Abstract
Rhythmic changes in histone acetylation at circadian clock genes suggest that temporal modulation of gene expression is regulated by chromatin modifications1–3. Furthermore, recent studies demonstrate a critical relationship between circadian and metabolic physiology4–7. The Nuclear Receptor Co-Repressor 1 (NCoR) functions as an activating subunit for the chromatin modifying enzyme histone deacetylase 3 (HDAC3)8. Lack of NCoR is incompatible with life, and hence it is unknown whether NCoR, and particularly its regulation of HDAC3, is critical for adult mammalian physiology9. Here we show that specific, genetic disruption of the NCoR-HDAC3 interaction in mice causes aberrant regulation of clock genes and results in abnormal circadian behavior. These mice are also leaner and more insulin sensitive due to increased energy expenditure. Unexpectedly, loss of a functional NCoR-HDAC3 complex in vivo does not lead to sustained elevations of known catabolic genes, but rather significantly alters the oscillatory patterns of several metabolic genes, demonstrating that circadian regulation of metabolism is critical for normal energy balance. These findings indicate that activation of HDAC3 by NCoR is a nodal point in the epigenetic regulation of circadian and metabolic physiology.
Mammals display circadian rhythms in behavioral and physiologic processes, such as sleep, feeding, blood pressure, and metabolism10–12, guided by external light-dark signals that are integrated through intrinsic central and peripheral molecular clocks13, 14. Several critical clock and clock output genes display daily cycling of histone acetylation, suggesting that epigenetic regulation of chromatin plays a central role in circadian regulation1–3. Nuclear receptors (NRs) regulate circadian rhythm and metabolism by interacting with cofactor complexes that act at the level of chromatin15–17.
NCoR is a large, multi-domain protein that is recruited by NRs to mediate transcriptional repression. NCoR stably associates with HDAC3 through its deacetylase activation domain (DAD), which is conserved in the highly related NCoR2 (SMRT)8. HDAC3 deacetylase activity requires association with the NCoR or SMRT DAD8, and DAD mutations significantly inhibit NR-mediated repression18. Both HDAC3 and NCoR knockout mice die early in embryogenesis9,19, indicating that these proteins are essential for normal development and postnatal survival.
The function of NCoR in the adult is unknown, as is the physiological significance of the NCoR-HDAC3 association. We utilized a homologous recombination strategy to generate C57Bl/6 mutant mice (referred to as DADm) with a single amino acid substitution (Y478A) within the NCoR DAD, creating a mutant protein that is stable but unable to associate with or activate HDAC318 (Supplementary Fig.1, 2a,b). Levels of NCoR and SMRT were similar in DADm and wildtype littermates (Supplementary Fig.2c,d), and the HDAC3 interaction with NCoR but not SMRT was lost in the DADm mice (Supplementary Fig. 2e,f)18. DADm mice were viable, born at normal Mendelian frequencies (Supplementary Table 1), and morphologically indistinguishable from wildtype littermates at birth. Thus, NCoR binding to HDAC3 is not required for normal development, and the embyronic defects of mice lacking NCoR are due to factors other than, or in addition to, HDAC3 recruitment by NCoR.
