Summary
Like many viruses, HIV can rapidly evolve when placed under selective pressure, including immune surveillance or the administration of antiretroviral drugs. The virus typically acquires resistance by mutating the targeted proteins. Accordingly, when HIV has been suppressed with RNA interference (RNAi) directed against viral RNAs, it has escaped by acquiring mutations at the target region that circumvent RNAi-mediated inhibition while conserving necessary viral functions. However, when we directed RNAi against a novel target in the viral TAR hairpin, which cannot be altered without severely impairing viral replication, HIV did not mutate the target site. Instead several mutations that indirectly compensated for the antiviral activity through upregulation of viral transcription were isolated. This represents a novel mechanism by which viruses can tune viral transcriptional regulation as an indirect mechanism to compensate for viral suppression.
Introduction
Human immunodeficiency virus (HIV) possesses a remarkable ability to adapt and evade both host immune responses and suppression by antiviral drugs. This capacity is driven by the rapid genetic diversity retroviruses acquire via reverse transcription. During this process, HIV Reverse Transcriptase (RT) introduces novel point mutations (~0.2 errors per genome, per replication cycle) and recombines sequences from both parental RNA strands by stochastically switching between RNA templates. Since 1010–1012 new viruses may be produced per day in vivo, HIV samples many possible genetic configurations, some of which may imbue progeny viruses with selective advantages (reviewed in (Rambaut et al., 2004))
Highly active anti-retroviral therapy (HAART) has extended the lives of HIV-positive patients; however, this approach suffers many shortcomings. In particular, rapid retroviral mutation has produced many drug-resistant strains of HIV (reviewed in (Clavel and Hance, 2004)), and patient compliance is challenging given the drugs’ many side effects (reviewed in (Carr, 2003)). Consequently, an urgent need exists for new HIV therapies that are less prone to the generation of resistant viral strains.
An emerging and promising alternative to HAART is the therapeutic induction of RNA interference (RNAi), a highly-conserved cellular mechanism for suppressing gene expression. Briefly, the cellular ribonuclease Dicer cleaves double-stranded RNAs or short hairpin RNAs (shRNA) to create ~21 nucleotide (nt) short interfering RNA (siRNA) duplexes. The antisense strand of these duplexes are used by the cellular RNA-Induced Silencing Complex (RISC) to sample a cell’s mRNAs and actively cleave messages that are complementary to this guide strand (reviewed in (McManus and Sharp, 2002)). Alternatively, guide strands complementary to sequences within promoter regions have been shown to downregulate gene expression through transcriptional silencing (Castanotto et al., 2005). It is thus possible to suppress HIV gene expression in a sequence-specific manner using shRNAs targeting the viral genome.
HIV gag, pol, tat, rev, env, vif and nef have been successfully targeted for RNAi-mediated inhibition of viral replication in cell culture (Jacque et al., 2002; ter Brake et al., 2006), and an anti-HIV RNAi therapy is currently in clinical trials to evaluate its safety and efficacy (Li et al., 2006; Rossi et al., 2007). However, since one or two nucleotide mismatches between the antisense guide strand and the target RNA can disrupt RISC-mediated cleavage (Jacque et al., 2002), HIV’s capacity for mutation also threatens the long-term efficacy of RNAi-based therapies (Haasnoot et al., 2007). In cell culture, HIV has already been shown to evolve resistance to RNAi directed against tat, gag, nef , or pol via direct point mutation of the target sequences (Boden et al., 2003; Das et al., 2004; ter Brake et al., 2006). Similarly, HIV evolved resistance to RNAi directed against nef by mutating or deleting the target, which is dispensable in cell culture (Das et al., 2004), or by a local structural rearrangement of the target mRNA that likely results in the exclusion of RISC (Westerhout et al., 2005). It has been proposed that directing RNAi against HIV sequences that cannot be mutated without compromising viral functionality may preclude viral escape. While highly-conserved sites within HIV transcripts have been targeted (Lee et al., 2005; ter Brake et al., 2006), escape from RNAi directed against these targets has still occurred in 10–20 days (ter Brake et al., 2006; von Eije et al., 2008). Combinations of shRNAs may also suppress viral replication for extended periods without escape (ter Brake et al., 2006). In practice, however, this combinatorial RNAi approach has constraints, because the total siRNA dose must be limited to avoid inducing interferon-mediated responses (Reynolds et al., 2006), and ineffective siRNAs can compete with effective ones for incorporation into RISC and thereby decrease overall RNAi efficacy (Castanotto et al., 2007). Alternatively, RNAi may be coupled with other RNA-based strategies to enhance suppression (Li et al., 2005). However, in all cases, RNAi targets must be selected carefully to both maximize inhibition and prevent viral escape.
