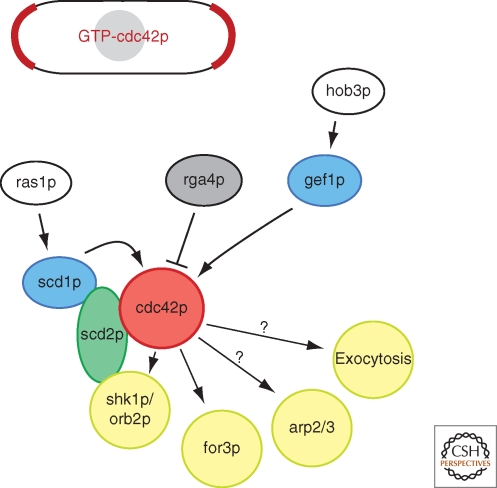
As in many organisms, the small GTPase cdc42p is at the core of cell polarization pathways in the fission yeast (Fig. 3). Regulatory factors, cdc42p, and its effectors form a signaling module that bears many similarities to that of other cell types, especially Saccharomyces cerevisiae (refer to other articles of this collection). In fission yeast, cdc42 is an essential gene: Deletion of cdc42 produces small, round, dense cells, whereas overexpression of constitutively active forms leads to large round cells (Miller and Johnson 1994). Until recently, investigation of cdc42p-dependent processes in fission yeast has been hindered by lack of good conditional cdc42 mutants, which are just beginning to be characterized.
Figure 3.
The cdc42p module. Schematic depiction of cdc42p signaling in fission yeast. Active, GTP-bound cdc42p localizes to sites of active growth and to the division site.
Activation of cdc42p is controlled by two distinct guanine exchange factors (GEFs), scd1p and gef1p (Chang et al. 1994; Coll et al. 2003; Hirota et al. 2003). A double mutant is lethal, whereas single deletions reveal that scd1p is necessary primarily for polarized growth, whereas gef1p is required for the process of NETO. The Ras homolog ras1p activates the cdc42p module through scd1p (Chang et al. 1994). Although ras1p is nonessential, it is necessary for mating and for polarized growth—S. pombe ras1 mutants are round—and is in turn activated by distinct GEFs for each function, ste6p and efc25p, respectively (Hughes et al. 1990; Tratner et al. 1997; Papadaki et al. 2002). This ras1p-scd1p-cdc42p pathway is analogous to the Bud1p-Cdc24p-Cdc42p pathway used in the budding yeast for cell polarization and bud site selection. There is some question whether ras1 itself provides a spatial cue: ras1p is present both at the plasma membrane and on internal membranes, but mutants in ras1 that perturb plasma membrane localization suggest that endomembrane localization is sufficient for ras1p to control the scd1-cdc42p pathway and polarized cell growth (Onken et al. 2006). The key spatial determinants may include cdc42p itself or its GEFs, which localize to the plasma membrane at cell tips and septum (Coll et al. 2003; Hirota et al. 2003). Cdc42p may also be activated in ras1p-independent ways. A Bar-domain protein hob3p contributes to cdc42p activation by gef1p (Coll et al. 2007). A negative regulator, rga4p, a cdc42p GTPase activating protein (GAP), is localized in cortical dots along the sides of cells, at nongrowing regions of the cell surface, and thus may spatially restrict cdc42p activity zones (Das et al. 2007; Tatebe et al. 2008).
One well-characterized, conserved effector of cdc42p is the formin for3p, an actin nucleator responsible for the assembly of actin filaments for actin cables (Feierbach and Chang 2001; Nakano et al. 2002). Like many formins, for3p adopts an autoinhibited conformation that prevents accessibility of its localization domain or its active site. Cdc42p binding, together with another formin-binding protein bud6p, competes with intramolecular interaction to relieve autoinhibition (Martin et al. 2007). Activation of for3p allows for for3p localization to the cell tip and actin cable assembly. Cdc42p may also activate Arp2/3 actin nucleation at actin patches to regulate endocytosis. However, although mammalian pathways show that cdc42p regulates Arp2/3 through the nucleation activator WASp protein, the yeast orthologues (wsp1p) lacks the predicted cdc42p interaction domains. It is also unknown whether and how cdc42p regulates exocytosis in S. pombe: The exocyst complex, a well-characterized Cdc42 effector that marks sites of exocytosis, is not essential for cell polarization in the fission yeast (Wang et al. 2002).
Additional polarity modules are represented by the orb kinases. These were identified in forward genetic screens as round-shaped mutants (Verde et al. 1995), and interestingly all encode kinases or their regulators (orb2/shk1, orb3/nak1, orb4/sts5, orb5/casein kinase, and orb6/ndr). One key cdc42p effector is the essential Ste20-like kinase orb2p/shk1p, which forms a complex with scd1p and cdc42p by association with the scaffolding protein scd2p (Chang et al. 1994). Potential downstream substrates of the orb2p/shk1p kinase include the microtubule-associated protein tea1p (see the following section) and the rho GAP rga8p, whose function is not clear (Kim et al. 2003; Yang et al. 2003). Orb6p, a homolog of the NDR/Tricornered kinase, forms complexes with mob2p and the furry-related protein mor2p and interacts with Lkb1/Par-4-related kinase orb3p and its cofactor mo25p (Verde et al. 1998; Hirata et al. 2002; Hou et al. 2003; Kanai et al. 2005; Mendoza et al. 2005). Although substrates of orb kinases remain to be fully characterized, it is likely that these kinases form important regulatory networks that ultimately modulate cdc42p activity or regulate downstream effectors such as formin activity and membrane trafficking.