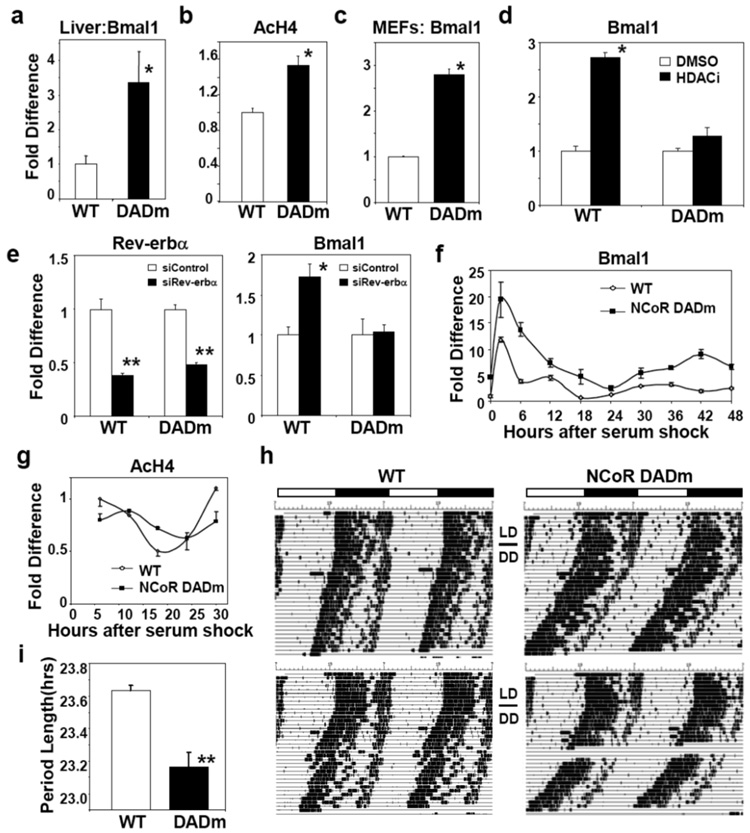
NCoR serves as corepressor for Rev-erbα (NR1D1), a NR that represses the clock gene, Bmal120, 21. Bmal1 is normally expressed at its lowest levels from ZT7-9. Consistent with an in vivo role for repression by NCoR-HDAC3, Bmal1 mRNA was increased from ZT7-9 in DADm mice (Fig. 1a), although its levels continued to cycle (data not shown). Histone H4 acetylation was increased at the Rev-erb response element (RORE) of the gene during this period (Fig. 1b)20. DADm mouse embryonic fibroblasts (MEFs) also displayed higher levels of Bmal1 (Fig. 1c). HDAC inhibition increased Bmal1 expression in wildtype but not DADm MEFs (Fig.1d), suggesting that derepression of Bmal1 in the DADm MEFs is epistatic with loss of HDAC3 activity. Moreover, depletion of Rev-erbα, or its heme ligand, derepressed Bmal1 in wildtype not DADm MEFs (Fig. 1e, Supplementary Fig. 3a), indicating that Rev-erbα is a key endogenous target of NCoR-HDAC3. Although cyclic expression of a Bmal1-luciferase reporter requires Rev-erbα and β22, Bmal1 expression remained rhythmic in DADm MEFs (Fig. 1f), potentially due to compensation by SMRT-HDAC3 or by regions of the endogenous gene not contained in the reporter construct. Nevertheless, the rhythmic expression of Bmal1 and Rev-erbα was abnormal in the DADm MEFs, demonstrating a cell-autonomous role for the NCoR-HDAC3 complex in maintaining normal circadian rhythm (Fig. 1f, Supplementary Fig. 3b). Cyclic histone aceylation was likewise altered in the DADm cells (Fig. 1g). NCoR did not oscillate and was recruited similarly to the Bmal1 RORE in the DADm and wildtype MEFs (Supplementary Fig. 3c, d) while, as predicted, HDAC3 recruitment was markedly reduced at this site in the DADm cells (Supplementary Fig. 3d).
Figure 1. NCoR-HDAC3 regulates peripheral clock and circadian physiology.
(a) Bmal1 expression in WT and DADm livers during ZT 7–9 (WT: n=6, DADm: n=4). (b) ChIP for acetylated histone H4 from WT and DADm livers at the Bmal1 RORE (n=3). (c) Bmal1 expression in immortalized mouse embryonic fibroblasts (MEFs) (n=3). (d) Bmal1 expression in MEFS after treatment with MS-275 (n=3). (e) Effect of Rev-erbα knockdown on MEF Rev-erbα and Bmal1 expression (n=3). (f) Bmal1 expression in MEFs following cell synchronization (fold difference relative to WT, time 0; n=3 per time point). (g) ChIP of acetylated histone H4 at the Bmal1 RORE following 50% serum shock. Mean +/− s.e.m of duplicate samples. Independent experiments gave similar results. (h) Voluntary locomotor wheel running activity, double plotted in each panel. (i) Average free running period (n=3). Three independent experiments gave similar results. Data are presented as mean +/− s.e.m. *p<0.05, **p<.01.
To evaluate whether the molecular dysregulation of the clock corresponds with circadian behavioral abnormalities, we monitored locomotor activity in constant darkness. DADm mice demonstrated a free running period of ~23.2h compared with 23.6h in wildtype mice (Fig. 1h,i). This decrease in the average period length of the DADm mice is highly significant and coincides with the period change reported in Rev-erbα knockout mice 21. These findings identify a critical role for activation of HDAC3 by NCoR in regulating normal circadian rhythm and demonstrate the dependence of circadian behavior on dynamic epigenetic modifications.
