Abstract
During Drosophila development, neuroblasts divide to generate progeny with two different fates. One daughter cell self-renews to maintain the neuroblast pool, whereas the other differentiates to populate the central nervous system. The difference in fate arises from the asymmetric distribution of proteins that specify either self-renewal or differentiation, which is brought about by their polarization into separate apical and basal cortical domains during mitosis. Neuroblast symmetry breaking is regulated by numerous proteins, many of which have only recently been discovered. The atypical protein kinase C (aPKC) is a broad regulator of polarity that localizes to the neuroblast apical cortical region and directs the polarization of the basal domain. Recent work suggests that polarity can be explained in large part by the mechanisms that restrict aPKC activity to the apical domain and those that couple asymmetric aPKC activity to the polarization of downstream factors. Polarized aPKC activity is created by a network of regulatory molecules, including Bazooka/Par-3, Cdc42, and the tumor suppressor Lgl, which represses basal recruitment. Direct phosphorylation by aPKC leads to cortical release of basal domain factors, preventing them from occupying the apical domain. In this framework, neuroblast polarity arises from a complex system that orchestrates robust aPKC polarity, which in turn polarizes substrates by coupling phosphorylation to cortical release.
Drosophila neuroblasts divide asymmetrically to give daughter cells with different fates. The apical kinase aPKC ensures that differentiation factors are restricted to the basal daughter cell.
Cells use polarity for remarkably diverse functions. In this article, I discuss a polarity that is harnessed to generate daughter cells with different fates. Using polarity to divide asymmetrically addresses several challenges that complex organisms face. The diversification of cell types and tissues that occurs during the development of complex organisms is one such challenge. Drosophila neuroblasts, the subject of this article, undergo repeated symmetry breaking asymmetric cell divisions (ACDs) to populate the central nervous system. In a similar manner in adult organisms, ACDs are important for adult homeostasis, replenishing cells that are turned over during the course of normal physiology (Betschinger and Knoblich 2004).
A fundamental aspect of ACD is the production of daughter cells containing distinct fate determinants. To segregate fate determinants, the cell becomes polarized to form mutually exclusive cortical domains, each with a set of fate determinants appropriate for one of the two daughter cells. The cleavage furrow forms at the interface of the two domains, partitioning the fate determinants into the two daughter cells where they function to either self-renew (to keep the progenitor population) or to differentiate (e.g., by changing the pattern of gene expression). One of the unique features of the symmetry breaking that occurs during ACD, at least as implemented by the neuroblast, is that it is remarkably dynamic, developing early in mitosis and depolarizing following the completion of cytokinesis.
Since the discovery of the first polarized components, neuroblasts have been an excellent model system for investigating the mechanisms of cell polarization and have been extensively analyzed. Although aspects of neuroblast polarity remain unclear, a core framework for how polarity is created and maintained is emerging. In this article, I focus on neuroblast polarity as centered around the activity of atypical protein kinase C, which has emerged as a key regulator of the process. In this framework, neuroblast polarity can be explained by events that polarize aPKC and those that couple aPKC activity to the polarization of fate determinants.
MAIN TEXT
Neuroblasts as a Model System for Investigating Asymmetric Cell Division
Neuroblasts are initially specified in the embryonic epithelium (the neuroectoderm), where they delaminate into the interior and undergo repeated asymmetric divisions to generate the central nervous system (Fig. 1). After a short period of quiescence, they continue neuron production in the larval brain. Intensive study of the molecular control of neuroblast cell divisions over the last decade has led to significant new insights into the mechanisms used to generate cell polarity and how polarity is used in ACD.
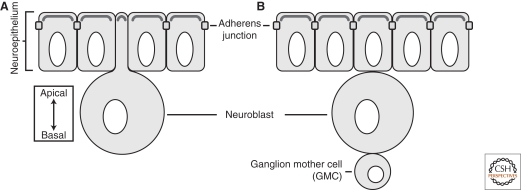
Figure 1.
The Drosophila neuroblast. (A) Embryonic neuroblasts form from the neuroepithelium where they delaminate, inheriting the polarity from the tissue from which they are born. (B) Once delaminated, neuroblasts round up, grow larger, and undergo repeated asymmetric cell divisions. Each division yields a self-renewed neuroblast and ganglion mother cell (GMC), which divides once more to populate the central nervous system.
Asymmetric division of a neuroblast yields a large, self-renewed neuroblast and a smaller intermediate progenitor called a ganglion mother cell (GMC). GMCs undergo one more division that gives rise to two postmitotic cells that become neurons or glial cells. The self-renewed neuroblast undergoes additional divisions such that each generates approximately 16 postmitotic progeny by the end of embryogenesis (Skeath and Thor 2003).
