Abstract
How are the asymmetric distributions of proteins, lipids, and RNAs established and maintained in various cell types? Studies from diverse organisms show that Par proteins, GTPases, kinases, and phosphoinositides participate in conserved signaling pathways to establish and maintain cell polarity.
An evolutionarily conserved signaling pathway that includes Par proteins, small GTPases, kinases, and phosphoinositides establishes and maintains cell polarity in metazoans.
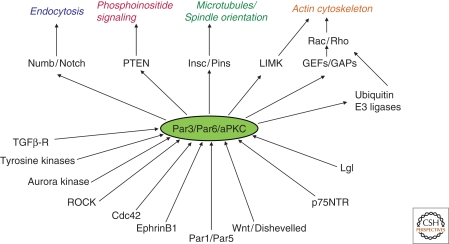
The asymmetric distribution of proteins, lipids, and RNAs is necessary for cell fate determination, differentiation, and specialized cell functions that underlie morphogenesis (St Johnston 2005; Gonczy 2008; Knoblich 2008; Macara and Mili 2008; Martin-Belmonte and Mostov 2008). A fundamental question is how this asymmetric distribution is established and maintained in different types of cells and tissues. The formation of a specialized apical surface on an epithelial cell seems quite different from the specification of axons versus dendrites in a neuron, or the asymmetric division of a nematode zygote. Yet, remarkably, a conserved molecular toolbox is used throughout the metazoa to establish and maintain cell polarity in these and many other contexts. This toolbox consists of proteins that are components of signal transduction pathways (Goldstein and Macara 2007; Assemat et al. 2008; Yamanaka and Ohno 2008). However, our understanding of these pathways, and their intersection with other signaling networks, remains incomplete. Moreover, the regulation and cross talk between the polarity proteins and other signaling components varies from one context to another, which complicates the task of dissecting polarity protein function. Nonetheless, rapid progress is being made in our understanding of polarity signaling, which is outlined in this article, with an emphasis on the Par proteins, because these proteins play major roles integrating diverse signals that regulate cell polarity (Fig. 1) (see Munro and Bowerman 2009; Prehoda 2009; Nelson 2009).
Figure 1.
An overview of Par complex signaling, showing inputs (bottom) and outputs (top) with cellular functions that are targeted by these pathways (italics).
PAR PROTEINS: INTERPRETERS OF CELL POLARITY
The establishment of cell polarity can be dissected into four primary components: (1) breaking symmetry; (2) establishing cortical landmarks; (3) polarizing the cytoskeleton; and (4) amplifying and maintaining the polarized state. In many different systems, the Par proteins, small GTPases, and the phosphoinositides and their regulators are among the conserved factors that play fundamental roles in each of these stages. The Par (partition defective) genes were initially identified in an elegant screen by Jim Priess and Ken Kemphues for maternal-effect genes that are embryonic lethal in Caenorhabditis elegans (Kemphues et al. 1988). Eventually, seven genes required for asymmetric cell division of the zygote were identified. The products of the Par genes are quite diverse in function. Par-1 and Par-4 encode serine/threonine-directed protein kinases (Guo and Kemphues 1995; Watts et al. 2000); Par-2 is a RING finger domain protein that may function as an E3 ubiquitin ligase (Levitan et al. 1994); Par-3 and Par-6 are PDZ (PSD95/Dlg/ZO1) domain-containing proteins with scaffolding or adaptor functions (Etemad-Moghadam et al. 1995; Hung and Kemphues 1999); Par-5 is a 14-3-3 protein that binds to phosphorylated serine and threonine residues (Morton et al. 2002); and PKC-3 is an atypical protein kinase C (aPKC). With the exception of Par-2, all of the Par proteins and aPKC are conserved throughout the Metazoa. Strikingly, most of these polarity proteins show a polarized distribution within the zygote, and are dependent on one another for their localization (Tabuse et al. 1998). Par-1 and Par-2 are restricted to the posterior of the zygote cortex, whereas Par-3, Par-6, and aPKC are restricted to the anterior cortex (although they are also present in the cytoplasm) (Schneider and Bowerman 2003; Munro 2006).
Par-3, Par-6, and aPKC can form a physical complex, sometimes called the Par complex (Joberty et al. 2000; Lin et al. 2000; Wodarz et al. 2000), which has been identified in all animal cells that have been examined (Goldstein and Macara 2007). Par-6 and aPKC associate through their amino-terminal PB1 domains (Hirano et al. 2005), and Par-6 inhibits the basal activity of aPKC, but also can act as a targeting subunit for the kinase, recruiting substrates for phosphorylation (Yamanaka et al. 2001). One of these substrates is Par-3. Par-6 binds to Par-3, through a PDZ–PDZ interaction, but aPKC can also bind through its kinase domain directly to the carboxy-terminal half Par-3, and phosphorylate a Ser residue (Nagai-Tamai et al. 2002). Importantly, Par-3, Par-6, and aPKC do not form a constitutive complex. Their interactions are regulated by multiple protein kinases, by small GTPases, and by competition for other binding partners (Fig. 1). These regulators can alter the subcellular distribution of the polarity proteins, and their function.
FACTORS THAT CONTROL PAR-3 LOCALIZATION
It is likely that the Par proteins provide critical spatial information during cell polarization. A key question, therefore, concerns how the Par proteins themselves are localized. It is probably fair to say that we do not yet know the complete answer to this question in any cell type, or for any polarity protein. Moreover, the mechanisms may be partly conserved and partly dependent on context. For example, in the C. elegans zygote, Par-3 association with the anterior cortex requires the expression of Par-6, aPKC, and Par-2 (Etemad-Moghadam et al. 1995; Hung and Kemphues 1999); but in Drosophila neuroblasts and embryonic epithelial cells, Par-3 apical localization is independent of aPKC (Harris and Peifer 2005). And in mammalian epithelial cells, Par-3 is not apical but is associated with tight junctions (Izumi et al. 1998; Joberty et al. 2000).
Protein localization often involves distinct steps that can include transport, delivery, anchoring at the destination, and active exclusion from other areas of the cell. Transport can simply involve passive diffusion, or directed movement along the cytoskeleton. Alternatively, the mRNA for the protein might be transported to the destination site, where it is locally translated. Anchors can include phospholipids, cytoskeletal elements, or more specific protein complexes.
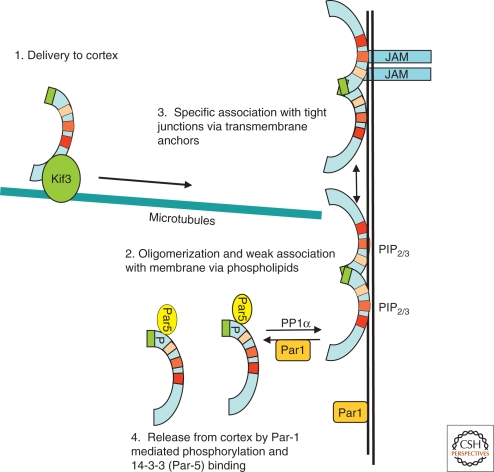
Par-3 transport: In the case of Par-3, transport and anchoring both appear to involve the cytoskeleton in at least certain cell types (Fig. 2). For instance, in the Drosophila embryo, Par-3 is transported in a basal-to-apical direction by dynein, presumably along microtubules, and is anchored in an actin-dependent manner to apical adherens junctions (Harris and Peifer 2005). Par-3 in turn recruits the Par-6/aPKC complex to the cortex at the apical surface, where another polarity protein, Crumbs, separates Par-3—which remains at the adherens junctions—from Par-6 and aPKC, which assume a more apical localization (Harris and Peifer 2005). In mammalian neurons, microtubules are also important for the movement of Par-3 to the growing tips of axons. Par-3 interacts both with the kinesin Kif3a and the microtubule-associated protein APC (adenomatous polyposis coli) (Nishimura et al. 2004; Shi et al. 2004). Neuronal microtubules are oriented with their plus ends toward the tips of nascent neurites. In undifferentiated neurites, APC is phosphorylated by GSK3-β, which decreases the affinity of APC for microtubules. However, PI3-kinase is localized at the tips of axons, where it can phosphorylate and inactivate GSK3β, allowing APC to positively regulate microtubule growth and bundling and provide a route for Kif3a-mediated transport of Par-3 along the microtubule tracks to the tips of axons (Shi et al. 2004). In the C. elegans zygote, in contrast, centrosomes and microtubules may play early roles in triggering Par asymmetry (Tsai and Ahringer 2007), but a flow of cortical actin is more directly involved in segregating the initial, uniformly cortical anterior Par protein complex to one pole (Munro et al. 2004; Munro and Bowerman 2009).