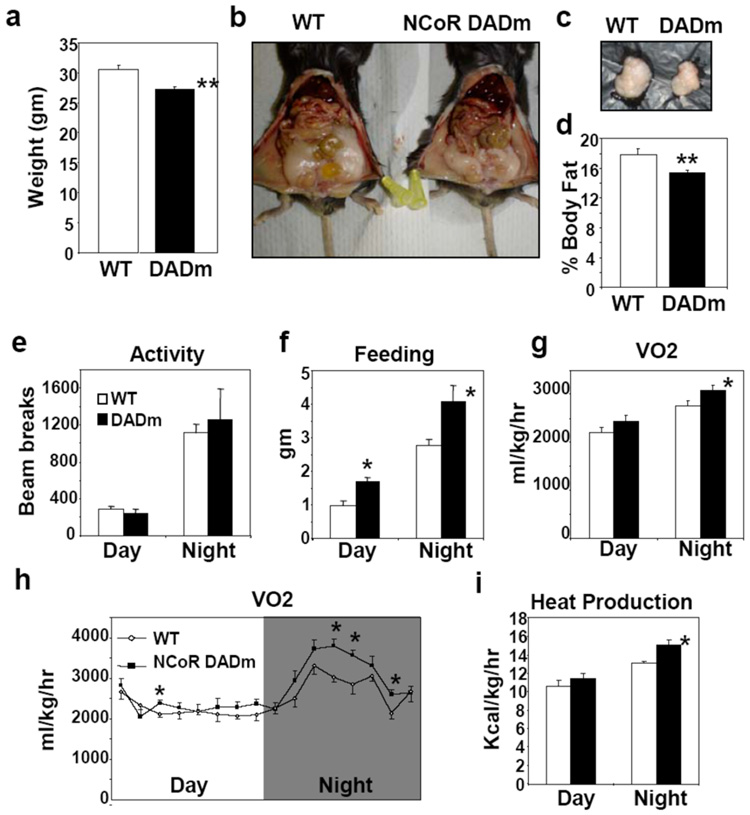
Due to the strong links between circadian and metabolic physiology, as well as the role of NRs in both systems, we also examined metabolic parameters. The NCoR DADm mice weighed the same as wildtype littermates at birth, but began to weigh significantly less between 4–6 weeks of age and maintained this difference throughout adulthood (Fig. 2a). Perigonadal fat pad weight (Fig. 2b,c) and whole body fat measured by nuclear magnetic resonance (Fig. 2d) were decreased in DADm mice. Histologic evaluation of NCoR DADm adipose tissue showed normal architecture, with a trend towards smaller adipocytes (Supplementary Fig. 4a). Consistent with previous reports that NCoR inhibits adipogenesis, adipocyte differentiation was modestly increased in DADm MEFs, indicating that the decrease in fat tissue is not due to impaired adipogenesis (Supplementary Fig. 4b,c)23.
Figure 2. DADm mice exhibit increased energy expenditure.
(a) Body weight of 24 week old male WT and DADm mice (n=10) (b) Representative abdominal images of WT and DADm mice. (c) Perigonadal fat pads harvested from mice in (b). (d) % Body fat composition measured by nuclear magnetic resonance for cohort presented in (a). (e–i) Effect of DADm on (e) locomotor activity measured by photobeam breaks, (f) food intake, (g,h) oxygen consumption (VO2), and (i) heat. Average VO2 for 3 consecutive measurements taken every 27 minutes is shown in (h) (n=4). Data is presented as mean +/− s.e.m. *p<0.05.
We next utilized metabolic cages to determine why the NCoR DADm mice are leaner. DADm mice demonstrated similar levels of locomotor activity to the wildtype mice and food intake was increased, indicating that their decreased weight did not result from increased activity or decreased feeding (Fig. 2e,f). Rather, DADm mice exhibited elevated oxygen consumption and heat measured by indirect calorimetry, particularly during the wakeful dark cycle (Fig. 2g,h, i). Therefore, these mice likely ate more food to compensate for increased catabolism. The increase in heat production is unlikely to be a primary consequence of brown adipose UCP expression which was unaltered in DADm mice (Supplementary Fig. 4d).