Embryonic neuroblasts give rise to the simple larval nervous system, whereas larval neuroblasts generate the thousands of neurons present in the adult organism (Campos-Ortega and Hartenstein 1997). Larval brain neuroblasts are very similar to their embryonic counterparts but there are several differences. Larval neuroblasts replenish their cytoplasm after each division, whereas the embryonic variants divide so rapidly that they shrink over the course of embryogenesis (Ito and Hotta 1992). Not surprisingly, larval neuroblasts can divide many more times because of this. Larval neuroblasts also comprise a less homogenous population. For example, certain subgroups generate GMCs that divide more than once analogous to the transit-amplifying cells generated by mammalian stem cells (Bowman et al. 2008; Boone and Doe 2008; also see Yamashita et al. 2009). Most larval neuroblasts are born as embryonic neuroblasts, although those present in the optic lobe originate from a larval epithelium (Egger et al. 2007).
The study of neuroblasts has benefited from the powerful genetic tools available in Drosophila. Genetic screens have identified many new regulatory factors, and lineage-specific expression using the UAS-GAL4 system has made it possible to analyze structure–function and overexpression effects specifically in neuroblasts without secondary effects from other cells and tissues.
Neuroblast Polarity
In metaphase neuroblasts, factors that specify GMC fate form a cortical domain that occupies roughly half the cell's surface area (Fig. 2). Opposite this area, another domain of equal size contains proteins that are ultimately partitioned into the self-renewed neuroblast. Embryonic neuroblasts are polarized along the apicobasal axis such that the apical domain is closest to the neuroepithelium and contains factors that are segregated into the self-renewed neuroblast. The basal domain lies deeper into the embryo and contains GMC fate determinants. Although larval neuroblasts are not uniformly oriented relative to the organismal axis, I will refer to the cortical domain that becomes the GMC as basal and the other as apical. As discussed in the following section, apical factors are not necessarily considered fate determinants, but instead are responsible for regulating basal polarity.
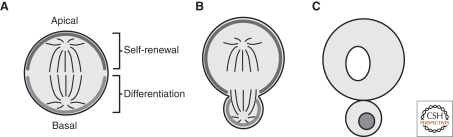
Figure 2.
Neuroblast polarity. (A) At metaphase, neuroblasts localize factors that specify differentiation to the basal cortical domain. The apical domain contains factors that keep the GMC fate determinants in the basal domain and ensure self-renewal. (B) By telophase, each cortical domain extends to the cleavage furrow, but does not pass, ensuring proper fate determinant segregation. (C) Following division, fate determinants such as Prospero are released from the cortex and enter the nucleus to change the pattern of gene expression.
The polarity present at metaphase is not present throughout the cell cycle but begins to develop at prophase. At this point, GMC fate determinants become displaced from the apical cortex such that by metaphase they are restricted to the basal cortex and by telophase they are found solely on the side of the cleavage furrow of the daughter cell that will become the GMC. Along the same timeline, several proteins localize to the apical cortex such that they are segregated into the self-renewed neuroblast. Cortical photobleaching experiments have revealed that basal domain formation takes place by exchange between the cortex and cytoplasm and not lateral transfer along the cortex (Mayer et al. 2005).
Does neuroblast polarity require extrinsic cues or is it cell autonomous? This question has been addressed by examining the polarization of cultured embryonic neuroblasts (Siegrist and Doe 2006). Cultured neuroblasts behave identically to those in situ as long as they are clustered with epithelial cells. Free, unclustered cells ultimately polarize but do so later in the cell cycle. In addition to a delay in polarization, the polarity axis is randomized after each division instead of being constantly aligned along a single axis. Thus, neuroblast polarity can be generated in the absence of extrinsic signals, but orientation of the polarity axis and robust polarization require extrinsic cues.
The identity of the external cues that coordinate neuroblast polarity is not known but because neuroblasts derive from the neuroepithelium, which is itself polarized, they are thought to inherit their polarity from their time as an epithelial cell. However, the inherited polarity does not persist as polarized epithelial factors become depolarized following cytokinesis and there are no factors known to remain polarized throughout the cell cycle. Thus, neuroblasts break symmetry repeatedly as they populate the central nervous system.
In addition to polarization, proper completion of ACD requires alignment of the mitotic spindle with the polarity axis, as the spindle specifies the cleavage furrow position (Fig. 2B) (Piekny et al. 2005; Siller and Doe 2009). Alignment of the spindle along the polarity axis ensures that the cleavage furrow forms between the two cortical domains, correctly partitioning fate determinants. As polarity must be coupled to spindle positioning, certain factors that regulate spindle orientation also influence polarity (see the following section).
What happens when neuroblast polarity is not properly established? The consequences can go beyond improper cell fate specification as polarity defects can be accompanied by overproliferation and tumor formation (Caussinus and Gonzalez 2005; Lee et al. 2006a; Bello et al. 2006). Normally, neuroblasts become mitotically inactive during the pupal stages, and mitotic exit occurs even when brain tissue is transplanted to adult flies. Proliferation control is disrupted in mutants for GMC fate determinants or in factors whose activity is required for fate determinant polarity, however. This leads to tumors that can be propagated to adult tissue in which they form large, metastasizing tumors (Caussinus and Gonzalez 2005).