One attractive RNAi target is the trans-activation response (TAR) hairpin, an untranslated and highly-structured sequence that is present in every viral RNA and is characterized by a high degree of nucleotide sequence conservation resulting from the indispensable role that TAR plays in viral transcription. Briefly, following reverse transcription and semi-random integration into a host cell chromosome (Schroder et al., 2002), transcription of all viral RNA from an HIV provirus is driven by the 5’ long terminal repeat (LTR). Initially, basal transcription is relatively inefficient and is governed by cellular transcription factors through a balance of positive and negative regulators that bind to cis regulatory elements within the 5’ LTR (Imai and Okamoto, 2006; Kato et al., 1991; Margolis et al., 1994; Nabel and Baltimore, 1987). Although this basal transcription is relatively inefficient, it leads to the accumulation of the HIV transcriptional transactivator, Tat. Tat binds to the “bulge” of TAR (Roy et al., 1990), which is present at the 5’ end of every nascent viral RNA transcript, and recruits the cellular factors cyclin T1 and cyclin-dependent kinase 9 (Cdk9) (Zhou et al., 1998) to lead to a greatly increased viral RNA transcription rate (Feinberg et al., 1991). The resulting Tat/TAR positive feedback loop is essential for producing the viral regulatory and structural proteins and genomic RNA required for assembling progeny virions (Sodroski et al., 1986). Furthermore, the TAR hairpin is present at both the 5’ and 3’ ends of both spliced and full-length HIV transcripts, such that every HIV RNA molecule contains two copies of this potential RNAi target.
Previous efforts to inhibit HIV with RNAi directed against TAR have been met with mixed success. It was reported that TAR’s extensive secondary structure precluded cleavage by RISC (Yoshinari et al., 2004); however, recently it was shown that RISC is able to efficiently cleave imperfect hairpins resembling TAR, given the appropriate selection of an antisense guide strand (Ameres et al., 2007), and some inhibition of HIV replication was observed using transient transfection of anti-TAR siRNAs in cell culture (Jacque et al., 2002). However, it is unknown whether a sustained induction of RNAi against TAR, such as would be required for therapeutic applications, can suppress HIV replication.
We have identified a novel RNAi target within TAR, and viral replication is efficiently blocked in cells constitutively expressing shRNAs targeting this sequence. However, by using high initial viral loads or including unprotected cells in the cultures, some persistent viral replication could be maintained, and under several such conditions, some viral populations rebounded in a stochastic manner. However, when these viral strains that “evaded” RNAi were sequenced and characterized, not one contained mutations in the RNAi-targeted region of TAR or was predicted to significantly alter RNA secondary structure in a way that would make it less susceptible to RNAi, indicating that resistance to RNAi was not acquired via previously described mechanisms. Instead, HIV accumulated promoter mutations and sequence duplications that appeared to compensate for RNAi-mediated inhibition, rather than escape it outright, by upregulating viral gene transcription. This represents a previously uncharacterized mode of viral evolution in response to inhibition by RNAi.
Results
A Novel anti-TAR shRNA Conferring Long-term Suppression of HIV Replication
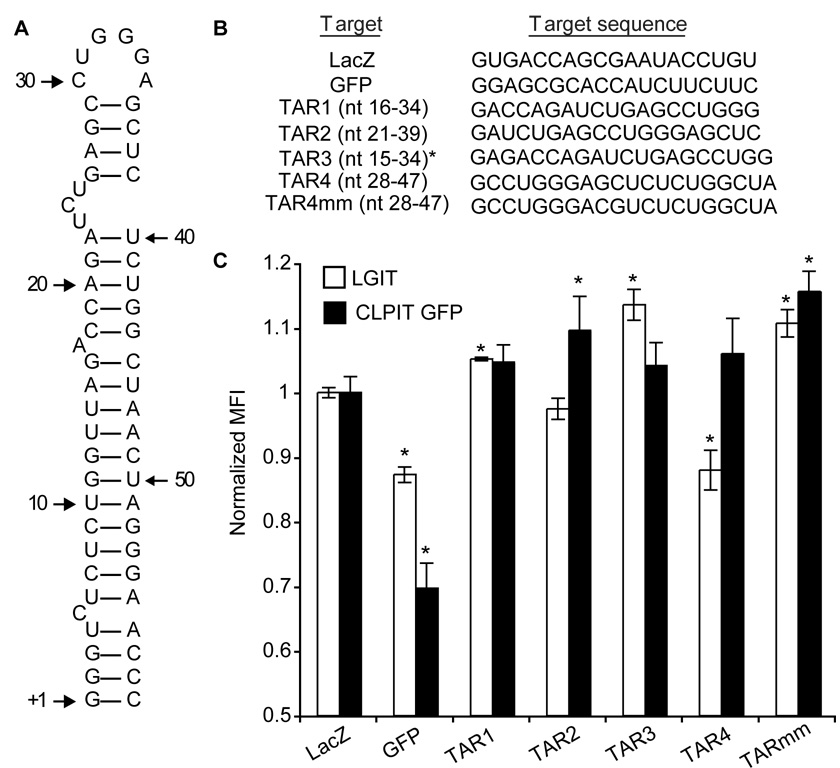