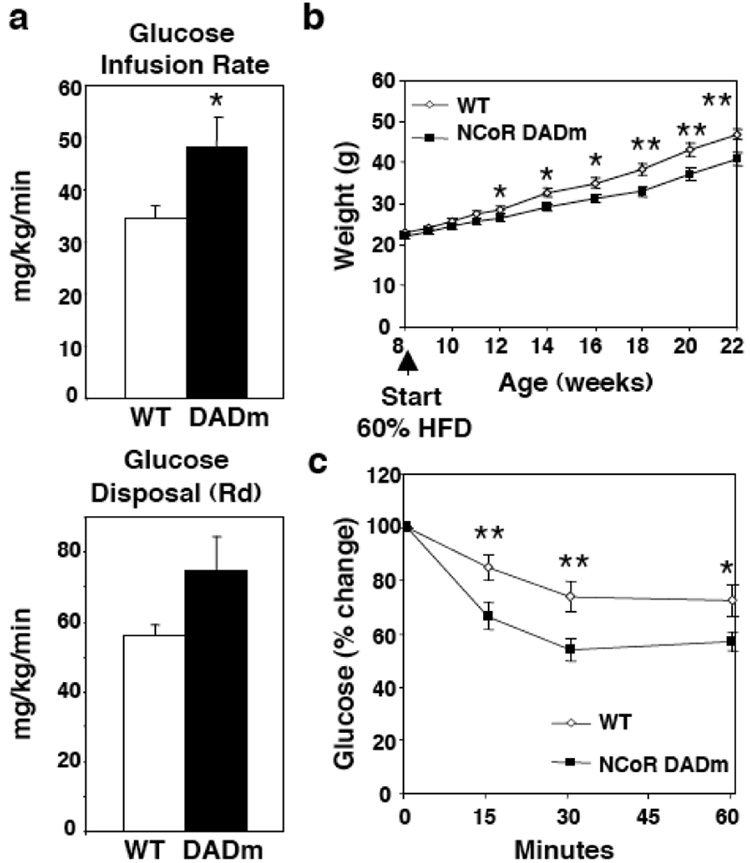
Hyperinsulinemic euglycemic clamp studies revealed increased insulin sensitivity of the DADm mice on a normal chow diet (Fig. 3a). On a high fat diet (HFD), DADm mice were resistant to diet-induced obesity and were protected from developing insulin resistance (Fig. 3b,c, Supplementary Fig. 5a), with reduced hepatic glucose production in the setting of the hyperinsulinemic clamp (Supplementary Fig. 5b). This was somewhat surprising because Rev-erbα recruits NCoR-HDAC3 to gluconeogenic genes to repress hepatic glucose production17. However, on normal chow prior to the development of large differences in insulin sensitivity, hepatic glucose output was indeed upregulated in the DADm mice (Supplementary Fig. 5c). Also, expression of the gluconeogenic PEPCK gene was increased in the DADm livers (Supplementary Fig. 5d), and the ability of the Rev-erbα ligand heme to repress PEPCK gene expression was abrogated in DADm primary hepatocytes (Supplementary Fig. 5e). Apparently, in the high fat diet model, the gluconeogenic effects of reduced Rev-erbα function were more than counterbalanced by the markedly reduced insulin resistance of the DADm mice. Although this insulin sensitivity was consistent with the leaner phenotype of the mice, weight matched DADm mice fed a HFD were also more insulin sensitive (Supplementary Fig. 6), suggesting that their improved insulin tolerance mice may not be entirely secondary to leanness.
Figure 3. DADm mice are resistant to diet-induced obesity.
(a) Hyperinsulinemic-euglycemic clamp measurements for 16 week old male WT and DADm mice fed a normal chow diet (n=4). (b) Weight curve for age matched WT and DADm mice started on 60% high fat diet (HFD) at 8 weeks. (c) Insulin tolerance test for mice in (b) at ZT8 after 15 weeks on HFD (WT, n=13 , DADm, n=12). Data is presented as mean +/− s.e.m. *p<0.05, **p<0.01.
To further understand the altered circadian and metabolic phenotypes of the DADm mice, ketone and free fatty acid levels were measured regularly during a 24 hour cycle. Serum ketones and fatty acids were significantly increased in the DADm mice at specific times during the cycle (Supplementary Fig. 7a,b), suggesting temporal dysregulation of fat metabolism such that cumulative increases in hepatic beta oxidation drive increased lipolysis. Importantly, in contrast to mice lacking HDAC3 in liver24, there was no evidence of hepatic steatosis in the DADm mice (Supplementary Fig. 7c), suggesting a potential protective effect of residual SMRT-dependent HDAC3 activity.