Thus, neuroblast polarization is a dynamic process that results in the formation of two mutually exclusive cortical domains, a basal domain that contains fate determinants that specify differentiation and an apical domain that contains polarity regulatory factors. Polarization is intrinsic, although in the absence of external cues, the polarity axis fails to persist and becomes randomized across multiple divisions. When neuroblasts are depolarized, proliferation can become deregulated, leading to tumor formation.
Specifying the Differentiated Fate: The Basal Domain
Several factors that are part of the basal neuroblast domain confer GMC fate to the basal daughter cell. Prospero (Pros) is a homeodomain-containing transcription factor that is inactive in the neuroblast but enters the nucleus of the basal daughter cell on division, with nuclear Pros being an early distinguishing marker for the GMC (Doe et al. 1991; Chu-Lagraff et al. 1991; Spana and Doe 1995; Knoblich et al. 1995). Over 700 genes have Pros binding sites near their coding sequences and many potential targets are involved in self-renewal or cell-cycle control (Choksi et al. 2006). In pros mutants, the basal daughter cell continues to express neuroblast markers and to proliferate (Li and Vaessin 2000).
Another basal domain component, Numb, was the first segregating determinant to be identified (Rhyu et al. 1994), albeit in sensory organ precursor (SOP) cells, which are peripheral nervous system progenitors. Numb is a well-studied repressor of the Notch pathway that is thought to function by regulating transport of Notch intermediates (Schweisguth 2004). In the neuroblast lineage, Numb is important for discrimination between GMC daughter cells. In numb mutants, larval neuroblasts overproliferate because, like their pros counterparts, they divide symmetrically (Lee et al. 2006b; Wang et al. 2006a).
Brain tumor (Brat) is the most recently identified basal component (Lee et al. 2006a; Bello et al. 2006; Betschinger et al. 2006). As its name suggests, Brat is a tumor suppressor like Pros and Numb. Brat binds Argonaute-1 (Schwamborn et al. 2009), a component of the RISC pathway, and can inhibit cell growth and ribosome biogenesis (Frank et al. 2002). Recent evidence suggests that it functions by regulating the production of microRNAs, which are known to be important for many developmental processes (Schwamborn et al. 2009).
What are the targeting mechanisms that lead to Numb, Pros, and Brat recruitment to the cortex? Pros and Brat are not able to directly associate with the cortex but require the adaptor protein Miranda for this function (Fig. 3) (Ikeshima-Kataoka et al. 1997; Shen et al. 1997). Miranda is a coiled-coil protein that interacts with both Pros and Brat. In miranda mutants, Pros and Brat are cytoplasmic. The reverse is not true, however: Miranda basal localization is unaffected in pros or brat mutants. Miranda sequesters these factors to the basal cortex until cytokinesis is complete, whereupon it is degraded and they are released (Fuerstenberg et al. 1998). Interestingly, whereas Prospero and Brat have mammalian orthologues, there is no known Miranda ortholog. Although Prospero and Brat are recruited to the cortex through interactions with Miranda, the mechanism by which Miranda itself associates with the cortex is unknown. Interestingly, the activity of the microfilament pointed end-directed motor protein Myosin VI (Jaguar) is required (Petritsch et al., 2003). Myosin VI accumulates in the basal region of the cell and interacts directly with Miranda. Although Myosin VI is likely to be involved in transporting Miranda to the cortex, the physical association mechanism that maintains it there is unknown.
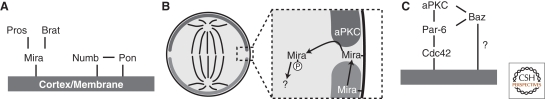
Figure 3.
Cortical targeting of apical and basal domain constituents. (A) Hierarchy of cortical localization for basal domain proteins. (B) Phosphorylation-mediated cortical displacement model for polarization of fate determinants by aPKC. In this model, the apical aPKC domain phosphorylates basal domain factors that enter the apical domain (e.g., by diffusion), causing them to be released from the cortex. The mechanism by which this cycle is completed is unknown. (C) Hierarchy of cortical localization for apical domain proteins.
Numb does not strictly require any known factors for cortical association, although the protein Partner of numb (Pon) allows for more efficient polarization (Lu et al. 1998). In pon mutants, Numb is not properly localized in metaphase, but ultimately localizes correctly in anaphase and telophase. As with Miranda, the precise cortical association mechanism of Numb is unknown.
These observations lead to a model in which differentiation factors are sequestered to the basal cortex in an inactive form. The differentiated fate is specified by the combined action of direct transcriptional regulation, a putative microRNA regulator, and Notch signaling. Cortical localization of the factors that drive these changes is mediated by Miranda and Numb (with Pon), although the mechanism of cortical targeting remains to be determined.
Cortical Displacement by Direct aPKC Phosphorylation Excludes Differentiation Factors from the Apical Domain
Given that Numb and Miranda (through its cargo proteins) specify GMC fate, how are they restricted to the basal domain? Extensive analysis has revealed that apical domain proteins are responsible for restricting Numb and Miranda to the basal domain. In the absence of specific apically localized proteins, basal fate determinants become unpolarized, localizing around the entire cortex, with a concomitant loss in asymmetric fate of the resulting daughter cells. Furthermore, when these apical factors are depolarized and targeted to the cortex uniformly (e.g., by overexpression), the polarity of basal fate determinants is lost as they become displaced into the cytoplasm.