Figure 2.
Mechanisms for the transport, cortical association, and anchoring of Par3 in mammalian cells. It is not yet known if all of these mechanisms operate in any one cell type, and additional processes, such as RNA localization, might conceivably play roles in certain circumstances. Junctional adhesion molecule (JAM) is shown as an example of a transmembrane protein to which Par-3 can be anchored, but others undoubtedly exist, such as the neurotrophin receptor, p75NTR, in mammalian Schwann cells (Chan et al. 2006).
Membrane attachment via phospholipids: How is Par-3 maintained at the plasma membrane once delivered? Although this question has not yet been definitively answered, multiple distinct mechanisms are probably involved, some that are general and some that are more specific. So far, they all appear to involve the PDZ domains of Par-3. First, Par-3 has been reported to bind directly to phosphoinositides through its PDZ-2 domain, which is essential for membrane localization (Wu et al. 2007). This phospholipid association is unusual because most PDZ domains recognize specific carboxy-terminal peptides, or loops, within polypeptides. Also, the interaction is not specific for PIP2 or PIP3, so it is unable to make use of spatial information in the asymmetric distribution of these phospholipids. The association might, however, provide a general, low-affinity cortical targeting function, which can be refined by further interactions with specific proteins (Fig. 2). It is interesting that Par-3 also interacts with the phosphoinositide phosphatase PTEN, which generates PIP2 from PIP3, but whether this somehow regulates the lipid/Par-3 association is not known (Wu et al. 2007; Feng et al. 2008).
A role for oligomerization: Oligomerization of Par3 is also important for cortical targeting. The amino-terminal conserved region 1 (CR1) is necessary for self-association of Par-3 into higher-order complexes (Benton and Johnston 2003a; Mizuno et al. 2003; Feng et al. 2007). The CR1 domain is not sufficient for membrane attachment, because the isolated fragment is diffuse throughout the cytoplasm. However, it is essential for membrane localization and function of Par-3 in both mammalian epithelial cells and Drosophila. It is also not clear how oligomerization of Par-3 maintains it at the plasma membrane, but oligomerization might act to complement a weak phosphoinositide binding (Fig. 2).
Anchoring to membrane proteins: Enrichment in a specific region of the cell cortex is mediated by direct interactions with proteins via the PDZ domains of Par-3. We refer to this as anchoring, although of course the individual protein molecules can be associating and disassociating from their attachment site in a highly dynamic fashion. In mammalian epithelia, cell–cell junctions can act as a primary landmark that precedes the establishment of polarity. Initial contacts between adjacent cells form through the Ca2+-dependent dimerization of E-cadherin, as well as via nectin, to form primordial junctions. Through ZO-1, nectin recruits another Ca2+-independent adhesion molecule, junction adhesion molecule (JAM), to junctions (Fukuhara et al. 2002). Par-3 is recruited to junctions through a direct interaction between the first PDZ domain of Par-3 with the carboxy-terminal PDZ-binding motif of JAM (Fig. 2) (Ebnet et al. 2000; Itoh et al. 2001). As the primordial junctions mature, they separate into distinct junctions, lateral adherens junctions, and more apical tight junctions. Because Par-6 also binds to the first PDZ domain of Par-3, its association is presumably mutually exclusive with that of JAM, but no competition has been reported. A similar mechanism occurs in the mouse neuroepithelium, where Par-3 is recruited to early junctions; however, in this case, Par-3 is apparently recruited through a direct interaction with nectin-1 or nectin-3, and Par-3 does not bind to JAM (Takekuni et al. 2003). In the Drosophila wing disc epithelium, Par-3 is also recruited to junctions through its PDZ domain, by interacting with the adherens junction proteins β-catenin and Echinoid, an immunoglobulin-domain transmembrane protein (Wei et al. 2005).
Localized mRNA translation: To date, there are no reports of localized Par-3 mRNA, but the transcripts for two other polarity genes, Crumbs and Stardust, are enriched in the apical region of Drosophila epithelial cells (Horne-Badovinac and Bilder 2008; Li et al. 2008). Both proteins are exclusively apical, are required for epithelial apical/basal polarity, and form a complex with one another. It is of particular interest that the Crumbs transcript is localized because Crumbs is a transmembrane protein that would normally have to transit the Golgi to be delivered in vesicles to the apical surface. An interesting speculation is that Crumbs is delivered by a different mechanism, to initiate apicalization, and that it subsequently directs the delivery of other apical proteins to maintain apical identity.
Stardust is a scaffold protein that can associate with Par-6 and a poly-PDZ domain protein called Patj. The mammalian homolog is called Pals1 (Roh et al. 2002). An alternatively spliced coding exon of Stardust contains the mRNA localization signal, which enables the transcript to be transported in a dynein-dependent manner to the apical region (Horne-Badovinac and Bilder 2008). Dynein is also important for Crumbs mRNA localization, and is essential for apico–basal polarization in the follicle epithelium. Strikingly, the Stardust splice variant is only produced early in development, whereas in mature epithelia, the mRNA is not localized. The physiological significance of this change is not yet understood, and it is not known if similar mechanisms pertain to the mammalian homologs for these polarity proteins.
Active exclusion: None of the mechanisms described above stably anchors Par-3 at the plasma membrane. FRAP experiments show that the protein is highly dynamic in Drosophila embryonic cells (Mayer et al. 2005), and it is quickly released from tight junctions on digitonin permeabilization of MDCK cells (unpublished observation). Therefore, the targeting and retention mechanisms for Par-3 described above are insufficient to maintain its polarized distribution within the cell. Active mechanisms have been discovered, however, that prevent Par-3 from accumulating in inappropriate areas of the cortex. The Par complex, consisting of Par-3, Par-6, and aPKC is often localized in a complementary pattern to that of Par-1, and through mutual phosphorylations, the two kinases, aPKC and Par-1, exclude each other from the complementary region of the cortex. Par-1 directly phosphorylates Par-3 (Benton and Johnston 2003b), and these phosphorylated residues act as docking sites for 14-3-3 (also known as Par-5), which binds and destabilizes the membrane association of Par-3 (Fig. 2). This mechanism can therefore exclude Par-3 (and associated aPKC and Par-6) from cortical regions that contain Par-1. Mammalian Par-3 is phosphorylated by Par-1 on serine 144 (S144, which is the same as S151 in Drosophila) (Hurd et al. 2003a). S144/S151 are located near CR1, which is necessary for oligomerization of Par-3. Binding of 14-3-3 to S151 inhibits self-oligomerization (Benton and Johnston 2003b), which may reduce phosphoinositide association, causing the protein to dissociate from the plasma membrane (Fig. 2).
Conversely, aPKC can phosphorylate Par-1, which both inhibits Par-1 kinase activity and disassociates it from the plasma membrane. Two distinct mechanisms have been identified, one in which aPKC directly phosphorylates Par-1 on Thr 595 (T595) (Hurov et al. 2004), and an indirect mechanism by which aPKC activates protein kinase D (PKD), which phosphorylates Par-1 on Ser 400 (S400) (Watkins et al. 2008). Phosphorylation of T595 reduces the kinase activity and displaces Par-1 from the membranes. Furthermore, phosphorylation of S400 recruits 14-3-3, which displaces Par-1 from the membrane. The phosphorylation and binding of 14-3-3 to Par-3 can be reversed by protein phosphatase 1α (PP1α) (Traweger et al. 2008), allowing recycling of Par-3 to appropriate cortical sites. This spatial antagonism probably operates in the C. elegans zygote to maintain the distinct anterior–posterior distributions of Par-3/aPKC/Par-6 and Par-1, as well as in epithelial cells of Drosophila and mammals, and may be a conserved and widely applied method for excluding proteins from specific areas of the cell cortex. For example, the cell fate determinants Numb and Miranda, and the polarity protein Lgl, are also removed from the plasma membrane by aPKC-dependent phosphorylations (Betschinger et al. 2003; Smith et al. 2007; Atwood and Prehoda 2009; Prehoda 2009). An Arf-GAP that is involved in convergent extension during Xenopus gastrulation is phosphorylated by aPKC/Par-6, which may alter its subcellular distribution (Hyodo-Miura et al. 2006). We predict that many other examples of the same mechanism occur, involving not only aPKC and Par-1, but other protein kinases that target 14-3-3 consensus sites.