MICROTUBULES, THE TEA SYSTEM, AND NETO
Microtubules contribute to polarity regulation and shape control in fission yeast. Cells with an abnormal interphase microtubule cytoskeleton still grow in a polarized way, but at aberrant positions. For instance, mutations in tubulin genes or in genes necessary for microtubule biogenesis, organization, or dynamics lead to misshapen cells that are curved or T-shaped (Umesono et al. 1983; Hirata et al. 1998; Radcliffe et al. 1998; Zimmerman et al. 2004). The branched forms are especially predominant after stress or recovery from starvation (Sawin and Snaith 2004; Tatebe et al. 2005). Time- lapse studies using pharmacological inhibition of MTs showed that MTs are also needed for efficient NETO (unpublished). Thus, these types of studies led to a proposal that microtubules function in interphase to label cell ends as sites for cell growth.
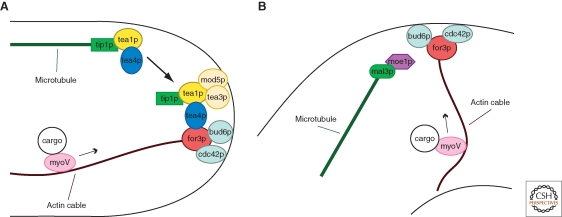
A substantial body of literature has now shown that one way that microtubules regulate cell polarization is through the “tea” system (Fig. 4A). tea1 was originally identified as a mutant that showed morphological and NETO defects, similar to those seen in MT mutants (Mata and Nurse 1997). Molecular and localization analysis revealed that tea1p, a kelch-repeat protein, localizes to the plus ends of MTs as well as to the cortex (Mata and Nurse 1997; Behrens and Nurse 2002; Feierbach et al. 2004). As tea1 mutants only have minor defects in MT organization, tea1p was shown to have more of a role in MT-regulation of polarity (Mata and Nurse 1997; Behrens and Nurse 2002). Subsequent biochemical studies identified that tea1p operates closely with a partner, tea4p (Martin et al. 2005; Tatebe et al. 2005).
Figure 4.
Microtubule-dependent polarization. (A) Model for how the tea1/4p complex is deposited at cell ends by microtubules and recruits the formin for3p for actin cable assembly. These in turn provide tracks for the delivery of myosin V-driven cargoes to cell tips. (B) Microtubule-induced tea1/4p-independent polarization at ectopic cortical sites.
These proteins are transported on growing microtubule plus ends to cell tips, where they are deposited as discrete “dots” at the cortex. MT plus end localization is dependent on the +TIP tip1p (CLIP170), which is transported to the microtubule end by the kinesin tea2p (Browning et al. 2000; Brunner and Nurse 2000; Browning et al. 2003; Busch et al. 2004). The growth of the MT delivers these proteins to the cell tip. When the microtubule ends reach the cell tip, tip1p, tea1p, and tea4p are somehow released at the cortex. These observations show how MT contact at the cell cortex directs the localization of these proteins. At the cortex, tea1p is anchored by at least two proteins, a prenylated membrane protein mod5p and the tea1p-like protein tea3p (Arellano et al. 2002; Snaith and Sawin 2003; Snaith et al. 2005). As mod5p and tea3p localization at the tip is dependent on tea1p itself, this suggests the existence of positive feedback regulation in assembling these complexes at the membrane. These tea1/4p dots at the cortex are stable and persist for many minutes or hours, and even in the absence of MTs (unpublished).
At the cortex, the tea1/4p complex is thought to recruit directly or indirectly other polarity factors, to initiate the construction of large protein complexes (the polarisome) that regulate cell polarization at the cell tips. Consistent with this idea, tea1p fractionates in large (45S and 75S) protein complexes in yeast extracts (Feierbach et al. 2004). These complexes may mediate actin assembly and other aspects of cell polarization, such as membrane trafficking. One downstream component is the pom1p kinase (a DYRK-family protein kinase), which shares similar functions as tea1p in cell polarization, but also has other roles in regulating spatial aspects of cytokinesis and cell cycle progression (see the following section) (Bähler and Pringle 1998; Behrens and Nurse 2002).
A critical target of the tea1/4p complex is the formin for3p itself. Tea4p binds for3p directly in vitro, and forms complexes with for3p in vivo (Martin et al. 2005). It may primarily act to recruit for3p to the cell tip, rather than regulating its autoinhibitory conformation. Remarkably, a tea1p-for3p fusion that targets the formin to the new end is sufficient to override cell cycle signals for inducing bipolar growth, suggesting that formin recruitment is a key event in establishment of a new site of polarized cell growth. These studies outline a simple model for NETO, in which microtubules transport tea1/tea4p to cell tips, where they recruit the formin to initiate actin assembly and cell polarization at the new cell end (Fig. 4).