Which apical domain components are responsible for preventing Numb and Miranda from occupying the apical domain? The factor most directly upstream of basal domain proteins is atypical protein kinase C (aPKC) (Rolls et al. 2003; Smith et al. 2007; Atwood and Prehoda 2009; see McCaffrey and Macara 2009). During mitosis, apical aPKC activity is required to restrict basal fate determinants from occupying the apical cortex. In aPKC mutants, basal components such as Miranda are no longer restricted basally and localize uniformly to the cortex. Expression of an aPKC variant with a lipid modification (aPKC-CAAX) so that it is recruited uniformly to the membrane leads to depolarization of the basal domain as factors within this domain are displaced into the cytoplasm (Lee et al. 2006c). Recent work has uncovered a direct mechanism for polarization of basal domain proteins by aPKC in which phosphorylation of Numb or Miranda leads to release from the cortex.
Recently, mammalian Numb was identified as a direct target of PKCζ (a mammalian aPKC isoform) (Smith et al. 2007). For mammalian Numb, phosphorylation at two specific serines by PKCζ regulates Numb cortical association. The same group found that Drosophila Numb is an aPKC target and that phosphorylation is required for Numb asymmetry in SOP cells. It has not yet been demonstrated that direct phosphorylation by aPKC polarizes Numb in neuroblasts, although the polarization mechanism of these two proteins appears to be the same in neuroblasts and SOPs.
In neuroblasts, phosphorylation by aPKC was recently shown to promote cortical displacement of Miranda (Atwood and Prehoda 2009). Although a complex model involving the tumor suppressor Lethal giant larvae (Lgl) and nonmuscle Myosin II had been proposed for aPKC-mediated Miranda polarization (described in the following section), Miranda phosphorylation appears to be necessary and sufficient for cortical displacement. Miranda contains an amino-terminal cortical localization domain that specifies cortical recruitment in neuroblasts and this domain is specifically phosphorylated by aPKC in vitro. Mutation of the phosphorylation sites to alanine causes Miranda to fail to respond to the presence of aPKC such that it is depolarized, overlapping with aPKC in the apical domain. When these sites are changed to phosphomimetic residues, Miranda remains in the cytoplasm, even in the absence of aPKC.
This leads to a model in which Numb and Miranda (and its fate determinant cargo, Pros and Brat) are restricted to the basal domain by direct phosphorylation by aPKC (Fig. 3B). At the basal domain, they are stably associated at the cortex through unknown mechanisms, but when the proteins diffuse into the apical domain, they become phosphorylated and released into the cytoplasm, consistent with the cortex–cytoplasm exchange of these factors observed in photobleaching experiments.
There are several aspects of the “polarity cycle” in this model that are unexplained. First, it is not known how phosphorylation leads to cortical displacement. For another aPKC substrate for which phosphorylation is known to regulate activity, Lethal giant larvae (Lgl), phosphorylation induces an intramolecular interaction that inhibits its activity (Betschinger et al. 2005), and Miranda and Numb may use a similar mechanism. Another important but unexplored aspect of the phosphorylation-induced cortical release model is what happens to the fate determinants once they are phosphorylated and displaced into the cytoplasm. Presumably, these factors are not competent to specify fate once they are in the cytoplasm, either through degradation or inhibition. Alternatively, they could be rapidly dephosphorylated to allow for reassociation at the basal cortex. Consistent with this possibility, a recent study has identified a protein phosphatase (PP4R3/Falafel) that is involved in Miranda cortical targeting (Sousa-Nunes et al. 2009).
Positive and Negative Signals Polarize aPKC Activity
As aPKC activity induces cortical release of Miranda and Numb, their segregation to the basal domain requires recruitment of aPKC to the apical domain and inhibition of its activity at the basal domain. Although cortical release of basal factors is a relatively simple matter of modulating Miranda and Numb phosphorylation states, the mechanisms for polarizing aPKC are significantly more complex. I will address aPKC polarization in two parts: how it is recruited to the apical domain and how it is inhibited from the basal domain. I will also discuss our understanding of the mechanisms of regulating aPKC enzymatic activity.
Proper recruitment of aPKC to the apical domain requires the activity of the so-called “Par complex” of which aPKC is a component but also includes Bazooka (Baz; aka Par-3) and Par-6 (Figs. 3C and 4A) (see McCaffrey and Macara 2009). Both Baz and Par-6 interact directly with aPKC: A PB1 domain in Par-6 interacts with the aPKC PB1 domain whereas Baz interacts with the aPKC kinase domain (and is an enzymatic substrate) (Suzuki and Ohno 2006; Goldstein and Macara 2007). Baz is the most upstream member of the complex as in baz mutants Par-6 and aPKC are cytoplasmic, whereas Baz remains polarized in par-6 or aPKC mutants (Rolls et al. 2003). Sizing chromatography analysis of embryonic extracts indicates that very little aPKC exists as a free monomer and most cofractionates with Par-6 (Atwood et al. 2007). The interaction with Par-6 appears to be the most persistent interaction with aPKC, whereas the Baz interaction is dynamic (Wirtz-Peitz et al. 2008).