POLARITY SIGNALING THROUGH PAR-3/PAR-6/APKC
Signaling through small GTPases: There are two intrinsically asymmetric components of cells: microtubules and actin filaments (see Mullins 2009). Both are vectorial polymers, and their organization is, therefore, of fundamental importance to cell polarization. This organization is dynamic and is highly regulated by signaling networks that respond to both external and internal cues. Central to these signaling networks are the Rho family GTPases, which cycle between GTP-bound (on) and GDP-bound (off) states and function as timed molecular switches. It is not too surprising, therefore, that polarity proteins both regulate and are regulated by these GTPases. Given the enormity of the cytoskeleton and Rho research fields, we will focus on those aspects that are related to Par proteins.
The Cdc42 GTPase was first identified in Saccharomyces cerevisiae, where it controls bud site establishment (Johnson 1999). Landmark proteins that localize to the future bud site recruit both Cdc42 and a guanine nucleotide exchange factor (GEF) that catalyzes the formation of the GTP-bound state of Cdc42 (Chang and Peter 2003; see Slaughter et al. 2009). In a classic feedback loop, Cdc42-GTP can in turn recruit an adapter protein that stabilizes the GEF and further enriches Cdc42 at the bud site (Butty et al. 2002). An unlocalized GTPase activating protein (GAP) switches off any Cdc42-GTP that diffuses away from the bud site, helping to maintain its polarized localization. Additionally, the Cdc42-GTP stimulates actin polymerization at the bud site, which can also recruit more Cdc42 (Wedlich-Soldner et al. 2003). The anchored actin filaments then transport vesicles that deliver membrane proteins for growth of the new bud.
Cdc42 as an upstream regulator of Par-6/aPKC: Cdc42 is a pivotal component of the polarity machinery in yeast, and its conservation throughout the evolution of the metazoa led to the expectation that it would play similar roles in animal development. Indeed, the injection of dominant–negative Cdc42 mutants disrupted polarized migration in mammalian fibroblasts, and the discovery that Cdc42-GTP binds to Par-6 provided a likely mechanism by which the GTPase might control cell polarization. Par-6 contains a partial CRIB domain, which is conserved among most of the downstream targets of Cdc42, and interacts with a region of the GTPase that undergoes a GTP-dependent switch in conformation (Garrard et al. 2003). It is still unclear, however, exactly how Par-6 behaves as an effector for Cdc42. One function is to relieve the inhibition on aPKC activity exerted by Par-6. Cdc42 can in this way activate aPKC. Recently, EphrinB1 was found to compete with Cdc42 for binding to Par-6, blocking tight junction formation (Lee et al. 2008). Tyrosine phosphorylation of the Ephrin releases it from Par-6. In this way, signaling through tyrosine kinase receptors could impact aPKC activity and consequent cell polarity decisions.
In addition to counteracting the inhibition of aPKC by Par-6, Cdc42 can also recruit the Par-6/aPKC complex to specific regions of the cell cortex where the GTPase is activated (see Prehoda 2009). For example, in Drosophila neuroblasts, Cdc42 mutants mislocalize Par-6/aPKC to the cytoplasm (Atwood et al. 2007), and in mammalian epithelial cells, knockdown of Cdc42 can partially mislocalize aPKC from the apical cortex (Martin-Belmonte et al. 2007). However, robust Cdc42 localization in neuroblasts also requires Par-6, suggesting the existence of a positive feedback loop that reinforces their positioning. Moreover, there are other mechanisms for aPKC and Par-6 localization: A dynamin-associated protein, Dap160, can bind to aPKC and stimulate its kinase activity, and in neuroblasts dap160 mutants, the aPKC is delocalized from the apical cortex (Chabu and Doe 2008). In the C. elegans zygote, Cdc42 is not essential for the initial anterior enrichment of Par-6, though the asymmetry is lost later during the first cell division, suggesting that other factors set up the polarity (Aceto et al. 2006); whereas in mammalian epithelial cells, Par-6 localization to the apical surface probably involves its association with Pals1 and/or Crumbs, rather than Cdc42 (Gao et al. 2002a; Hurd et al. 2003b).
Another member of the GTPase family, Rho-1, has been implicated in this process. Following fertilization of the C. elegans oocyte, a GAP called CYK-4 has been reported to associate with the male pronucleus at the presumptive posterior cortex and locally inactivates Rho-1 (Jenkins et al. 2006). However, CYK-4 is also a component of a complex involved in cytokinesis, and in this situation, at least, it is specific for Rac inactivation rather than Rho-1 (Canman et al. 2008). Two other RhoGAPs are present in C. elegans but seem to have a distinct function from CYK-4 because they control the ratio of the anterior/posterior domain rather than its formation (Schmutz et al. 2007; Schonegg et al. 2007). Rho-1 GTP normally activates a downstream kinase that phosphorylates myosin light chain, increasing contractility, but the accumulation of the GAP depletes Rho-GTP and relaxes actomyosin at the posterior cortex. In addition, a GEF for the Rho-1 GTPase is excluded from the posterior cortex, restricting Rho-GTP production to the anterior end of the zygote (Motegi and Sugimoto 2006). The Rho-GTP stimulates anterior myosin contractility, which generates a cortical actin flow, translocating Par-6, aPKC, Par-3, and Cdc42 to the anterior end of the zygote. Cdc42 then maintains this distribution of the Par proteins.
The Rho GTPase may play a distinct role in mammalian cells by controlling the association of Par-3 with Par-6/aPKC. Nakayama et al. found that a protein kinase downstream of RhoA (ROCK) can phosphorylate Par-3 on a threonine residue (T833) adjacent to the aPKC-binding site in the carboxyl terminus of Par-3, and that this phosphorylation blocks the association with aPKC and Par-6 (Nakayama et al. 2008). However, this site is not conserved in C. elegans.
A key remaining question concerns how Cdc42-GTP is localized. Presumably, a GEF activates the GTPase in a temporally or spatially restricted manner to ensure that Par-6 recruitment is localized correctly. Spatially restricted GAPs could also function to exclude Cdc42-GTP from certain regions of the cell, by converting it to the GDP-bound state. Indeed, during radial polarization in the early C. elegans embryo, a GAP called PAC-1 localizes to the basolateral membranes, whereas Par-6 is restricted to the contact-free cell surfaces, most likely because Cdc42-GTP is destroyed by the PAC-1 at other surfaces (Anderson et al. 2008; see Munro and Bowerman 2009). In MDCK cells, annexin2, a Ca-dependent regulator of actin dynamics, has been proposed to recruit Cdc42-GTP to the apical surface (Martin-Belmonte et al. 2007), but it remains unclear how the annexin-bound Cdc42 could also bind Par-6. Nonetheless, this model could potentially dispense with a localized GEF or GAP. So far, no Cdc42-specific GEF has been implicated in Par-6 recruitment for any organism, but RNAi screens are likely to resolve this issue in the near future.