In addition to actin, many other factors are at play for cell polarization. It is thus likely that the tea1/4p complex has inputs into cdc42p regulation and in aspects of membrane trafficking, including endocytosis, exocytosis, and membrane organization. Toward this point, two other genes implicated in NETO are arf6, which encodes a member of the Arf GTPases involved in vesicle trafficking, and sla2/end4, which is required for endocytosis (Castagnetti et al. 2005; Fujita 2008). There are also likely to be feedback loops that reinforce initial polarities.
How the timing of the NETO transition in G2 phase is regulated remains unknown. This is unlikely to be directly controlled by the tea1/4p complex as tea1/4p localize to both cell ends in pre-NETO and post-NETO cells. One hypothesis is that the formation of a complex between the tea pathways and downstream components, such as the formin for3p is a rate-limiting step. This activation certainly could involve signaling mechanisms downstream of the cell cycle machinery. A requirement for completion of DNA synthesis suggests that some aspect of DNA checkpoint control could be involved. One interesting observation is that tea4p associates with type I phosphatases and recruits them to cell tips (Alvarez-Tabares et al. 2007). Further, partial loss of function of many of the kinases fundamental for general cell polarization leads to monopolar growth, suggesting that NETO may be triggered by phosphorylation events. Two kinases in particular stand out as good candidates for this regulatory step. First, the pom1p kinase may be under cell cycle control, as its activity is low in pre-NETO G1 cells and increases in G2 cells, but its substrate(s) for cell morphogenesis are unknown (Bähler and Pringle 1998; Bähler and Nurse 2001). Second, the shk1p protein kinase can phosphorylate tea1p directly and a hypomorphic mutation, orb2-34, displays NETO defect similar to tea1Δ (Kim et al. 2003). These two kinases provide good entry points to study the cell cycle regulation of NETO.
POLARITY INITIATION AT AN ECTOPIC SITE
The study of NETO has been instrumental in defining the links between microtubules and the polarity machinery, but admittedly, this line of research does not really inform on whether microtubules break symmetry. As NETO is a process in which a cell end is activated for polarization, one could argue that this “new end” could be already marked from its history as a site of previous division. Thus, there has been a question of whether microtubules can initiate polarization at the cortex at a site that has never been a site of growth or division.
Recent studies have addressed this problem by physically manipulating the shape of S. pombe cells. The cell wall of fission yeast cells is actually elastic, and thus these cells can be bent using external forces. In these studies, yeast cells are bent by introducing them into small round wells or into curved chambers in microfluidic devices (Terenna et al. 2008; Minc et al. 2009). On the change in cell shape, microtubule ends begin to contact the cell sides. At these MT contact sides, polarity factors such as tea1p or bud6p accumulate in a microtubule-dependent manner within seconds of MT contact. Within about 10 minutes, a mature “patch” of polarity factors is formed. Many polarity proteins, including the formin for3p, activated cdc42p, and actin cables, appear in these cortical zones. Interestingly, this effect is independent of the tea1 pathway, but rather depends on a pathway defined by the +TIP mal3p (EB1) and moe1p, a protein that binds to mal3p and the cdc42p GEF scd1p (Chen et al. 1999; Chen et al. 2000). One plausible model is that the MTs induce polarization through mal3p and moe1p-dependent activation of scd1p and cdc42p, leading to the activation of the formin for3p. These studies thus suggest the presence of a second pathway that connects the MTs with for3p (Fig. 4B).
COMPETITION BETWEEN PATHWAYS THAT POSITION SITES OF POLARIZATION
There appear to be at least three layer of mechanisms that regulate polarization site positioning: 1) the MT/tea1/tea4 pathway, which operates in NETO and promotes cell tip polarization (Mata and Nurse 1997; Martin et al. 2005; Tatebe et al. 2005); 2) the MT/mal3/moe1 pathway, identified as operating for polarization on the cell sides in bent cells, but not needed for NETO (Minc et al. 2009); and 3) a “nuclear pathway” that promotes polarization and branching off the cell side near the nucleus in the absence of MTs (Castagnetti et al. 2007). These competing activities are revealed when one or more of the others are deleted. In cells with tea1p and intact MTs, the tea pathway is predominant and promotes polarization from the cell ends. Growing cell ends also contribute to repress branching from the sides. In the absence of tea1, the mal3/moe1 pathway promotes MT dependent branching at the cell sides, at sites of MT contact. In the absence of MTs and/or the two pathways, the cell then branches near the nucleus using the third, as yet uncharacterized pathway.
As normal cells have tea1p, one might wonder why the other pathways are present. One explanation is that these alternate pathways for polarization could be used in particular environmental conditions. For instance, cells in nature that are bent by environmental forces or by their own growth in tight spaces could use the mal3/moe1 pathway to establish new “free” sites of growth. Cells that lose their MTs on cold temperatures or encounters with natural anti-MT agents in the environment could use the third pathway to initiate growth. Thus, these three layers of cell polarization mechanisms may favor yeast growth in more wild environments.