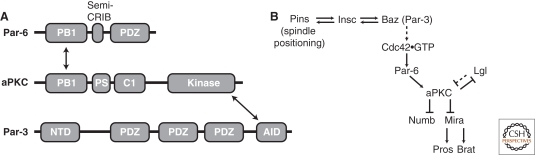
Figure 4.
Regulation of aPKC polarity and activity. (A) Domain structure of Par complex proteins. PDZ (PSD-95, Dlg, and ZO-1); PS (pseudosubstrate); NTD (amino-terminal domain); AID (aPKC interaction domain); C1 (cysteine-rich domain type 1); PB1 (Phox Bem1 domain). Arrows denote domain interactions that assemble the complex. (B) Pathway for regulation of aPKC polarity and effect on substrates. Solid lines denote direct interactions, whereas dashed lines indicate that intermediates may be present.
Both Baz and Par-6 are PDZ domain proteins (Fig. 4A). Baz contains three PDZ domains along with an oligomerization domain in its amino-terminal region (Benton and St Johnston 2003). For the mammalian Baz homolog, Par-3, the second PDZ domain binds phosphoinositides (Wu et al. 2007), providing a potential membrane recruitment mechanism, whereas the third PDZ domain binds the lipid phosphatase PTEN. Par-6 contains a single PDZ domain that binds the epithelial proteins Stardust and Crumbs but no neuroblast ligands have been identified.
How is the Par-6/aPKC complex recruited to the cortex? Par-6 binds to the Rho GTPase Cdc42 and this is a potential mechanism for cortical recruitment (Fig. 4B) (Atwood et al. 2007). Cdc42 binds to a short sequence within Par-6, termed the “semi-CRIB” domain (“semi” because the amino-terminal sequence of the Par-6 domain differs significantly from canonical CRIB domains). Like other CRIB sequences, the Par-6 CRIB selectively binds to the activated, GTP-bound form of Cdc42. Cdc42 binding activates the Par-6 PDZ domain (Peterson et al. 2004) and, as Rho GTPases are lipid modified, Cdc42 is a possible direct link to the membrane. There are conflicting reports of the function of the interaction of Par-6 with Cdc42 in neuroblasts. An earlier study that expressed Cdc42 dominant negative and constitutively active variants found no effect on neuroblast polarity (Hutterer et al. 2004). However, using a different driver to express the same Cdc42 variants does induce polarity defects, as does loss of cdc42 activity (Atwood et al. 2007). These defects are consistent with Cdc42 · GTP recruiting Par-6/aPKC through a direct interaction with Par-6. Interestingly, Baz localization is only slightly perturbed in neuroblasts lacking Cdc42, indicating that Baz is upstream of Cdc42. Cdc42 itself is slightly enriched at the apical cortex, although it is not known how the nucleotide state of Cdc42 might be polarized. In other systems, Rho GTPase nucleotide exchange factors are known to associate with Par-3, providing a possible link between Baz and Cdc42 nucleotide state asymmetry (Chen and Macara 2005; Nishimura et al. 2005).
Recent work has led to the identification of several new factors involved in neuroblast polarity, emphasizing the continued utility of this system. These include factors that regulate aPKC localization and/or activity at various points along the pathway described above, such as DAP160 (Chabu and Doe 2008), Canoe (Speicher et al. 2008), TNF/TNFR (Wang et al. 2006b), and the anaphase promoting complex (APC) (Slack et al. 2007).
How is aPKC prevented from occupying the basal domain? One of the most studied yet enigmatic polarity factors is Lethal giant larvae (Lgl). In larval neuroblasts from lgl mutants, Miranda is cytoplasmic (Betschinger et al. 2003; Lee et al. 2006c). This was originally interpreted to mean that Lgl is a cortical targeting factor for Miranda. Lgl is an aPKC substrate and expression of an unphosphorylatable Lgl, Lgl3A, leads to uniformly cortical Miranda, consistent with Lgl targeting Miranda to the cortex. However, it is now known that Lgl is a potent inhibitor of aPKC activity, so the Lgl3A phenotype can be interpreted as arising from loss of apical aPKC activity (Lee et al. 2006c; Wirtz-Peitz et al. 2008; Atwood and Prehoda 2009). Furthermore, in lgl mutants, aPKC can be detected in the basal domain, suggesting a possible mechanism for the cytoplasmic Miranda observed in these mutants. Thus, Lgl appears to be required to prevent aPKC from entering the basal domain but the mechanism for this activity is unknown. Given the requirements for aPKC cortical recruitment enumerated above, there are several possible points at which Lgl could impinge on the pathway. For example, Lgl binds both Par-6 and aPKC (Betschinger et al. 2003), so it could directly inhibit their cortical association.