Rho GTPases as downstream effectors of Par-3/Par-6/aPKC: Because actin dynamics plays a key role in cell polarization and is regulated by Rho GTPases, one might expect these GTPases to be modulated in some way by Par proteins. At least in mammalian cells, this idea has been validated by several laboratories, which have identified a number of different mechanisms (Fig. 1). First, the carboxy-terminal region of Par-3 has been shown to bind to a Rac GEF, called Tiam1 (Chen and Macara 2005; Mertens et al. 2005; Nishimura et al. 2005; Zhang and Macara 2006). Under some circumstances, including the formation of tight junctions by MDCK epithelial cells, and the formation of excitatory synapses by hippocampal neurons, Par-3 sequesters Tiam1 to prevent its inappropriate activation of Rac (Chen and Macara 2005; Zhang and Macara 2006). The silencing of Par-3 in these contexts causes a constitutive increase in Rac-GTP levels, which in turn disrupts junction and synapse formation. Both of these processes are actin-mediated, and presumably the inappropriate activation of Rac causes misorganization of actin filaments at the cell cortex. In other cell types, however, Par-3 might recruit Tiam1 to sites within the cell where Rac needs to be activated. In such cases, loss of Par-3 might reduce Rac activation at these sites. For example, silencing of Par-3 or Tiam1 expression in keratinocytes reduces their ability to sustain polarized migration (Pegtel et al. 2007). Cdc42 might regulate Rac activation through association with the Par-6/aPKC/Par-3 complex, perhaps by activating aPKC to phosphorylate Par-3 or Tiam1. Whatever the case, Par-3 can be defined as a scaffold protein that recruits signaling components to specific locations in the cell, enabling spatial control of the signal output.
Through quite distinct mechanisms, Par-6 can regulate RhoA activity. First, via its association with aPKC, Par-6 can activate a RhoGAP called p190, thereby reducing Rho-GTP levels (Zhang and Macara 2008). This pathway is important in controlling synapse density in hippocampal neurons. Elevated expression of Par-6 increases dendritic spine density, whereas silencing of Par-6 reduces spine density. However, the molecular linkage between Par-6/aPKC and the p190 RhoGAP is still unknown.
A second, different mechanism by which Par-6 can affect Rho is through association with Smad-ubiquitin regulatory factor 1 (Smurf1), an E3 ligase that down-regulates the TGFβ signaling pathway (Fig. 1). In addition to targeting Smads for degradation, Smurf1 can also ubiquinate RhoA (Wang et al. 2003). TGFβ type II receptor can—at least in some cell types—bind to and phosphorylate Par-6 on a conserved carboxy-terminal serine residue (S345) (Ozdamar et al. 2005). This phosphorylation appears to stimulate the association of Par-6 (or aPKC) with Smurf1, which mediates the localized ubiquitination and destruction of RhoA. In NMuMG cells, loss of RhoA can cause the dissolution of tight junctions and an epithelial–mesenchymal transition. However, these effects might be cell-type specific because, for example in MDCK cells, dominant–negative RhoA expression had no effect on tight junctions (Bruewer et al. 2004).
Cross talk between Wnt signaling and Par proteins: Planar cell polarity signaling is stimulated by Wnt ligands that activate seven transmembrane receptors in the Frizzled family (Simons and Mlodzik 2008; see Vladar et al. 2009). Downstream of these receptors, an adapter protein called Dishevelled (Dvl) can activate RhoA and the downstream kinase ROCK, which, as described above, phosphorylates Par-3. In addition, however, Dvl is a target for phosphorylation by Par-1, and can directly associate with aPKC (Sun et al. 2001; Ossipova et al. 2005). The association with aPKC stabilizes and activates the kinase, and has been implicated in axonal differentiation and polarized migration of mammalian cells (Schlessinger et al. 2007; Zhang et al. 2007). Moreover, the Drosophila Frizzled receptor is phosphorylated and inhibited by aPKC, which is recruited to Frizzled through another polarity protein, Patj (Djiane et al. 2005). During planar cell polarity signaling, aPKC is down-regulated, whereas Par-3 expression is elevated. Clearly, there are multiple mechanisms for cross talk between planar cell polarity and Par polarity signaling pathways, and a deeper understanding of these interactions will be an important focus for future work.
Signaling through protein kinases: Most signaling pathways use protein kinases either to amplify, attenuate, or direct information. It is not surprising, therefore, to find that phosphorylation plays an important role in regulating the association of Par-3 and Par-6 with their effectors and with the cell cortex (Figs. 1–3).
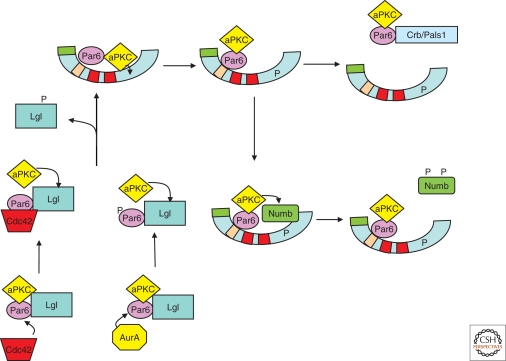
Figure 3.
A model for the interactions of the Par complex with other polarity proteins and protein kinases. This model synthesizes data from a number of laboratories, and attempts to reconcile observations that aPKC can bind directly to Par-3, but also can bind indirectly through Par-6. The direct association is inhibited by phosphorylation; whereas the indirect association is inhibited by competition for Par-6 by the Crumbs/Pals1 complex and by the Lgl polarity protein. In addition, the schematic shows Par-3 functioning to recruit other targets for aPKC, such as Numb, and the regulation of Par-6 by the Cdc42 GTPase and the Aurora A kinase.
Several serine/threonine kinases target Par-3, and many of these phosphorylations are conserved throughout the animal kingdom. As discussed above, Par-1 phosphorylates a conserved Serine in the carboxyl terminus of Par-3, preventing oligomerization and inducing disassociation from the membrane (Fig. 2) (Benton and Johnston 2003b); and aPKC phosphorylates a conserved Serine in the carboxyl terminus of Par-3, releasing aPKC from its adjacent binding site (Nagai-Tamai et al. 2002). Until recently, the function of this latter phosphorylation was obscure, but recent studies in mammalian cells, using knockdown and replacement of Par-3 with nonphosphorylatable mutants, has shown that the Par-3/aPKC interaction is essential for Par-3 function (Horikoshi et al. 2009; McCaffrey and Macara 2009). In mammary morphogenesis, a Par-3 mutant that is unable to bind aPKC is mislocalized and cannot rescue normal ductal growth in the absence of the endogenous Par-3 (McCaffrey and Macara 2009). In addition, the Rhoactivated kinase, ROCK, can phosphorylate a nearby Threonine, also disrupting aPKC binding (Nakayama et al. 2008). However, an interaction between Par-6/aPKC is still possible with Par-3 through its PDZ domains (Fig. 3). We speculate that after Par-3 has recruited Par-6/aPKC—to the tight junction in mammalian epithelial cells—the phosphorylation and release somehow enables Par-6/aPKC to be handed off to an apical polarity complex consisting of Crumbs, Pals1, and Patj (Fig. 3). This complex is essential for apical membrane establishment, and for tight junction formation (Roh et al. 2003; Straight et al. 2004; Shin et al. 2005). Par-6 can interact directly with all three members of the complex in a mutually exclusive manner. The interaction between Par-6 with Pals1 inhibits the interaction of Pals1 with Patj; however, the biological significance of this effect has not yet been established.
Importantly, the mitotic regulatory kinase Aurora-A can phosphorylate Par-6 (Fig. 3) (Wirtz-Peitz et al. 2008). This is a key step in ensuring asymmetric stem cell divisions in the Drosophila embryo. At the onset of mitosis, Aurora-A phosphorylates Par-6 within the amino-terminal PB1 domain that binds to aPKC, releasing Par-6 and relieving its inhibition of aPKC (see Prehoda 2009). The activated aPKC phosphorylates Lgl, which causes it to dissociate from the Par-6/aPKC complex and allows the interaction of the complex with Par-3. Par-3 then acts to recruit Numb as a substrate for aPKC. Phosphorylation of Numb by aPKC causes it to be released from the cell cortex (Smith et al. 2007; Wirtz-Peitz et al. 2008). Because the Par complex is initially asymmetric, the loss of Numb occurs at only one side of the mitotic cell, so one daughter will inherit Numb whereas the other does not. Numb negatively regulates the cell fate-determining transcription factor Notch, ensuring that only one daughter cell responds to Notch signaling. Interestingly, the Lgl polarity protein seems here to function as a buffer, to suppress the phosphorylation of Numb until the appropriate time in the cell cycle. The recruitment of Numb by Par-3 seems to be a conserved function, because in migrating mammalian fibroblasts the same mechanism is used to release Numb from the cell cortex, thereby regulating the internalization of integrins (Nishimura and Kaibuchi 2007). It will be interesting to know whether buffering of signaling components is the principal function of Lgl and if it is important for controlling Numb phosphorylation during cell migration.