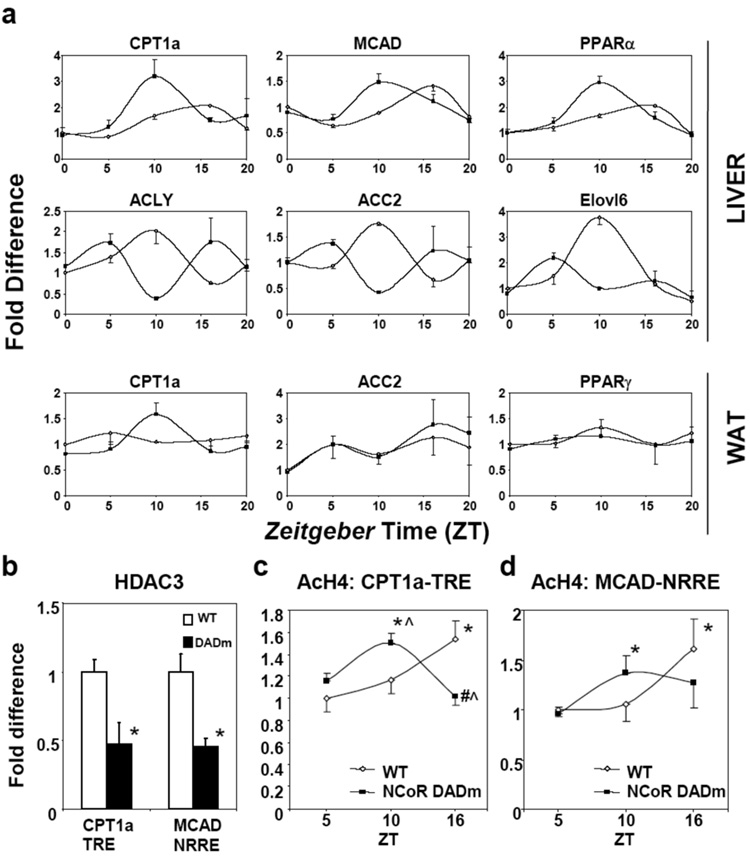
Microarray analysis at a single time point demonstrated modest changes in hepatic gene expression between the DADm and wildtype mice, although both circadian and metabolic pathways were enriched (data not shown). However, 24 hour studies revealed dramatic alterations in cyclic expression of several critical genes involved in lipid metabolism in the liver (Fig. 4a). DADm mice displayed phase shifts in the expression of genes involved in fat breakdown, such as carnitine palmitotransferase 1a (CPT1a), medium chain acyl-CoA dehydrogenase (MCAD), and their transcriptional regulator peroxisome proliferator activated receptor α (PPARα). There was a trend toward increased induction of CPT1a and MCAD after treatment of DADm hepatocytes with PPARα agonist (Supplementary Fig. 8). ATP citrate lyase (ACLY), which produces acetyl coA, and acetyl coA carboxylase 2 (ACC2), whose malonyl CoA product allosterically inhibits CPT1a, were also dysregulated but anti-phase with CPT1a (Fig. 4a). Elongation of long-chain fatty acids family member 6 (Elovl6), whose deficiency favors leanness and insulin sensitivity25, exhibited 4 fold lower expression at ZT10 in DADm liver. In white adipose tissue, modest circadian changes in CPT1a were noted but, unlike in the liver, ACC2 and PPARα expression were not altered (Fig. 4a). Taken together, NCoR-HDAC3 is required for normal circadian regulation of genes involved in the breakdown, biogenesis, and modification of lipids in the liver.
Figure 4. Activation of HDAC3 by NCoR regulates circadian metabolic gene expression in the liver.
(a) Diurnal expression of lipid metabolic genes (WT, open diamonds; DADm, black squares relative to WT, ZT0. Lights on and off at ZT0 and ZT12, respectively. (n=2–3 per genotype per ZT). (b) ChIP for HDAC3 from WT and DADm MEFs (n=3). (c, d) ChIP for acetylated histone H4 from livers at (c) CPT1a TRE and (d) MCAD NRRE (n=3 per genotype per ZT). qPCR results are relative to WT, ZT5. Data is presented as mean +/− s.e.m. *p<0.05 relative to ZT5, ^p<0.05 relative to WT, #p<.05 relative to ZT10.
To determine the mechanism of altered beta oxidation cycling in DADm mice, we examined cofactor and histone aceytlation at well-characterized nuclear receptor response elements in the CPT1a and MCAD genes. As expected, NCoR was similarly recruited to the nuclear receptor responsive elements in the CPT1a and MCAD genes in wildtype and DADm cells (Supplementary Fig. 9a)26, 27. However, HDAC3 recruitment was markedly reduced at these same genomic sites (Fig. 4b). Moreover, circadian oscillation at these response elements was disturbed in the DADm livers in a pattern that paralleled the alterations in gene expression (Fig. 4c,d) while NCoR recruitment remained unchanged (Supplementary Fig. 9b). An intronic region of the CPT1a gene did not exhibit oscillation of histone acetylation, demonstrating site specificity of circadian histone acetylation (Supplementary Fig. 9c).