Lgl also binds the motor protein Myosin II (Strand et al. 1994). Although one study implicated Myosin II in aPKC-mediated cortical displacement of Miranda (Barros et al. 2003), these observations were based on chemical inhibition of Rho Kinase, which phosphorylates and activates Myosin II. However, this compound potently inhibits aPKC (Atwood and Prehoda 2009), such that the inclusion of Myosin II in the pathway may be artifactual.
How is the enzymatic activity of aPKC regulated? The atypical PKCs are named as such because, unlike canonical PKCs, they are not activated by Ca2+ and diacylglycerol, but contain a similar serine/threonine kinase domain (Newton 2001). Given this functional difference, it is not surprising that aPKCs contain distinct regulatory domain structure compared to their canonical relatives. At its amino terminus, aPKC contains a PB1 domain that binds to the polarity protein Par-6. This is followed by a pseudosubstrate motif that is a sequence similar to the enzymatic substrate but without a phosphorylatable residue. In other kinases, the pseudosubstrate acts as a competitive inhibitor in cis to regulate enzymatic activity. A C1 domain follows the pseudosubstrate, which binds lipids such as diacylglycerol in other PKCs. Ceramide is an activator of mammalian aPKCs (Wang et al. 1999), although the significance of this interaction for neuroblast polarity, if any, is not known.
Regulation of aPKC by polarity proteins appears to be linked to the cell cycle. Par-6 is a potent repressor of aPKC kinase activity (Yamanaka et al. 2001; Atwood et al. 2007). This repression is partially relieved when Cdc42 binds to Par-6, providing a possible mechanism for coupling localization to activation. Par-6 repression of aPKC can also be relieved by phosphorylation of the Par-6 PB1 domain by the mitotic kinase Aurora A (Wirtz-Peitz et al. 2008). This appears to cause a dynamic rearrangement of the Par-6/aPKC complex. PB1 domain phosphorylation causes Par-6 to dissociate from aPKC (it remains with aPKC through bridging interactions with Lgl, however). As Par-6 represses aPKC kinase activity, AurA phosphorylation leads to an increase in aPKC activity, allowing it to phosphorylate Lgl and release it from the complex. The Par complex protein Baz can now bridge Par-6 and aPKC, and phosphorylate the cell fate determinant Numb (and perhaps others). This intriguing model provides a compelling link between a central regulator of polarity (the Par complex) and the cell cycle.
Several possible feedback mechanisms are possible involving aPKC. A positive feedback loop may exist between aPKC and Baz. As discussed above, Baz is a positive regulator of aPKC localization and activity, presumably through Cdc42. Baz is also an aPKC substrate, and although Baz is found in an apical crescent in aPKC mutants, these crescents are weaker than in wild-type, suggesting that robust Baz localization and activity requires aPKC activity. Lgl and aPKC form another feedback loop, but in this case, it is mutually antagonistic. Lgl inhibits aPKC activity and perhaps localization, as discussed previously. Lgl is also an aPKC substrate, which leads to its own inactivation and possibly cortical release. What role feedback plays in neuroblast polarity will require further investigation, but it would not be surprising if robust symmetry breaking was the result of the combined action of positive and negative feedback loops.
Coupling Cortical Polarity to Spindle Positioning
Proper segregation of fate determinants during asymmetric cell division requires that the mitotic spindle aligns with the polarity axis. Spindle alignment is important so that the cleavage furrow forms between the two cortical domains, properly segregating their contents into the two daughter cells. The cellular machinery that controls spindle orientation must interface with cortical polarity regulatory factors. The primary connection between the spindle and cortical polarity is the protein Inscuteable, which links the two by binding Baz and the adapter protein Partner of Inscuteable (Pins) (Kraut et al. 1996; Shober et al. 1999; Yu et al. 2000). Pins links the heterotrimeric G-protein subunit Gαi to the microtubule and Dynein-binding protein Mud (NuMA in mammals) (Siller et al. 2006; Izumi et al. 2006; Bowman et al. 2006; Nipper et al. 2007). In mud mutants, cortical polarity and spindle positioning are unlinked as the spindle position becomes less correlated with the polarity axis. Because Inscuteable connects these two complexes, loss of certain components in the spindle orienting pathway can influence polarity. For example, Gαi mutants have spindle position defects but can also influence polarity (Yu et al. 2003). Additionally, it has been found that astral microtubules can induce polarity through the kinesin Khc73 and the tumor suppressor Discs large (Dlg) (Siegrist and Doe 2005). Dlg binds to Pins, providing the connection to Inscuteable and cortical polarization. Although many of the players have been identified, the exact nature of the mechanical linkage between the spindle apparatus and cortical organization will require further investigation.