The Par complex can also regulate downstream protein kinases. As discussed previously, Par-6 inhibits aPKC, whereas aPKC phosphorylates and inhibits the Par-1 kinase, and triggers its disassociation from the cell cortex, thereby excluding it from the apical domain (Hurov et al. 2004; Kusakabe and Nishida 2004). A region in the carboxyl terminus of Par-3 binds to the LIMK2 protein kinase, and attenuates LIMK2 kinase activity (Chen and Macara 2006). The principal function of LIMK2 is to phosphorylate and inhibit cofilin, an actin-severing protein that is essential for actin remodeling during junction formation (Ivanov et al. 2004, MBC), so Par-3 can locally alter actin dynamics through this mechanism.
Tyrosine kinases in particular have diversified in higher organisms and undertake many roles throughout development. Par-3 is a target of Src family tyrosine kinases, in response to epidermal growth factor (EGF) receptor activation (Wang et al. 2006). Phosphorylation of Par-3 on a tyrosine residue in the carboxy-terminal region (Y1127) delays tight junction assembly in mammalian epithelial cells. This effect has been ascribed to a reduction in the association of Par-3 with LIMK2 (Wang et al. 2006). It is not yet clear how universal this mechanism is, however, because constitutive Src activation, or EGF treatment, causes an epithelial–mesenchymal transition and the loss of tight junctions, which is the opposite of what would be predicted from these data. Nonetheless, high-throughput phosphoproteomics screens have identified multiple tyrosine phosphorylation sites in the carboxyl terminus of Par-3, and Par-6 can associate (indirectly) with ErbB2 (Aranda et al. 2006), suggesting that there may be other inputs into the polarity complex that have yet to be understood.
In this regard, another study has also identified a link between the Par proteins and Src kinase, although with the opposite effect of destabilizing cell adhesions. In a stable blood-testis-barrier, Par-6 is in a complex with Pals1 and JAM-C at cell–cell adhesions (Wong et al. 2008). During adhesion remodeling, however, Pals1, presumably with Par-6, associates with Src and is sequestered away from JAM-C, which weakens the cell–cell junctions. In this system, Par-3 and Par-6 appear to cooperate in regulating some aspects of cell–cell adhesion. Depletion of Par-3, but not Par-6, disrupts tight junction formation, whereas depletion of Par-6 reduces N-cadherin levels. Depletion of either Par protein causes disruption of JAM-A at adhesions. It would be interesting to determine whether Par-3 is part of the Par-6/Pals1/Src complex and whether Par-3 is tyrosine phosphorylated in this system, which might weaken cell–cell adhesions during blood-testis-barrier remodeling.
Phosphoinositides as polarity signals: The first clear demonstrations that phosphoinositides function in cell polarity arose from studies on the chemotaxis of Dictyostelium and mammalian neutrophils (see Orlando and Guo 2009). These cells can translate extraordinarily shallow chemoattractant gradients into steep front–rear polarity, and phosphoinositides play a crucial role in establishing this polarity, with high levels of the lipid kinase PI-3K and its product, PIP3, accumulating at the leading edge where the highest chemoattractant concentration is sensed (Devreotes and Janetopoulos 2003; Charest and Firtel 2006). The lipid phosphatase PTEN is excluded from the leading edge, but is associated with the lateral and rear plasma membranes, leading to restricted PIP3 at the leading edge of migrating cells (Charest and Firtel 2006). Somewhat surprisingly, however, there is no evidence that the Par polarity proteins play any role in chemotaxis by these actively migrating cells. Yet, in other contexts, Par proteins do seem to interact with the phosphoinositide signaling pathway. For example, the PDZ domains of Par-3 bind directly to PTEN (Feng et al. 2008). In Drosophila photoreceptors, Par-3 recruits PTEN to the nascent zonula adherens and promotes conversion of PIP3 to PIP2 in that membrane domain (Pinal et al. 2006). It has been reported that mammalian Par-3 recruits PTEN to cell–cell contacts in MDCK cells (Feng et al. 2008), but Par-3 is almost exclusively localized to tight junctions in these cells, and PTEN is mostly apical. In C. elegans, the Par proteins mediate enrichment of a phosphoinositide kinase (PI-4P kinase) at the posterior of the one-cell embryo, which is required for normal spindle movements (Panbianco et al. 2008). Interestingly, this asymmetric localization also depends on casein kinase, which is enriched at the opposite, anterior end of the zygote. Presumably, the PI-4PK generates PIP2 at the posterior, which somehow interacts with the machinery involved in aster microtubule attachment to the cortex. However, the underlying mechanisms by which Par proteins regulate the PI-4K and casein kinase distributions remain to be discovered.
We should also consider the possibility that Par proteins operate downstream of phosphoinositide signaling. For example, Par-3 PDZ domain 2 can bind directly to phosphoinositides (Wu et al. 2007), and, in the Drosophila oocyte, loss of phosphatidylinositol-5 kinase activity, which produces PIP2 at the plasma membrane, causes mislocalization of Par-3, Par-1, and Lgl (Gervais et al. 2008). However, this effect might be quite indirect, because the microtubule and actin cytoskeletons are also disorganized (Gervais et al. 2008).
There is substantial evidence that the phosphoinositides play a central role in defining polarized membrane domains. Spatially restricted activation of PI3-kinase can locally generate its product, PIP3, which may recruit and regulate downstream effectors that can bind to this lipid through a plekstrin homology (PH) domain. PTEN can counteract the effect of PI3-K by dephosphorylating PIP3 to PIP2. In neurons, local activation of PI3-kinase at the growing tips of neurites induces the growth of axons (Shi et al. 2003; Menager et al. 2004), whereas overexpression of PTEN disrupts the polarization of the axon. Elegant experiments in MDCK epithelial cells have recently highlighted the key role played by phosphoinositides in specifying membrane identity (Gassama-Diagne et al. 2006; Martin-Belmonte et al. 2007; Martin-Belmonte and Mostov 2008). These cells display a highly asymmetric distribution of these lipids, in which PIP3 is completely excluded from the apical surface but is abundant on the basolateral surface, whereas PIP2 is present throughout the plasma membrane but is enriched at the apical surface. This spatial asymmetry results from the localization of PI3-kinase to adherens junctions, via binding to E-cadherin (Pece et al. 1999), whereas the phosphatase PTEN is restricted to the apical surface. Remarkably, the addition of exogenous PIP3 to the apical surface of such cells transiently induces a patch of membrane with basolateral characteristics, whereas apical markers are excluded (Gassama-Diagne et al. 2006). Conversely, addition of exogenous PIP2 directly to the basolateral surface of MDCK cysts relocalizes apical markers to this region. Mostov and colleagues have proposed that Annexin-2 binds to PIP2 at the apical surface and recruits or activates Cdc42, which in turn recruits Par-6/aPKC (Gassama-Diagne et al. 2006). However, as discussed previously, there are other mechanisms that recruit Par-6/aPKC to the apical surface independently of Cdc42, including Pals1 binding. Moreover, the proposed mechanism is apparently independent of Par-3, yet Par-3 is known to be required for apical localization of aPKC (Hirose et al. 2006; McCaffrey and Macara 2009), so there are key aspects to this system that have yet to be understood. An additional puzzle is that in Drosophila retinal epithelial cells, the phosphoinositide distribution is the inverse of that seen in MDCK cells, such that PIP3 is concentrated on the apical surface, where it activates the protein kinase Akt. Par-3 recruits PTEN to the cell–cell junctions, leading to destruction of PIP3 along the lateral membrane (Pinal et al. 2006). An important goal for future studies is to understand the cross talk between the Par proteins and localized phosphoinositide metabolism.