DEFINING THE CELL MIDDLE
Another example of symmetry breaking occurs in establishing the site of cell division for cytokinesis. In this case, a medial ring-shaped cortical band on the side of the rod-shaped cell is marked in interphase as the future division site. In mitosis, this band recruits contractile ring components such as myosin and actin to form the contractile ring.
Although elements of the mitotic spindle appear to specify the cleavage furrow position in animal cells, in fission yeast (as in some plant cells), the position of the nucleus functions as the primary spatial cue that specifies this site. Normally, the nucleus is positioned near the middle of the rod-shaped cells, and an actin-based contractile ring forms at cortex at the middle of the cell. When the nucleus is displaced, the site of ring assembly is set at the site of the displaced nucleus (Daga and Chang 2005; Tolic-Norrelykke et al. 2005). Interphase cytoplasmic MTs are indirectly involved in this process, as they are responsible for nuclear positioning (Tran et al. 2000; Tolic-Norrelykke et al. 2005; Daga et al. 2006b). MTs attach to the outer nuclear envelope and exert pushing forces on the nucleus when they contact the cell tips. It has been proposed that a balance of these pushing forces is responsible for nuclear centering.
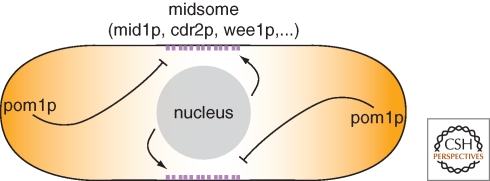
Mid1p, which has some similarities to the animal cytokinesis factor anillin, appears to be a molecular mark responsible for ring positioning. mid1 mutants have profound defects in positioning the ring (Sohrmann et al. 1996; Chang et al. 1997). Mid1p localizes to a series of >50 cortical dots overlying the position of the nucleus throughout interphase (Paoletti and Chang 2000). In early mitosis, myosin and other contractile ring components are recruited to these cortical structures to initiate ring assembly (Wu et al. 2003; Motegi et al. 2004). Recent work has revealed that mid1p actually resides at the cortex in a larger protein complex named the “midsome,” which functions not only in cytokinesis but also in cell cycle control (see the following section) (Fig. 5). The nucleus provides a positive spatial signal for positioning mid1p. One impressive demonstration of this link is that mid1p dots continually respond to nuclear position in a dynamic manner, and even move with the nucleus when the nucleus moves (Daga and Chang 2005). It has been proposed that the nucleus may affect mid1p position in part through a nuclear shuttling mechanism that could effectively increase the local concentration of mid1p in the perinuclear region (Paoletti and Chang 2000; Almonacid et al. 2009).
Figure 5.
Positive and negative signals for septum positioning and cell cycle regulation. The medially positioned nucleus provides positive signal for the localization of midsome component mid1p, whereas polarity regulators, including the kinase pom1p, inhibit mid1p localization and septum formation at the tips. Pom1p also negatively regulates the midsome component cdr2p to delay M phase entry until sufficient cell length has been reached.
In addition to the positive effect of the nucleus, it is now apparent that negative cues from the cell tips contribute to division site positioning, to prevent rings from forming at the very ends of cells (Fig. 5). In particular, the protein kinase pom1p is a candidate negative inhibitor that resides in a gradient emanating from the cell tips. Pom1p appears to inhibit mid1p distribution, although from only one of the cell ends (the nongrowing cell end) (Celton-Morizur et al. 2006; Padte et al. 2006). Pom1p also provides additional mid1-independent inhibitory function: pom1 single mutants display cytokinetic rings that are only slightly offset, whereas in a mid1 pom1 double mutant, these cells form rings at the very cell tips (Huang et al. 2007). Although this process is not strictly dependent on microtubules, pom1p is indirectly localized to cell tips via MT plus ends and the tea1/4p proteins (Bähler and Pringle 1998). Thus, MTs may contribute indirectly to both negative and positive signals operating in cytokinesis.
REGULATION OF THE CELL CYCLE FROM THE CELL TIPS
Polarity factors at cell tips also contribute to regulation of cell size and the cell cycle. As fission yeast cells grow through interphase, the length of these cells reflects the length of the interphase cell cycle period. It has been predicted that cells may somehow sense that they are big enough before entering into mitosis; however, the mechanisms for how cells sense their own size has been elusive. Recent findings show that the pom1p kinase, a polarity factor that is positioned by tea1/4p and MTs at cell tips, may act as a cell length sensor (Martin and Berthelot-Grosjean 2009; Moseley et al. 2009). In addition to regulating cell polarity and cytokinesis, pom1p is also a dose-dependent inhibitor of mitotic entry that negatively regulates cdr2p, an inhibitor of the cell cycle kinase wee1p. Pom1p forms gradients emanating from the cell tips, whereas wee1p and cdr2p are localized (with mid1p) to the medial midsome structures near the nucleus (Fig. 5). In small cells, pom1p is thought to inhibit cdr2p. As cells grow and progress through G2, the source of the pom1p gradient at cell ends are moved further apart and the concentration of pom1p at the cell middle is reduced, allowing cdr2p activation and cell cycle progression. The model posits that cells enter M phase at a critical cell length at which the cell tips and the gradient of pom1p are far enough away from the medial region.