To identify sequences within TAR susceptible to RNAi-mediated inhibition, we first tested four shRNAs targeting three different regions within TAR for their ability to inhibit HIV gene expression (Figure 1A and 1B). TAR1 and TAR3 were based on a single siRNA reported to have moderate success in suppressing short-term viral replication (Jacque et al., 2002), and the others targeted new sequences within TAR (Supplementary Table 1). When shRNA-encoding plasmids were transiently transfected (>90% transfection efficiency) into HEK 293T cells expressing the green fluorescent protein (GFP) from the HIV LTR (LGIT) (such that each mRNA contains TAR hairpins at both the 5’ and 3’ ends), TAR4 shRNA reduced GFP expression to an extent comparable to the potent anti-GFP control (Leirdal and Sioud, 2002) 3 days post-transfection (Figure 1C). By contrast, TAR4 shRNA did not inhibit GFP expression in HEK 293T cells expressing GFP from a murine retroviral vector (CLPIT GFP), indicating its specificity. GFP levels decreased between 1 and 4 days post-tranfection, however no additional knockdown was observed 5 days post-transfection (data not shown).
Figure 1. Identification of potent RNAi targets in the HIV TAR element.
(A) Secondary structure of HIV TAR RNA. (B) shRNA target sequences, with corresponding TAR nucleotide (nt) positions indicated in parentheses. *The initial G of TAR3 is not present in TAR. (C) Cells expressing GFP from either the HIV (LGIT) or murine retrovirus (CLPIT-GFP) LTR were transiently transfected with U6-shRNA expression plasmids (1.5 µg per 1 × 105 cells), and after 3 days mean fluorescence intensity (MFI) was quantified by flow cytometry (Supplementary Figure 1) and normalized to an empty vector-transfected control (LGIT Mock MFI = 83.4, CLPIT GFP Mock MFI = 40.9). Experiments were performed in biological triplicate and are representative of at least four independent experiments. Error bars indicate one standard deviation and (*) indicates a significantly different MFI as compared to the LacZ negative control in the same cell type (p<0.05).
It has been reported that TAR can be processed by Dicer and may act as a miRNA (Klase, 2007). To clarify the mechanism of gene knockdown, we also measured gene expression in the presence of a TAR4 shRNA with two central mismatches at nucleotides 9 and 10 (TAR4mm), which would be predicted to abrogate RNAi-mediated cleavage of a target (Yu et al., 2002), but not necessarily miRNA inhibition (Hutvagner and Zamore, 2002). The mismatched shRNA showed no significant knockdown in the LGIT cell line, which indicates that exact sequence identity is required for knockdown and supports a RNAi mechanism.
To test whether the TAR4 shRNA could suppress HIV replication, we constructed a stable SupT1 human T cell line that constitutively expresses this shRNA. These cells were infected with a “triple-deletion” strain of HIV pNL4-3, which contains deletions (in vpu, nef, and part of the U3 region) that do not impair replication in cell culture but enhance safety considerations (Deacon et al., 1995; Du et al., 1993). Unprotected SupT1 cells were also included in some cultures at several relative proportions to assess the impact that non-suppressive cells may have on viral replication and evolution. Eight replicates of each condition were used, as viral mutation and escape are stochastic processes. Cells were initially challenged at a ratio of infectious virus to cells (multiplicity of infection, MOI) of 0.015. This MOI was selected to induce robust viral replication in unprotected cells over an 8-day period, as higher MOIs rapidly killed cells and actually reduced the endpoint viral titer (Supplementary Figure 2). For long-term culturing, viral supernatant was transferred to fresh cell cultures (containing proportions of protected and unprotected cells matching the starting population) at 8-day intervals to compensate for the death of infected cells and the expansion of surviving cells. Viral titers were tracked using an indicator cell line (CEM GFP) that measures active viruses rather than the accumulation of viral proteins (Gervaix et al., 1997). Briefly, the indicator cells contain an integrated copy of the HIV LTR followed by GFP. In the presence of Tat, supplied by HIV infection, these cells express GFP and can be measured via flow cytometry to determine the infectious titer (Berthoux et al., 1999; Schumacher et al., 2008) (Supplementary Figure 3).