CONCLUDING REMARKS
Understanding cell polarization is one of the major goals of cell biology and will inevitably have a broad impact not only at the level of basic science but also in understanding diseases such as cancer and neurological degeneration. Work from many laboratories has uncovered a complicated web of signaling systems that surround and intersect with the Par proteins, yet we still understand very little about what the Par proteins do, how they are localized, how their various interactions are regulated, and which signaling components operate in which contexts. After all, the organization of a polarized cell is a formidably complicated process that involves cytoskeletal remodeling, membrane traffic, RNA localization, and protein complex assembly and disassembly, with feedback to gene expression and protein turnover. It is conceivable that the Par proteins participate in all of these processes, either directly or indirectly. It is worth remembering, however, that although the polarity machinery can work in a cell autonomous fashion, the Par genes do not exist, as far as is known, in any unicellular organism, which suggests that a key role for the Par proteins is to facilitate, mediate, or interpret cell–cell and cell-matrix interactions. The tissue context might, therefore, be expected to modulate Par protein functions. Even in vitro, the context can exert important influences. For example, Par-6 and aPKC are concentrated at tight junctions of epithelial cells grown in two-dimensional cultures, but are spread over the apical surface in three-dimensional cultures. Within the much more complicated environment of an organism, such influences are likely to be widespread and might alter signaling inputs to and outputs from the Par proteins. It will be of interest in the future to determine whether these effects control morphogenesis, and to what extent differential Par function contributes to phenotypic variation both between different cell types in one organism, and between species.
It will also be important to understand why Par genes have multiplied in the vertebrate lineage. For example, vertebrates possess three isoforms of Par-6 (Joberty et al. 2000). They all possess the same core properties, and—at least in cultured mammalian neurons—are interchangeable (Zhang and Macara 2008). Yet, they are differentially expressed, possess subtle biochemical distinctions (Gao and Macara 2004), and in zebrafish, are biologically unique (Munson et al. 2008). There are also multiple splice variants of the Par proteins that presumably operate differentially in certain contexts, but we are a long way from understanding their physiological significance (Gao et al. 2002b). As an example, the chimpanzee (and probably other mammals) expresses at least 24 splice variants of Par-3. Some of these lack only a few amino acids, whereas others lack entire domains. Various splice variants in the mouse are differentially expressed, but we do not know why. We lack sufficient data to fully understand Par proteins even in the simpler metazoa, let alone in vertebrates. For this reason, the use of diverse model organisms will continue to be essential into the foreseeable future.
ACKNOWLEDGMENTS
Work in our laboratory was supported by a research grant from the National Institutes of Health (GM090702) to I.M. L.M.M is the recipient of a Postdoctoral fellowship from the Canadian Institutes of Health Research.
Footnotes
Editors: Rong Li and Bruce Bowerman
Additional Perspectives on Symmetry Breaking in Biology available at www.cshperspectives.org
REFERENCES
- Aceto D, Beers M, Kemphues KJ 2006. Interaction of PAR-6 with CDC-42 is required for maintenance but not establishment of PAR asymmetry in C. elegans. Dev Biol 299:386–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DC, Gill JS, Cinalli RM, Nance J 2008. Polarization of the C. elegans embryo by ρGAP-mediated exclusion of PAR-6 from cell contacts. Science 320:1771–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, Pawson T, Muthuswamy SK 2006. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol 8:1235–1245 [DOI] [PubMed] [Google Scholar]
- Assemat E, Bazellieres E, Pallesi-Pocachard E, Le Bivic A, Massey-Harroche D 2008. Polarity complex proteins. Biochim Biophys Acta 1778:614–630 [DOI] [PubMed] [Google Scholar]
- Atwood SX, Prehoda KE 2009. aPKC Phosphorylates Miranda to Polarize Fate Determinants during Neuroblast Asymmetric Cell Division. Curr Biol 19:723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood SX, Chabu C, Penkert RR, Doe CQ, Prehoda KE 2007. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J Cell Sci 120:3200–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Johnston DS 2003a. A conserved oligomerization domain in Drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol 13:1330–1334 [DOI] [PubMed] [Google Scholar]
- Benton R, Johnston DS 2003b. Drosophila PAR-1 and 14-3-3 inhibit bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell 115:691–704 [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA 2003. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422:326–330 [DOI] [PubMed] [Google Scholar]
- Bruewer M, Hopkins AM, Hobert ME, Nusrat A, Madara JL 2004. ρA, Rac1, and Cdc42 exert distinct effects on epithelial barrier via selective structural and biochemical modulation of junctional proteins and F-actin. Am J Physiol Cell Physiol 287:327–335 [DOI] [PubMed] [Google Scholar]
- Butty AC, Perrinjaquet N, Petit A, Jaquenoud M, Segall JE, Hofmann K, Zwahlen C, Peter M 2002. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. Embo J 21:1565–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman JC, Lewellyn L, Laband K, Smerdon SJ, Desai A, Bowerman B, Oegema K 2008. Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science 322:1543–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabu C, Doe CQ 2008. Dap160/intersectin binds and activates aPKC to regulate cell polarity and cell cycle progression. Development 135:2739–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JR, Jolicoeur C, Yamauchi J, Elliott J, Fawcett JP, Ng BK, Cayouette M 2006. The polarity protein Par-3 directly interacts with p75NTR to regulate myelination. Science 314:832–836 [DOI] [PubMed] [Google Scholar]
- Chang F, Peter M 2003. Yeasts make their mark. Nat Cell Biol 5:294–299 [DOI] [PubMed] [Google Scholar]
- Charest PG, Firtel RA 2006. Feedback signaling controls leading-edge formation during chemotaxis. Curr Opin Genet Dev 16:339–347 [DOI] [PubMed] [Google Scholar]
- Chen X, Macara IG 2005. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol 7:262–269 [DOI] [PubMed] [Google Scholar]
- Chen X, Macara IG 2006. Par-3 mediates the inhibition of LIM kinase 2 to regulate cofilin phosphorylation and tight junction assembly. J Cell Biol 172:671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P, Janetopoulos C 2003. Eukaryotic chemotaxis: Distinctions between directional sensing and polarization. J Biol Chem 278:20445–20448 [DOI] [PubMed] [Google Scholar]
- Djiane A, Yogev S, Mlodzik M 2005. The apical determinants aPKC and dPatj regulate Frizzled-dependent planar cell polarity in the Drosophila eye. Cell 121:621–631 [DOI] [PubMed] [Google Scholar]
- Ebnet K, Schulz CU, Meyer Zu Brickwedde MK, Pendl GG, Vestweber D 2000. Junctional adhesion molecule interacts with the PDZ domain-containing proteins AF-6 and ZO-1. J Biol Chem 275:27979–27988 [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ 1995. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell 83:743–752 [DOI] [PubMed] [Google Scholar]
- Feng W, Wu H, Chan LN, Zhang M 2007. The Par-3 NTD adopts a PB1-like structure required for Par-3 oligomerization and membrane localization. Embo J 26:2786–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Wu H, Chan LN, Zhang M 2008. Par-3-mediated junctional localization of the lipid phosphatase PTEN is required for cell polarity establishment. J Biol Chem 283:23440–23449 [DOI] [PubMed] [Google Scholar]
- Fukuhara A, Irie K, Yamada A, Katata T, Honda T, Shimizu K, Nakanishi H, Takai Y 2002. Role of nectin in organization of tight junctions in epithelial cells. Genes Cells 7:1059–1072 [DOI] [PubMed] [Google Scholar]
- Gao L, Macara IG 2004. Isoforms of the polarity protein par6 have distinct functions. J Biol Chem 279:41557–41562 [DOI] [PubMed] [Google Scholar]
- Gao L, Joberty G, Macara IG 2002a. Assembly of epithelial tight junctions is negatively regulated by Par6. Curr Biol 12:221–225 [DOI] [PubMed] [Google Scholar]
- Gao L, Macara IG, Joberty G 2002b. Multiple splice variants of Par3 and of a novel related gene, Par3L, produce proteins with different binding properties. Gene 294:99–107 [DOI] [PubMed] [Google Scholar]