CONCLUDING REMARKS
Using the fission yeast, major progress has been made in understanding the molecular basis of spatial regulation. Functional cortical zones may be delineated by the collective dynamic interactions of microtubules, actin, and a large number of protein complexes (as well as membranes and organelles). Changes in these factors choreograph spatial reorganization during the different phases of the cell cycle. In these studies, a number of basic concepts involved in symmetry breaking have emerged:
Memory. Cells are rarely entirely “symmetric.” It is clear that fission yeast cells remember where previous sites of growth (at cell tips) and division sites are located, even those from previous generations, suggesting that there are hard-wired “historical landmarks” that help to guide polarization. However, cells can lose this memory in certain conditions, for instance on starvation, when they then become reliant on other spatial cues such as microtubules.
Microtubules. MTs modulate polarity by transporting factors on their plus ends and depositing them onto the cortex. MT-associated factors then recruit the polarity machinery, such as cdc42p and formins, for cell polarization. These MT-based systems are critical for maintaining cell shape and regulating bipolar growth. At least two distinct pathways have been identified that connect MTs with actin assembly through formin recruitment—the tea1/4p complex and a novel moe1p-dependent pathway.
Positive feedback. There is an important positive feedback loop between the mechanisms that shape the cell and cell shape itself, such that fission yeast cells rarely need to break symmetry during vegetative growth. The rod shape of the cell helps to orient the MTs along the long axis of the cell. This directs the MT plus ends and its cargoes to the cell tips, which in turn induces polarized growth that maintains cell shape. Additional feedbacks also certainly exist, for instance, to amplify the signal from the MT ends to establish a zone of polarization at the cortex or to maintain actin organization at an established growth site.
Inhibitory modulators. Proper spatial organization also requires that the cortical zones be contained by inhibitory mechanisms from neighboring domains. This is observed in the case of tip growth, where growing cell tips provide inhibitory signals preventing growth at cell sides, or in the case of cell division positioning, where cell tips form an exclusion zone for septum assembly.
Shape and size control. The basic concepts of how shapes and sizes are determined are still largely unknown, even in this simple rod. However, recent evidence is beginning to suggest how gradients of key proteins can provide a morphogen field for size control, even in these small single cells.
The genetic dissection and physical manipulation of fission yeast’s simple rods has led to the discovery of basic modules and principles of cell morphogenesis that are also used to shape more complex cells. For instance, microtubules provide spatial information for cell migration or placement of the division site in mammalian cells; positive feedback mechanisms are known to reinforce polarity establishment in multiple systems; and distinct cortical domains in epithelial cells and worm embryos form thanks to reciprocal negative regulations. Further dissection of the fission yeast cell is likely to provide important novel insights in cell morphogenesis.
ACKNOWLEDGMENTS
Research in SGM laboratory is supported by a Swiss National Science Foundation (SNF) Professorship grant and a Human Frontiers Science Program (HFSP) Career Development Award. FC is supported by NIH GM56836 and GM069670, and an Ellison Medical Foundation Senior Scholar Award.
Footnotes
Editors: Rong Li and Bruce Bowerman
Additional Perspectives on Symmetry Breaking in Biology available at www.cshperspectives.org
REFERENCES
- Almonacid M, Moseley JB, Janvore J, Mayeux A, Fraisier V, Nurse P, Paoletti A 2009. Spatial control of cytokinesis by Cdr2 kinase and mid1/anillin nuclear export. Curr Biol (in press). [DOI] [PubMed] [Google Scholar]
- Alvarez-Tabares I, Grallert A, Ortiz JM, Hagan IM 2007. Schizosaccharomyces pombe protein phosphatase 1 in mitosis, endocytosis and a partnership with Wsh3/Tea4 to control polarised growth. J Cell Sci 120:3589–3601 [DOI] [PubMed] [Google Scholar]
- Arellano M, Niccoli T, Nurse P 2002. Tea3p is a cell end marker activating polarized growth in Schizosaccharomyces pombe. Curr Biol 12:751–756 [DOI] [PubMed] [Google Scholar]
- Bähler J, Nurse P 2001. Fission yeast Pom1p kinase activity is cell cycle regulated and essential for cellular symmetry during growth and division. Embo J 20:1064–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Pringle JR 1998. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev 12:1356–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens R, Nurse P 2002. Roles of fission yeast tea1p in the localization of polarity factors and in organizing the microtubular cytoskeleton. J Cell Biol 157:783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratman SV, Chang F 2008. Mechanisms for maintaining microtubule bundles. Trends Cell Biol 18:580–586 [DOI] [PubMed] [Google Scholar]