We have demonstrated that the association between HDAC3 and the transcriptional corepressor NCoR regulates circadian behavior and metabolism. A transcriptional coactivator recently identified as a regulator of circadian and metabolic pathways, PGC-1α, does not possess intrinsic histone acetylase (HAT) activity, but interacts with HAT-containing coactivators7, and the Clock gene was recently shown to possess HAT activity2. These findings emphasize that circadian metabolic transcriptional regulation is regulated at the level of histone acetylation and that the cycling of epigenetic modifications is critical to maintaining normal energy homeostasis. Loss of NCoR-dependent HDAC3 activity did not lead to constant elevations of gene expression, as might have been predicted, likely due to complex interplay with other histone-modifying circadian factors. Indeed, dysregulation of the molecular clock and metabolic gene expression in the DADm mice undoubtedly reflects alterations in several interconnected pathways including the molecular clock as well as signaling by multiple nuclear receptors.
Recent studies have demonstrated critical links between circadian regulation and normal health, longevity, and diet4, 28, 29. The DADm mice exhibit dramatically altered metabolic oscillatory expression, with a net effect of increased lipid consumption and a lean, insulin sensitive, obesity-resistant metabolic phenotype. Thus, although shift work and other disruptions of normal rhythms can be metabolically deleterious30, alteration of normal circadian physiology can also be associated with a favorable metabolic profile. Targeting the NCoR-HDAC3 deacetylase enzyme would be a highly specific means of combating diseases of nutritional excess including obesity, diabetes, and the metabolic syndrome.
Methods summary
Generation of DADm mice
The Y478A mutation was introduced into exon 13 of the NCoR gene using a targeting vector containing a loxP flanked NeoR gene downstream from exon 13. The targeting vector was electroporated into C57BL/6 ES cells and a clone confirmed to contain the homologous targeted mutation was injected into C57BL/6 blastocysts. Blastocysts were implanted in pseudopregnant female mice and germline transmission led to the generation of C57Bl/6 F1 NCoRY478A Neo/+ mice. These mice were mated to C57Bl/6 TgEIIa-Cre mice to remove the floxed NeoR cassette, generating C57BL/6 NCoRY478A/+ mice which were bred to wildtype mice in order to obtain (Cre-)NCoRY478A/+ mice. Experimental cohorts were generated from (Cre-)NCoRY478A/+x(Cre-)NCoRY478A/+ breedings.
Mice
Age-matched male mice were used for all experiments. Animals were housed up to 5 per cage in a ventilated isolator cage system in a 12 hr light/dark cycle, with free access to water and chow or a high-fat diet (60 kcal% fat). Mice used for circadian gene expression experiments were housed under strict light/dark regulation and minimal disturbance for 3 weeks prior to sacrifice. For metabolic experiments, mice were placed on a high fat diet at 8 weeks and insulin tolerance tests were performed following 16 weeks on the diet using 0.75U/kg insulin. Mice underwent body composition analysis by nuclear magnetic resonance (NMR) and monitoring of feeding, locomotor activity and indirect calorimetry. For hyperinsulinemic-euglycemic clamp studies, 2.5 mU/kg/min insulin or 5 mU/kg/min was administered in chow fed mice and high fat diet mice, respectively.
Methods
Generation of DADm mice
The targeting vector containing 3.5 kb arms was generated using RP23 NCoR1 containing BAC clone templates (Invitrogen) followed by subcloning of PCR products into a pBluescript KS backbone (Stratagene). The Y478A mutation was introduced into exon 13 of the NCoR gene using site directed mutagenesis (Stratagene). A cassette containing a loxP flanked NeoR gene expressed from the phosphoglycerate kinase promoter was cloned within an intron approximately 500bp downstream from exon 13. The negative selection diphtheria toxin gene was cloned downstream of the 3’ arm. The targeting vector was linearized with XhoI and SacII (New England Bioloabs) and electroporated into C57BL/6 ES cells (Chemicon). A clone confirmed to contain the homologous targeted mutation was injected into C57BL/6 blastocysts and these blastocysts were implanted in pseudopregnant female mice. Germline transmission led to the generation of C57Bl/6 F1 NCoRY478A Neo/+ mice. These mice were mated to C57Bl/6 TgEIIa-Cre mice (Jackson Laboratory) to remove the floxed NeoR cassette, generating C57BL/6 NCoRY478A/+ mice which were bred to wildtype mice in order to obtain (Cre-)NCoRY478A/+ mice. Experimental cohorts were generated from (Cre-)NCoRY478A/+x(Cre-)NCoRY478A/+ breedings.