CONCLUDING REMARKS
Neuroblasts have been a remarkably fruitful model system for investigating symmetry breaking. After a period of rapid progress in identifying the central players, the field is now poised to further uncover the detailed mechanism by which the neuroblast cortex is dynamically organized during mitosis. In this review, I have focused on aPKC as a central component of the polarity machinery. Further mechanistic understanding will require determining how aPKC phosphorylation of downstream targets leads to cortical release and the fate of these factors once they are displaced into the cytoplasm. More enigmatic is how aPKC itself becomes polarized, including the role of Cdc42 polarization in this process and the mechanism by which Lgl inhibits basal aPKC activity. Importantly, as aPKC has emerged as a key component of many polarized systems, it is likely that the lessons learned in neuroblasts will contribute to our understanding of polarity in general. Beyond its use as a polarity model system, the neuroblast is now contributing to broader questions in cell biology, such as its use as a cancer stem cell model. Given the ever-growing set of tools available in Drosophila, it is likely that the contributions to our understanding of symmetry breaking events from the neuroblast will come at an even more rapid pace than seen over the last decade.
Footnotes
Editors: Rong Li and Bruce Bowerman
Additional Perspectives on Symmetry Breaking in Biology available at www.cshperspectives.org
REFERENCES
- Atwood SX, Chabu C, Penkert RR, Doe CQ, Prehoda KE 2007. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J Cell Sci 120:3200–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood SX, Prehoda KE 2009. aPKC Phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr Biol 19:723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros CS, Phelps CB, Brand AH 2003. Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Develop Cell 5:829–840 [DOI] [PubMed] [Google Scholar]
- Bello B, Reichert H, Hirth F 2006. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development 133:2639–2648 [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D 2003. A conserved oligomerization domain in Drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol 13:1330–1334 [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA 2003. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422:326–330 [DOI] [PubMed] [Google Scholar]
- Betschinger J, Knoblich JA 2004. Dare to be different: Asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr Biol 14:R674–685 [DOI] [PubMed] [Google Scholar]
- Betschinger J, Eisenhaber F, Knoblich JA 2005. Phosphorylation-induced autoinhibition regulates the cytoskeletal protein Lethal (2) giant larvae. Curr Biol 15:276–282 [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA 2006. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124:1241–1253 [DOI] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ 2008. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Develop Neurobiol 68:1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA 2006. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Develop Cell 10:731–742 [DOI] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA 2008. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Develop Cell 14:535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega J, Hartenstein V 1997. The embryonic development of Drosophila melanogaster Springer, Heidelberg, Germany [Google Scholar]
- Caussinus E, Gonzalez C 2005. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat Gen 37:1125–1129 [DOI] [PubMed] [Google Scholar]
- Chabu C, Doe CQ 2008. Dap160/intersectin binds and activates aPKC to regulate cell polarity and cell cycle progression. Development 135:2739–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Macara IG 2005. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol 7:262–269 [DOI] [PubMed] [Google Scholar]
- Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH 2006. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Develop Cell 11:775–789 [DOI] [PubMed] [Google Scholar]
- Chu-Lagraff Q, Wright DM, McNeil LK, Doe CQ 1991. The prospero gene encodes a divergent homeodomain protein that controls neuronal identity in Drosophila. Development 2:79–85 [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP 1991. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell 65:451–464 [DOI] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ 2007. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Develop 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DJ, Edgar BA, Roth MB 2002. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development 129:399–407 [DOI] [PubMed] [Google Scholar]
- Fuerstenberg S, Peng CY, Alvarez-Ortiz P, Hor T, Doe CQ 1998. Identification of Miranda protein domains regulating asymmetric cortical localization, cargo binding, and cortical release. Mol Cell Neurosci 12:325–339 [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG 2007. The PAR proteins: Fundamental players in animal cell polarization. Develop Cell 13:609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer A, Betschinger J, Petronczki M, Knoblich JA 2004. Sequential roles of Cdc42, Par-6, aPKC, and Lgl in the establishment of epithelial polarity during Drosophila embryogenesis. Develop Cell 6:845–854 [DOI] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F 1997. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390:625–629 [DOI] [PubMed] [Google Scholar]
- Ito K, Hotta Y 1992. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Develop Biol 149:134–148 [DOI] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F 2006. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol 8:586–593 [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN 1995. Asymmetric segregation of Numb and Prospero during cell division. Nature 377:624–627 [DOI] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA 1996. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature 383:50–55 [DOI] [PubMed] [Google Scholar]
- Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ 2006a. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Develop Cell 10:441–449 [DOI] [PubMed] [Google Scholar]
- Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, Doe CQ 2006b. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Develop 20:3464–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Robinson KJ, Doe CQ 2006c. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature 439:594–598 [DOI] [PubMed] [Google Scholar]