- Browning H, Hackney DD, Nurse P 2003. Targeted movement of cell end factors in fission yeast. Nat Cell Biol 5:812–818 [DOI] [PubMed] [Google Scholar]
- Browning H, Hayles J, Mata J, Aveline L, Nurse P, McIntosh JR 2000. Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J Cell Biol 151:15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D, Nurse P 2000. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102:695–704 [DOI] [PubMed] [Google Scholar]
- Busch KE, Hayles J, Nurse P, Brunner D 2004. Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev Cell 6:831–843 [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Nurse P 2006. Self-organization of interphase microtubule arrays in fission yeast. Nat Cell Biol 8:1102–1107 [DOI] [PubMed] [Google Scholar]
- Castagnetti S, Behrens R, Nurse P 2005. End4/Sla2 is involved in establishment of a new growth zone in Schizosaccharomyces pombe. J Cell Sci 120:2205–2213 [DOI] [PubMed] [Google Scholar]
- Castagnetti S, Novak B, Nurse P 2007. Microtubules offset growth site from the cell centre in fission yeast. J Cell Sci 120:2205–2213 [DOI] [PubMed] [Google Scholar]
- Celton-Morizur S, Racine V, Sibarita JB, Paoletti A 2006. Pom1 kinase links division plane position to cell polarity by regulating Mid1p cortical distribution. J Cell Sci 119:4710–4718 [DOI] [PubMed] [Google Scholar]
- Chang EC, Barr M, Wang Y, Jung V, Xu HP, Wigler MH 1994. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell 79:131–141 [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P 1997. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol 137:169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CR, Chen J, Chang EC 2000. A conserved interaction between Moe1 and Mal3 is important for proper spindle formation in Schizosaccharomyces pombe. Mol Biol Cell 11:4067–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CR, Li YC, Chen J, Hou MC, Papadaki P, Chang EC 1999. Moe1, a conserved protein in Schizosaccharomyces pombe, interacts with a Ras effector, Scd1, to affect proper spindle formation. Proc Natl Acad Sci 96:517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll PM, Rincon SA, Izquierdo RA, Perez P 2007. Hob3p, the fission yeast ortholog of human BIN3, localizes Cdc42p to the division site and regulates cytokinesis. Embo J 26:1865–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll PM, Trillo Y, Ametzazurra A, Perez P 2003. Gef1p, a new guanine nucleotide exchange factor for Cdc42p, regulates polarity in Schizosaccharomyces pombe. Mol Biol Cell 14:313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes JC, Carnero E, Ishiguro J, Sanchez Y, Duran A, Ribas JC 2005. The novel fission yeast (1,3)beta-D-glucan synthase catalytic subunit Bgs4p is essential during both cytokinesis and polarized growth. J Cell Sci 118:157–174 [DOI] [PubMed] [Google Scholar]
- Cortes JC, Ishiguro J, Duran A, Ribas JC 2002. Localization of the (1,3)beta-D-glucan synthase catalytic subunit homologue Bgs1p/Cps1p from fission yeast suggests that it is involved in septation, polarized growth, mating, spore wall formation and spore germination. J Cell Sci 115:4081–4096 [DOI] [PubMed] [Google Scholar]
- Daga RR, Chang F 2005. Dynamic positioning of the fission yeast cell division plane. Proc Natl Acad Sci 102:8228–8232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga RR, Lee KG, Bratman S, Salas-Pino S, Chang F 2006a. Self-organization of microtubule bundles in anucleate fission yeast cells. Nat Cell Biol 8:1108–1113 [DOI] [PubMed] [Google Scholar]
- Daga RR, Yonetani A, Chang F 2006b. Asymmetric microtubule pushing forces in nuclear centering. Curr Biol 16:1544–1550 [DOI] [PubMed] [Google Scholar]
- Das M, Wiley DJ, Medina S, Vincent HA, Larrea M, Oriolo A, Verde F 2007. Regulation of cell diameter, For3p localization, and cell symmetry by fission yeast Rho-GAP Rga4p. Mol Biol Cell 18:2090–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DR, Cross RA 2000. Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr Biol 10:766–775 [DOI] [PubMed] [Google Scholar]
- Feierbach B, Chang F 2001. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol 11:1656–1665 [DOI] [PubMed] [Google Scholar]
- Feierbach B, Verde F, Chang F 2004. Regulation of a formin complex by the microtubule plus end protein tea1p. J Cell Biol 165:697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A 2008. ADP-ribosylation factor arf6p may function as a molecular switch of new end take off in fission yeast. Biochem Biophys Res Commun 366:193–198 [DOI] [PubMed] [Google Scholar]
- Gachet Y, Hyams JS 2005. Endocytosis in fission yeast is spatially associated with the actin cytoskeleton during polarised cell growth and cytokinesis. J Cell Sci 118:4231–4242 [DOI] [PubMed] [Google Scholar]