Mice
Age-matched male mice were used for all experiments. Genotyping was performed at weaning on genomic tail DNA. Animals were housed up to 5 per cage in a ventilated isolator cage system in a 12 hr light/dark cycle, with free access to water and chow or a high-fat diet (60 kcal% fat)(Research Diets). Wheel running studies were conducted as described previously31. Mice used for circadian gene expression experiments were housed under strict light/dark regulation and minimal disturbance for 3 weeks prior to sacrifice. Mice were placed on a high fat diet at 8 weeks and insulin tolerance tests were performed following 16 weeks on the diet using 0.75U/kg insulin. Mice that had been on normal chow or high fat diet for 16 weeks underwent body composition analysis by nuclear magnetic resonance (NMR) and dual emission xray absorptiometry (DEXA), and monitoring of feeding, locomotor activity and indirect calorimetry32. Hyperinsulinemic-euglycemic clamp was performed as previously described, except 2.5 mU/kg/min insulin was administered in chow fed mice and 5 mU/kg/min in mice on a high fat diet 32. Serum concentrations of β-hydroxybutyric acid (Stanbio) and NEFA (Wako) were measured using enzymatic colorimetric assays. Statistical analysis was performed using student’s t-test. All studies were approved by the University of Pennsylvania School of Medicine Institutional Animal Care and Use Committee.
Tissue and cell harvesting
Tissue samples were harvested following euthanasia and immediately frozen in liquid nitrogen. MEFs were harvested from day 12.5 embryos and immortalized by repeated passages. Primary hepatocytes were harvested as described previously20. Cells were grown at 37°C in 5% CO2. Primary MEFs were treated with 0.6uM dexamethasone, 10ug/ml insulin, and 500uM 3-isobutyl-1-methylxanthine (no thiazolidinedione) to induce adipocyte differentiation. Cells were harvested and stained with Oil Red O on day 5 following addition of the differentiation cocktail. Immortalized MEF lines were treated with MS-275 for 24 hours at 1uM prior to harvest. For cell synchronization studies, MEFs were grown in serum free media for 24 hours prior to synchronization with 50% horse serum for 2 hours after which cells were switched back to serum free media33. Cells were treated with 5uM succinylacetone for 12 hours, 6uM heme for 6 hours, or 50uM Wy-14,643 for 48 hours. 1uM of the siControl or siReverbα(Dharmacon) double stranded oligos were prepared with Nucleofector Solution V (Amaxa biosystems) and 1 × 106 cells were electroporated according to the manufacturer’s protocol. After 48 h, media was changed and 24h later, cells were harvested.
Immunoprecipitation and Immunoblotting
Samples for immunoprecipitation experiments were homogenized in a modified RIPA buffer containing protease inhibitor cocktail (Roche). Lysates were precleared with Protein G agarose beads and then incubated with mouse HDAC3 (Upstate) or mouse IgG (Santa Cruz) antibodies at 4 °C overnight followed by 1 h incubation with protein G agarose beads. Immunoprecipitates were washed five times with modified RIPA, eluted, and subjected to immunoblot analysis. Blots were probed with the following primary antibodies: rabbit NCoR 34, rabbit SMRT (Bethyl), rabbit HDAC3 (Abcam). For liver chromatin immunoprecipitations, 50 mg of tissue from each mouse was minced and crosslinked in 1% formaldehyde for 15 minutes. After 2 washes, cells were manually lysed using a dounce and then subjected to nuclear lysis and sonication. ChIP assays were performed according to the protocol of Upstate Biotechnology with minor modifications and the following antibodies: rabbit HDAC3 (Santa Cruz/Abcam), rabbit acetylated histone H4 (Upstate), rabbit NCoR (Abcam). The following primer pairs were used for ChIP experiments: Bmal1: 5'-agcctaacgcagagcagaac-3', 5'-gccaatcagagagagcgaac-3', MCAD: 5’-cactgggcacacagtcttcttc-3’, 5’-ccttgcccgagcctaaact-3’, CPT1a TRE: 5’-ggtgacgttggctgagcaa-3’, 5’-tgagcccctgtacacgttttg-3’, CPT1a Intron: 5’-cagcgccttgaacttgca-3’, 5’-caaacggtcaaagtacaggaaagtc-3’.
RNA isolation and Real-time PCR analysis
Tissue samples were homogenized using a TissueLyser (Qiagen). RNA was isolated using a RNeasy Lipid Tissue Kit (Qiagen) then subjected to reverse transcription (Applied Biosystems). mRNA transcripts were quantified by real-time PCR analysis using TaqMan (Applied Biosystems) or Sybr (Applied Biosystems). Samples were analyzed using a Prism 7900 thermal cycler and sequence detector (Perkin Elmer/ABI). Sybr dissociation curves always indicated the formation of a single PCR product. Data was analyzed with a threshold set in the linear range of amplification and processed based on a standard curve of serial ten fold dilutions for each primer set. The gene of interest was normalized to an unaffected endogenous control gene (GAPDH or 36B4) and plotted as mean fold change (+/− s.e.m.). Statistical analysis was performed using student’s t-test.