- Garrard SM, Capaldo CT, Gao L, Rosen MK, Macara IG, Tomchick DR 2003. Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6. Embo J 22:1125–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassama-Diagne A, Yu W, ter Beest M, Martin-Belmonte F, Kierbel A, Engel J, Mostov K 2006. Phosphatidylinositol-3,4,5-trisphosphate regulates the formation of the basolateral plasma membrane in epithelial cells. Nat Cell Biol 8:963–970 [DOI] [PubMed] [Google Scholar]
- Gervais L, Claret S, Januschke J, Roth S, Guichet A 2008. PIP5K-dependent production of PIP2 sustains microtubule organization to establish polarized transport in the Drosophila oocyte. Development 135:3829–3838 [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG 2007. The PAR proteins: Fundamental players in animal cell polarization. Dev Cell 13:609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P 2008. Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat Rev Mol Cell Biol 9:355–366 [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81:611–620 [DOI] [PubMed] [Google Scholar]
- Harris TJ, Peifer M 2005. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J Cell Biol 170:813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Yoshinaga S, Takeya R, Suzuki NN, Horiuchi M, Kohjima M, Sumimoto H, Inagaki F 2005. Structure of a cell polarity regulator, a complex between atypical PKC and Par6 PB1 domains. J Biol Chem 280:9653–9661 [DOI] [PubMed] [Google Scholar]
- Hirose T, Karasawa M, Sugitani Y, Fujisawa M, Akimoto K, Ohno S, Noda T 2006. PAR3 is essential for cyst-mediated epicardial development by establishing apical cortical domains. Development 133:1389–1398 [DOI] [PubMed] [Google Scholar]
- Horikoshi Y, Suzuki A, Yamanaka T, Sasaki K, Mizuno K, Sawada H, Yonemura S, Ohno S 2009. Interaction between PAR-3 and the aPKC-PAR-6 complex is indispensable for apical domain development of epithelial cells. J Cell Sci 122:1595–1606 [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Bilder D 2008. Dynein regulates epithelial polarity and the apical localization of stardust A mRNA. PLoS Genet 4:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung TJ, Kemphues KJ 1999. PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development 126:127–135 [DOI] [PubMed] [Google Scholar]
- Hurd TW, Fan S, Liu C-J, Kweon HK, Hakansson K, Margolis B 2003a. Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Current Biology 13:2082–2090 [DOI] [PubMed] [Google Scholar]
- Hurd TW, Gao L, Roh MH, Macara IG, Margolis B 2003b. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol 5:137–142 [DOI] [PubMed] [Google Scholar]
- Hurov JB, Watkins JL, Piwnica-Worms H 2004. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr Biol 14:736–741 [DOI] [PubMed] [Google Scholar]
- Hyodo-Miura J, Yamamoto TS, Hyodo AC, Iemura S, Kusakabe M, Nishida E, Natsume T, Ueno N 2006. XGAP, an ArfGAP, is required for polarized localization of PAR proteins and cell polarity in Xenopus gastrulation. Dev Cell 11:69–79 [DOI] [PubMed] [Google Scholar]
- Itoh M, Sasaki H, Furuse M, Ozaki H, Kita T, Tsukita S 2001. Junctional adhesion molecule (JAM) binds to PAR-3: A possible mechanism for the recruitment of PAR-3 to tight junctions. J Cell Biol 154:491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Hirose T, Tamai Y, Hirai S, Nagashima Y, Fujimoto T, Tabuse Y, Kemphues KJ, Ohno S 1998. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J Cell Biol 143:95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N, Saam JR, Mango SE 2006. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science 313:1298–1301 [DOI] [PubMed] [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol 2:531–539 [DOI] [PubMed] [Google Scholar]
- Johnson DI 1999. Cdc42: An essential ρ-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev 63:54–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS 1988. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52:311–320 [DOI] [PubMed] [Google Scholar]
- Knoblich JA 2008. Mechanisms of asymmetric stem cell division. Cell 132:583–597 [DOI] [PubMed] [Google Scholar]
- Kusakabe M, Nishida E 2004. The polarity-inducing kinase Par-1 controls Xenopus gastrulation in cooperation with 14-3-3 and aPKC. Embo J 23:4190–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Nishanian TG, Mood K, Bong YS, Daar IO 2008. EphrinB1 controls cell-cell junctions through the Par polarity complex. Nat Cell Biol 10:979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan DJ, Boyd L, Mello CC, Kemphues KJ, Stinchcomb DT 1994. par-2, a gene required for blastomere asymmetry in Caenorhabditis elegans, encodes zinc-finger and ATP-binding motifs. Proc Natl Acad Sci 91:6108–6112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang L, Hays TS, Cai Y 2008. Dynein-mediated apical localization of crumbs transcripts is required for Crumbs activity in epithelial polarity. J Cell Biol 180:31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol 2:540–547 [DOI] [PubMed] [Google Scholar]
- Macara IG, Mili S 2008. Polarity and differential inheritance–universal attributes of life? Cell 135:801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Belmonte F, Mostov K 2008. Regulation of cell polarity during epithelial morphogenesis. Curr Opin Cell Biol 20:227–234 [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F, Gassama A, Datta A, Yu W, Rescher U, Gerke V, Mostov K 2007. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell 128:383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer B, Emery G, Berdnik D, Wirtz-Peitz F, Knoblich JA 2005. Quantitative analysis of protein dynamics during asymmetric cell division. Curr Biol 15:1847–1854 [DOI] [PubMed] [Google Scholar]
- McCaffrey LM, Macara IG 2009. The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev 23:1450–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menager C, Arimura N, Fukata Y, Kaibuchi K 2004. PIP3 is involved in neuronal polarization and axon formation. J Neurochem 89:109–118 [DOI] [PubMed] [Google Scholar]
- Mertens AE, Rygiel TP, Olivo C, van der Kammen R, Collard JG 2005. The Rac activator Tiam1 controls tight junction biogenesis in keratinocytes through binding to and activation of the Par polarity complex. J Cell Biol 170:1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Suzuki A, Hirose T, Kitamura K, Kutsuzawa K, Futaki M, Amano Y, Ohno S 2003. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J Biol Chem 278:31240–31250 [DOI] [PubMed] [Google Scholar]
- Morton DG, Shakes DC, Nugent S, Dichoso D, Wang W, Golden A, Kemphues KJ 2002. The Caenorhabditis elegans par-5 gene encodes a 14-3-3 protein required for cellular asymmetry in the early embryo. Dev Biol 241:47–58 [DOI] [PubMed] [Google Scholar]
- Motegi F, Sugimoto A 2006. Sequential functioning of the ECT-2 ρGEF, ρ-1 and CDC-42 establishes cell polarity in Caenorhabditis elegans embryos. Nat Cell Biol 8:978–985 [DOI] [PubMed] [Google Scholar]
- Mullins RD 2009. Cytoskeletal mechanisms for breaking cellular symmetry. Cold Spring Harb Perspect Biol 2:a003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro EM 2006. PAR proteins and the cytoskeleton: A marriage of equals. Curr Opin Cell Biol 18:86–94 [DOI] [PubMed] [Google Scholar]
- Munro E, Bowerman B 2009. Cellular symmetry breaking during C. elegans development. Cold Spring Harb Perspect Biol 1:a003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR 2004. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell 7:413–424 [DOI] [PubMed] [Google Scholar]
- Munson C, Huisken J, Bit-Avragim N, Kuo T, Dong PD, Ober EA, Verkade H, Abdelilah-Seyfried S, Stainier DY 2008. Regulation of neurocoel morphogenesis by Pard6 γ b. Dev Biol 324:41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai-Tamai Y, Mizuno K, Hirose T, Suzuki A, Ohno S 2002. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells 7:1161–1171 [DOI] [PubMed] [Google Scholar]
- Nakayama M, Goto TM, Sugimoto M, Nishimura T, Shinagawa T, Ohno S, Amano M, Kaibuchi K 2008. ρ-kinase phosphorylates PAR-3 and disrupts PAR complex formation. Dev Cell 14:205–215 [DOI] [PubMed] [Google Scholar]
- Nelson WJ 2009. Remodeling epithelial cell organization: Transitions between Front–Rear and Apical–Basal polarity. Cold Spring Harb Perspect Biol 1:a000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Kaibuchi K 2007. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell 13:15–28 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Kato K, Yamaguchi T, Fukata Y, Ohno S, Kaibuchi K 2004. Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat Cell Biol 6:328–334 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Yamaguchi T, Kato K, Yoshizawa M, Nabeshima YI, Ohno S, Hoshino M, Kaibuchi K 2005. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol 7:270–277 [DOI] [PubMed] [Google Scholar]