- Harold FM 1990. To shape a cell: An inquiry into the causes of morphogenesis of microorganisms. Microbiol Rev 54:381–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata D, Kishimoto N, Suda M, Sogabe Y, Nakagawa S, Yoshida Y, Sakai K, Mizunuma M, Miyakawa T, Ishiguro J, Toda T 2002. Fission yeast Mor2/Cps12, a protein similar to Drosophila Furry, is essential for cell morphogenesis and its mutation induces Wee1-dependent G(2) delay. Embo J 21:4863–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata D, Masuda H, Eddison M, Toda T 1998. Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. Embo J 17:658–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Tanaka K, Ohta K, Yamamoto M 2003. Gef1p and Scd1p, the Two GDP-GTP exchange factors for Cdc42p, form a ring structure that shrinks during cytokinesis in Schizosaccharomyces pombe. Mol Biol Cell 14:3617–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou MC, Wiley DJ, Verde F, McCollum D 2003. Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J Cell Sci 116:125–135 [DOI] [PubMed] [Google Scholar]
- Huang Y, Chew TG, Ge W, Balasubramanian MK 2007. Polarity determinants Tea1p, Tea4p, and Pom1p inhibit division-septum assembly at cell ends in fission yeast. Dev Cell 12:987–996 [DOI] [PubMed] [Google Scholar]
- Huckaba TM, Gay AC, Pantalena LF, Yang HC, Pon LA 2004. Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J Cell Biol 167:519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DA, Fukui Y, Yamamoto M 1990. Homologous activators of ras in fission and budding yeast. Nature 344:355–357 [DOI] [PubMed] [Google Scholar]
- Janson ME, Loughlin R, Loiodice I, Fu C, Brunner D, Nedelec FJ, Tran PT 2007. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell 128:357–368 [DOI] [PubMed] [Google Scholar]
- Janson ME, Setty TG, Paoletti A, Tran PT 2005. Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J Cell Biol 169:297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG 2003. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115:475–487 [DOI] [PubMed] [Google Scholar]
- Kamasaki T, Arai R, Osumi M, Mabuchi I 2005. Directionality of F-actin cables changes during the fission yeast cell cycle. Nat Cell Biol 7:916–917 [DOI] [PubMed] [Google Scholar]
- Kanai M, Kume K, Miyahara K, Sakai K, Nakamura K, Leonhard K, Wiley DJ, Verde F, Toda T, Hirata D 2005. Fission yeast MO25 protein is localized at SPB and septum and is essential for cell morphogenesis. Embo J 24:3012–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Yang P, Catanuto P, Verde F, Lai H, Du H, Chang F, Marcus S 2003. The Kelch repeat protein, Tea1, is a potential substrate target of the p21-activated kinase, Shk1, in the fission yeast, Schizosaccharomyces pombe. J Biol Chem 278:30074–30082 [DOI] [PubMed] [Google Scholar]
- Marks J, Hagan IM, Hyams JS 1986. Growth polarity and cytokinesis in fission yeast: The role of the cytoskeleton. J Cell Sci Suppl 5:229–241 [DOI] [PubMed] [Google Scholar]
- Martin SG, Berthelot-Grosjean M 2009. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature doi:10.1038/nature08054 [DOI] [PubMed] [Google Scholar]
- Martin SG, McDonald WH, Yates JR 3rd, Chang F 2005. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev Cell 8:479–491 [DOI] [PubMed] [Google Scholar]
- Martin SG, Rincon SA, Basu R, Perez P, Chang F 2007. Regulation of the formin for3p by cdc42p and bud6p. Mol Biol Cell 18:4155–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Nurse P 1997. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89:939–949 [DOI] [PubMed] [Google Scholar]
- Mendoza M, Redemann S, Brunner D 2005. The fission yeast MO25 protein functions in polar growth and cell separation. Eur J Cell Biol 84:915–926 [DOI] [PubMed] [Google Scholar]
- Miller PJ, Johnson DI 1994. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol Cell Biol 14:1075–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc N, Boudaoud A, Chang F 2009. Mechanical forces of fission yeast growth. Curr Biol (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc N, Bratman SV, Basu R, Chang F 2009. Establishing new sites of polarization by microtubules. Curr Biol 19:83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison JM, Nurse P 1985. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J Cell Sci 75:357–376 [DOI] [PubMed] [Google Scholar]
- Moseley JB, Mayeux A, Paoletti A, Nurse P 2009. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature doi:10.1038/nature08074 [DOI] [PubMed] [Google Scholar]
- Motegi F, Arai R, Mabuchi I 2001. Identification of two type V myosins in fission yeast, one of which functions in polarized cell growth and moves rapidly in the cell. Mol Biol Cell 12:1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi F, Mishra M, Balasubramanian MK, Mabuchi I 2004. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. J Cell Biol 165:685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill DP, Edwards SR, Hyams JS 2006. A critical role for the type V myosin, Myo52, in septum deposition and cell fission during cytokinesis in Schizosaccharomyces pombe. Cell Motil Cytoskeleton 63:149–161 [DOI] [PubMed] [Google Scholar]
- Nakano K, Imai J, Arai R, Toh EA, Matsui Y, Mabuchi I 2002. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J Cell Sci 115:4629–4639 [DOI] [PubMed] [Google Scholar]