- Li L, Vaessin H 2000. Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Develop 14:147–151 [PMC free article] [PubMed] [Google Scholar]
- Lu B, Rothenberg M, Jan LY, Jan YN 1998. Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell 95:225–235 [DOI] [PubMed] [Google Scholar]
- Mayer B, Emery G, Berdnik D, Wirtz-Peitz F, Knoblich JA 2005. Quantitative analysis of protein dynamics during asymmetric cell division. Curr Biol 15:1847–1854 [DOI] [PubMed] [Google Scholar]
- McCaffrey LM, Macara IG 2009. Widely conserved signaling pathways in the establishment of cell polarity. Cold Spring Harb Perspect 1:a001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC 2001. Protein kinase C: Structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem Rev 101:2353–2364 [DOI] [PubMed] [Google Scholar]
- Nipper RW, Siller KH, Smith NR, Doe CQ, Prehoda KE 2007. Gαi generates multiple Pins activation states to link cortical polarity and spindle orientation in Drosophila neuroblasts. Proc Natl Acad Sci 104:14306–14311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Yamaguchi T, Kato K, Yoshizawa M, Nabeshima Y, Ohno S, Hoshino M, Kaibuchi K 2005. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol 7:270–277 [DOI] [PubMed] [Google Scholar]
- Peterson FC, Penkert RR, Volkman BF, Prehoda KE 2004. Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Mol Cell 13:665–676 [DOI] [PubMed] [Google Scholar]
- Petritsch C, Tavosanis G, Turck CW, Jan LY, Jan YN 2003. The Drosophila myosin VI Jaguar is required for basal protein targeting and correct spindle orientation in mitotic neuroblasts. Develop Cell 4:273–281 [DOI] [PubMed] [Google Scholar]
- Piekny A, Werner M, Glotzer M 2005. Cytokinesis: Welcome to the Rho zone. Trends Cell Biol 15:651–658 [DOI] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN 1994. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76:477–491 [DOI] [PubMed] [Google Scholar]
- Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ 2003. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol 163:1089–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M, Schaefer M, Knoblich JA 1999. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature 402:548–551 [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136:913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F 2004. Regulation of notch signaling activity. Curr Biol 14:129–138 [PubMed] [Google Scholar]
- Shen CP, Jan LY, Jan YN 1997. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell 90:449–458 [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ 2005. Microtubule-induced Pins/Gαi cortical polarity in Drosophila neuroblasts. Cell 123:1323–1335 [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ 2006. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development 133:529–536 [DOI] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, Doe CQ 2006. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol 8:594–600 [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ 2009. Spindle orientation during asymmetric cell division. Nat Cell Biol 11:365–374 [DOI] [PubMed] [Google Scholar]
- Skeath JB, Thor S 2003. Genetic control of Drosophila nerve cord development. Curr Opinion Neurobiol 13:8–15 [DOI] [PubMed] [Google Scholar]
- Slack C, Overton PM, Tuxworth RI, Chia W 2007. Asymmetric localisation of Miranda and its cargo proteins during neuroblast division requires the anaphase-promoting complex/cyclosome. Development 134:3781–3787 [DOI] [PubMed] [Google Scholar]
- Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, She YM, Berry DM, Bonneil E, Thibault P, Schweisguth F, et al. 2007. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO J 26:468–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Nunes R, Chia W, Somers WG 2009. Protein phosphatase 4 mediates localization of the Miranda complex during Drosophila neuroblast asymmetric divisions. Genes Develop 23:359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana EP, Doe CQ 1995. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development 121:3187–3195 [DOI] [PubMed] [Google Scholar]
- Speicher S, Fischer A, Knoblich J, Carmena A 2008. The PDZ protein Canoe regulates the asymmetric division of Drosophila neuroblasts and muscle progenitors. Curr Biol 18:831–837 [DOI] [PubMed] [Google Scholar]
- Strand D, Jakobs R, Merdes G, Neumann B, Kalmes A, Heid HW, Husmann I, Mechler BM 1994. The Drosophila lethal(2)giant larvae tumor suppressor protein forms homo-oligomers and is associated with nonmuscle myosin II heavy chain. J Cell Biol 127:1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Ohno S 2006. The PAR-aPKC system: Lessons in polarity. J Cell Sci 119:979–987 [DOI] [PubMed] [Google Scholar]
- Wang YM, Seibenhener ML, Vandenplas ML, Wooten MW 1999. Atypical PKC zeta is activated by ceramide, resulting in coactivation of NF-kappaB/JNK kinase and cell survival. J Neurosci Res 55:293–302 [DOI] [PubMed] [Google Scholar]
- Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, Chia W 2006a. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Develop 20:3453–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cai Y, Chia W, Yang X 2006b. Drosophila homologs of mammalian TNF/TNFR-related molecules regulate segregation of Miranda/Prospero in neuroblasts. EMBO J 25:5783–5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblich JA 2008. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell 135:161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Feng W, Chen J, Chan LN, Huang S, Zhang M 2007. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol Cell 28:886–898 [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Horikoshi Y, Suzuki A, Sugiyama Y, Kitamura K, Maniwa R, Nagai Y, Yamashita A, Hirose T, Ishikawa H, et al. 2001. PAR-6 regulates aPKC activity in a novel way and mediates cell-cell contact-induced formation of the epithelial junctional complex. Genes Cells 6:721–731 [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Yuan H, Cheng J, Hunt AJ 2009. Polarity in stem cell division: Asymmetric stem cell division in tissue homeostasis. Cold Spring Harb Perspect Biol 2:a001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Cai Y, Kaushik R, Yang X, Chia W 2003. Distinct roles of Gαi and Gβ13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J Cell Biol 162:623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Morin X, Cai Y, Yang X, Chia W 2000. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100:399–409 [DOI] [PubMed] [Google Scholar]