- Orlando K, Guo W 2009. Membrane organization and dynamics in cell polarity. Cold Spring Harb Perspect Biol 1:a001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipova O, Dhawan S, Sokol S, Green JB 2005. Distinct PAR-1 proteins function in different branches of Wnt signaling during vertebrate development. Dev Cell 8:829–841 [DOI] [PubMed] [Google Scholar]
- Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL 2005. Regulation of the polarity protein Par6 by TGFβ receptors controls epithelial cell plasticity. Science 307:1603–1609 [DOI] [PubMed] [Google Scholar]
- Panbianco C, Weinkove D, Zanin E, Jones D, Divecha N, Gotta M, Ahringer J 2008. A casein kinase 1 and PAR proteins regulate asymmetry of a PIP(2) synthesis enzyme for asymmetric spindle positioning. Dev Cell 15:198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Chiariello M, Murga C, Gutkind JS 1999. Activation of the protein kinase Akt/PKB by the formation of E-cadherin-mediated cell-cell junctions. Evidence for the association of phosphatidylinositol 3-kinase with the E-cadherin adhesion complex. J Biol Chem 274:19347–19351 [DOI] [PubMed] [Google Scholar]
- Pegtel DM, Ellenbroek SI, Mertens AE, van der Kammen RA, de Rooij J, Collard JG 2007. The par-tiam1 complex controls persistent migration by stabilizing microtubule-dependent front-rear polarity. Curr Biol 17:1623–1634 [DOI] [PubMed] [Google Scholar]
- Pinal N, Goberdhan DC, Collinson L, Fujita Y, Cox IM, Wilson C, Pichaud F 2006. Regulated and polarized PtdIns(3,4,5)P3 accumulation is essential for apical membrane morphogenesis in photoreceptor epithelial cells. Curr Biol 16:140–149 [DOI] [PubMed] [Google Scholar]
- Prehoda KE 2009. Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harb Perspect Biol 1:a001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh MH, Fan S, Liu CJ, Margolis B 2003. The Crumbs3-Pals1 complex participates in the establishment of polarity in mammalian epithelial cells. J Cell Sci 116:2895–2906 [DOI] [PubMed] [Google Scholar]
- Roh MH, Makarova O, Liu CJ, Shin K, Lee S, Laurinec S, Goyal M, Wiggins R, Margolis B 2002. The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J Cell Biol 157:161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger K, McManus EJ, Hall A 2007. Cdc42 and noncanonical Wnt signal transduction pathways cooperate to promote cell polarity. J Cell Biol 178:355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz C, Stevens J, Spang A 2007. Functions of the novel ρGAP proteins RGA-3 and RGA-4 in the germ line and in the early embryo of C. elegans. Development 134:3495–3505 [DOI] [PubMed] [Google Scholar]
- Schneider SQ, Bowerman B 2003. Cell polarity and the cytoskeleton in the Caenorhabditis elegans zygote. Annu Rev Genet 37:221–249 [DOI] [PubMed] [Google Scholar]
- Schonegg S, Constantinescu AT, Hoege C, Hyman AA 2007. The ρ GTPase-activating proteins RGA-3 and RGA-4 are required to set the initial size of PAR domains in Caenorhabditis elegans one-cell embryos. Proc Natl Acad Sci 104:14976–14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SH, Jan LY, Jan YN 2003. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell 112:63–75 [DOI] [PubMed] [Google Scholar]
- Shi SH, Cheng T, Jan LY, Jan YN 2004. APC and GSK-3β are involved in mPar3 targeting to the nascent axon and establishment of neuronal polarity. Curr Biol 14:2025–2032 [DOI] [PubMed] [Google Scholar]
- Shin K, Straight S, Margolis B 2005. PATJ regulates tight junction formation and polarity in mammalian epithelial cells. J Cell Biol 168:705–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Mlodzik M 2008. Planar cell polarity signaling: From fly development to human disease. Annu Rev Genet 42:517–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter BD, Smith SE, Li R 2009. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb Perspect Biol 1:a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, She YM, Berry DM, Bonneil E, Thibault P, Schweisguth F, et al. 2007. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. Embo J 26:468–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D 2005. Moving messages: The intracellular localization of mRNAs. Nat Rev Mol Cell Biol 6:363–375 [DOI] [PubMed] [Google Scholar]
- Straight SW, Shin K, Fogg VC, Fan S, Liu CJ, Roh M, Margolis B 2004. Loss of PALS1 expression leads to tight junction and polarity defects. Mol Biol Cell 15:1981–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TQ, Lu B, Feng JJ, Reinhard C, Jan YN, Fantl WJ, Williams LT 2001. PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat Cell Biol 3:628–636 [DOI] [PubMed] [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S 1998. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development 125:3607–3614 [DOI] [PubMed] [Google Scholar]
- Takekuni K, Ikeda W, Fujito T, Morimoto K, Takeuchi M, Monden M, Takai Y 2003. Direct binding of cell polarity protein PAR-3 to cell-cell adhesion molecule nectin at neuroepithelial cells of developing mouse. J Biol Chem 278:5497–5500 [DOI] [PubMed] [Google Scholar]
- Traweger A, Wiggin G, Taylor L, Metalnikov P, Pawson T 2008. Protein phosphatase 1 regulates the phosphorylation state of the polarity scaffold Par-3. Proc Natl Acad Sci 105:10402–10407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Ahringer J 2007. Microtubules are involved in anterior-posterior axis formation in C. elegans embryos. J Cell Biol 179:397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar EK, Antic D, Axelrod JD 2009. Planar cell polarity signaling: The developing cell's compass. Cold Spring Harb Perspect Biol 1:a002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Du D, Fang L, Yang G, Zhang C, Zeng R, Ullrich A, Lottspeich F, Chen Z 2006. Tyrosine phosphorylated Par3 regulates epithelial tight junction assembly promoted by EGFR signaling. Embo J 25:5058–5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL 2003. Regulation of cell polarity and protrusion formation by targeting ρA for degradation. Science 302:1775–1779 [DOI] [PubMed] [Google Scholar]
- Watkins JL, Lewandowski KT, Meek SE, Storz P, Toker A, Piwnica-Worms H 2008. Phosphorylation of the Par-1 polarity kinase by protein kinase D regulates 14-3-3 binding and membrane association. Proc Natl Acad Sci 105:18378–18383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JL, Morton DG, Bestman J, Kemphues KJ 2000. The C. elegans par-4 gene encodes a putative serine-threonine kinase required for establishing embryonic asymmetry. Development 127:1467–1475 [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R, Altschuler S, Wu L, Li R 2003. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299:1231–1235 [DOI] [PubMed] [Google Scholar]
- Wei SY, Escudero LM, Yu F, Chang LH, Chen LY, Ho YH, Lin CM, Chou CS, Chia W, Modolell J, Hsu JC 2005. Echinoid is a component of adherens junctions that cooperates with DE-Cadherin to mediate cell adhesion. Dev Cell 8:493–504 [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblich JA 2008. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell 135:161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Grimm A, Knust E 2000. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J Cell Biol 150:1361–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong EW, Mruk DD, Lee WM, Cheng CY 2008. Par3/Par6 polarity complex coordinates apical ectoplasmic specialization and blood-testis barrier restructuring during spermatogenesis. Proc Natl Acad Sci 105:9657–9662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Feng W, Chen J, Chan LN, Huang S, Zhang M 2007. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol Cell 28:886–898 [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Ohno S 2008. Role of Lgl/Dlg/Scribble in the regulation of epithelial junction, polarity and growth. Front Biosci 13:6693–6707 [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Horikoshi Y, Suzuki A, Sugiyama Y, Kitamura K, Maniwa R, Nagai Y, Yamashita A, Hirose T, Ishikawa H, et al. 2001. PAR-6 regulates aPKC activity in a novel way and mediates cell-cell contact-induced formation of the epithelial junctional complex. Genes Cells 6:721–731 [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG 2006. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol 8:227–237 [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG 2008. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 ρGAP and the ρ GTPase. Dev Cell 14:216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhu J, Yang GY, Wang QJ, Qian L, Chen YM, Chen F, Tao Y, Hu HS, Wang T, Luo ZG 2007. Dishevelled promotes axon differentiation by regulating atypical protein kinase C. Nat Cell Biol 9:743–754 [DOI] [PubMed] [Google Scholar]