- Onken B, Wiener H, Philips MR, Chang EC 2006. Compartmentalized signaling of Ras in fission yeast. Proc Natl Acad Sci 103:9045–9050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padte NN, Martin SG, Howard M, Chang F 2006. The cell-end factor pom1p inhibits mid1p in specification of the cell division plane in fission yeast. Curr Biol 16:2480–2487 [DOI] [PubMed] [Google Scholar]
- Paoletti A, Chang F 2000. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol Biol Cell 11:2757–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadaki P, Pizon V, Onken B, Chang EC 2002. Two ras pathways in fission yeast are differentially regulated by two ras guanine nucleotide exchange factors. Mol Cell Biol 22:4598–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe P, Hirata D, Childs D, Vardy L, Toda T 1998. Identification of novel temperature-sensitive lethal alleles in essential beta-tubulin and nonessential alpha 2-tubulin genes as fission yeast polarity mutants. Mol Biol Cell 9:1757–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Snaith HA 2004. Role of microtubules and tea1p in establishment and maintenance of fission yeast cell polarity. J Cell Sci 117:689–700 [DOI] [PubMed] [Google Scholar]
- Sawin KE, Tran PT 2006. Cytoplasmic microtubule organization in fission yeast. Yeast 23:1001–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith HA, Samejima I, Sawin KE 2005. Multistep and multimode cortical anchoring of tea1p at cell tips in fission yeast. Embo J 24:3690–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith HA, Sawin KE 2003. Fission yeast mod5p regulates polarized growth through anchoring of tea1p at cell tips. Nature 423:647–651 [DOI] [PubMed] [Google Scholar]
- Sohrmann M, Fankhauser C, Brodbeck C, Simanis V 1996. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev 10:2707–2719 [DOI] [PubMed] [Google Scholar]
- Tatebe H, Nakano K, Maximo R, Shiozaki K 2008. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr Biol 18:322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Shimada K, Uzawa S, Morigasaki S, Shiozaki K 2005. Wsh3/Tea4 is a novel cell-end factor essential for bipolar distribution of Tea1 and protects cell polarity under environmental stress in S. pombe. Curr Biol 15:1006–1015 [DOI] [PubMed] [Google Scholar]
- Terenna CR, Makushok T, Velve-Casquillas G, Baigl D, Chen Y, Bornens M, Paoletti A, Piel M, Tran PT 2008. Physical mechanisms redirecting cell polarity and cell shape in fission yeast. Curr Biol 18:1748–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolic-Norrelykke IM, Sacconi L, Stringari C, Raabe I, Pavone FS 2005. Nuclear and division-plane positioning revealed by optical micromanipulation. Curr Biol 15:1212–1216 [DOI] [PubMed] [Google Scholar]
- Tran PT, Doye V, Chang F, Inoue S 2000. Microtubule-dependent nuclear positioning and nuclear-dependent septum positioning in the fission yeast Schizosaccharomyces [correction of Saccharomyces] pombe. Biol Bull 199:205–206 [DOI] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoue S, Chang F 2001. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol 153:397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tratner I, Fourticq-Esqueoute A, Tillit J, Baldacci G 1997. Cloning and characterization of the S. pombe gene efc25+, a new putative guanine nucleotide exchange factor. Gene 193:203–210 [DOI] [PubMed] [Google Scholar]
- Umesono K, Toda T, Hayashi S, Yanagida M 1983. Cell division cycle genes nda2 and nda3 of the fission yeast Schizosaccharomyces pombe control microtubular organization and sensitivity to anti-mitotic benzimidazole compounds. J Mol Biol 168:271–284 [DOI] [PubMed] [Google Scholar]
- Verde F, Mata J, Nurse P 1995. Fission yeast cell morphogenesis: Dentification of new genes and analysis of their role during the cell cycle. J Cell Biol 131:1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Wiley DJ, Nurse P 1998. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc Natl Acad Sci 95:7526–7531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtler V, Rajagopalan S, Balasubramanian MK 2003. Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J Cell Sci 116:867–874 [DOI] [PubMed] [Google Scholar]
- Wang H, Tang X, Liu J, Trautmann S, Balasundaram D, McCollum D, Balasubramanian MK 2002. The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe. Mol Biol Cell 13:515–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Kuhn JR, Kovar DR, Pollard TD 2003. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell 5:723–734 [DOI] [PubMed] [Google Scholar]
- Yang P, Qyang Y, Bartholomeusz G, Zhou X, Marcus S 2003. The novel Rho GTPase-activating protein family protein, Rga8, provides a potential link between Cdc42/p21-activated kinase and Rho signaling pathways in the fission yeast, Schizosaccharomyces pombe. J Biol Chem 278:48821–48830 [DOI] [PubMed] [Google Scholar]
- Zimmerman S, Tran PT, Daga RR, Niwa O, Chang F 2004. Rsp1p, a J domain protein required for disassembly and assembly of microtubule organizing centers during the fission yeast cell cycle. Dev Cell 6:497–509 [DOI] [PubMed] [Google Scholar]