Supplementary Material
Acknowledgements
We thank W. Pear (University of Pennsylvania) for providing EIIa-Cre C57BL/6 mice, G. Barnes and J. Rusche (Repligen) for providing MS-275, P. White and J. Tobias for bioinformatics assistance, and L. Yin, S.-H. You, M. Qatanani, and other members of the Lazar lab for helpful discussions. We also thank J. Richa and The Transgenic Mouse Core, H. Collins and the Radioimmunoassay/Biomarkers Core, R. Dhir and the Metabolic Phenotyping Core of the Penn Diabetes and Endocrinology Research Center (DK19525), H. Fu and the Mouse Embryonic Stem Cell Core (DK49210) and the Morphology Core of the Center for Molecular Studies in Digestive and Liver Disease (DK50306 and DK49210) for consultation and services. This work was supported by NIH grant DK43806 (to MAL), and TA was supported by a National Research Training Grant in Developmental Biology.
References
- 1.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 2.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 3.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 4.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 8.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jepsen K, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 10.Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- 11.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schultz TF, Kay SA. Circadian clocks in daily and seasonal control of development. Science. 2003;301:326–328. doi: 10.1126/science.1085935. [DOI] [PubMed] [Google Scholar]
- 13.Shearman LP, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 14.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 15.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 16.Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 17.Yin L, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 18.Ishizuka T, Lazar MA. The nuclear receptor corepressor deacetylase activating domain is essential for repression by thyroid hormone receptor. Mol Endocrinol. 2005;19:1443–1451. doi: 10.1210/me.2005-0009. [DOI] [PubMed] [Google Scholar]
- 19.Bhaskara S, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 21.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 22.Liu AC, et al. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu C, et al. The nuclear receptor corepressors NCoR and SMRT decrease peroxisome proliferator-activated receptor gamma transcriptional activity and repress 3T3-L1 adipogenesis. J Biol Chem. 2005;280:13600–13605. doi: 10.1074/jbc.M409468200. [DOI] [PubMed] [Google Scholar]
- 24.Knutson SK, et al. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. Embo J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuzaka T, et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat Med. 2007;13:1193–1202. doi: 10.1038/nm1662. [DOI] [PubMed] [Google Scholar]
- 26.Jansen MS, Cook GA, Song S, Park EA. Thyroid hormone regulates carnitine palmitoyltransferase Ialpha gene expression through elements in the promoter and first intron. J Biol Chem. 2000;275:34989–34997. doi: 10.1074/jbc.M001752200. [DOI] [PubMed] [Google Scholar]
- 27.Leone TC, et al. The human medium chain Acyl-CoA dehydrogenase gene promoter consists of a complex arrangement of nuclear receptor response elements and Sp1 binding sites. J Biol Chem. 1995;270:16308–16314. doi: 10.1074/jbc.270.27.16308. [DOI] [PubMed] [Google Scholar]
- 28.Martino TA, et al. Circadian rhythm disorganization produces profound cardiovascular and renal disease in hamsters. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1675–R1683. doi: 10.1152/ajpregu.00829.2007. [DOI] [PubMed] [Google Scholar]
- 29.Fuller PM, Lu J, Saper CB. Differential rescue of light- and food-entrainable circadian rhythms. Science. 2008;320:1074–1077. doi: 10.1126/science.1153277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramsey KM, Marcheva B, Kohsaka A, Bass J. The clockwork of metabolism. Annu Rev Nutr. 2007;27:219–240. doi: 10.1146/annurev.nutr.27.061406.093546. [DOI] [PubMed] [Google Scholar]
- 31.Kapfhamer D, et al. Mutations in Rab3a alter circadian period and homeostatic response to sleep loss in the mouse. Nat Genet. 2002;32:290–295. doi: 10.1038/ng991. [DOI] [PubMed] [Google Scholar]
- 32.Qi Y, et al. Loss of resistin improves glucose homeostasis in leptin deficiency. Diabetes. 2006;55:3083–3090. doi: 10.2337/db05-0615. [DOI] [PubMed] [Google Scholar]
- 33.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 34.Ishizuka T, Lazar MA. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol. 2003;23:5122–5131. doi: 10.1128/MCB.23.15.5122-5131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.