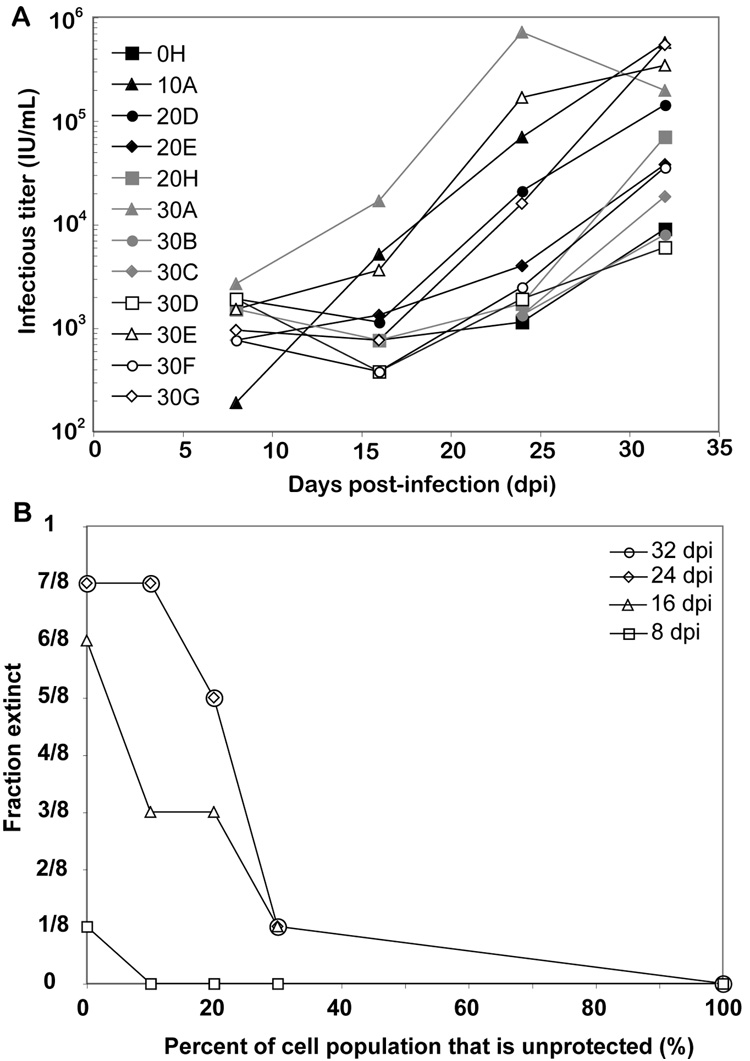
While viral replication in unprotected cells was robust, cultures containing protected cells showed considerably lower titers when first measured 8 days post-infection (dpi). This suppression of viral replication led some populations to become “extinct,” with undetectable active virus (data not shown). Despite this initial suppression, several cultures proceeded to rebound to high viral titers in the following 24 days (Figure 2A). The recovered viral populations were then cultured in the presence of purely protected cells for up to 18 days, allowing for the enrichment of adaptive mutations in these populations. The recovery of efficient viral replication was highly stochastic, such that cultures of identical initial cellular composition either eventually supported replication or extinguished the infection. However, the probability of recovering efficient replication was a strong function of the fraction of the cell population that was unprotected (Figure 2B). More specifically, in wells containing only protected cells, virus was extinguished in seven out of eight wells. However, consistent with our previous computational predictions (Leonard and Schaffer, 2005), less protected cultures were much more likely to support viral replication, and a relatively sharp viral replication “threshold” appeared as the percent of unprotected cells reached ~20%. Thus, even though evasion of RNAi is a stochastic process, key parameters (such as the fraction of the population that is unprotected) govern the likelihood that viral replication will recover.
Figure 2. Recovery of HIV replication in anti-TAR RNAi cells.
(A) TAR4 shRNA-expressing and unprotected SupT1 cells were mixed at a range of ratios denoted by the percentage of unprotected cells (0–30, 8 replicates - A thru H - of each ratio) and challenged with HIV at an MOI of 0.015. Every 8 days, supernatant was transferred to fresh cells, and infectious titers were calculated. Viral populations that went extinct are not shown. (B) The fraction of cultures in which the virus population went extinct, within each group of replicates having a given ratio of protected to unprotected cells, was determined at each time point.
HIV Evades anti-TAR RNAi by a Novel Compensatory Mechanism
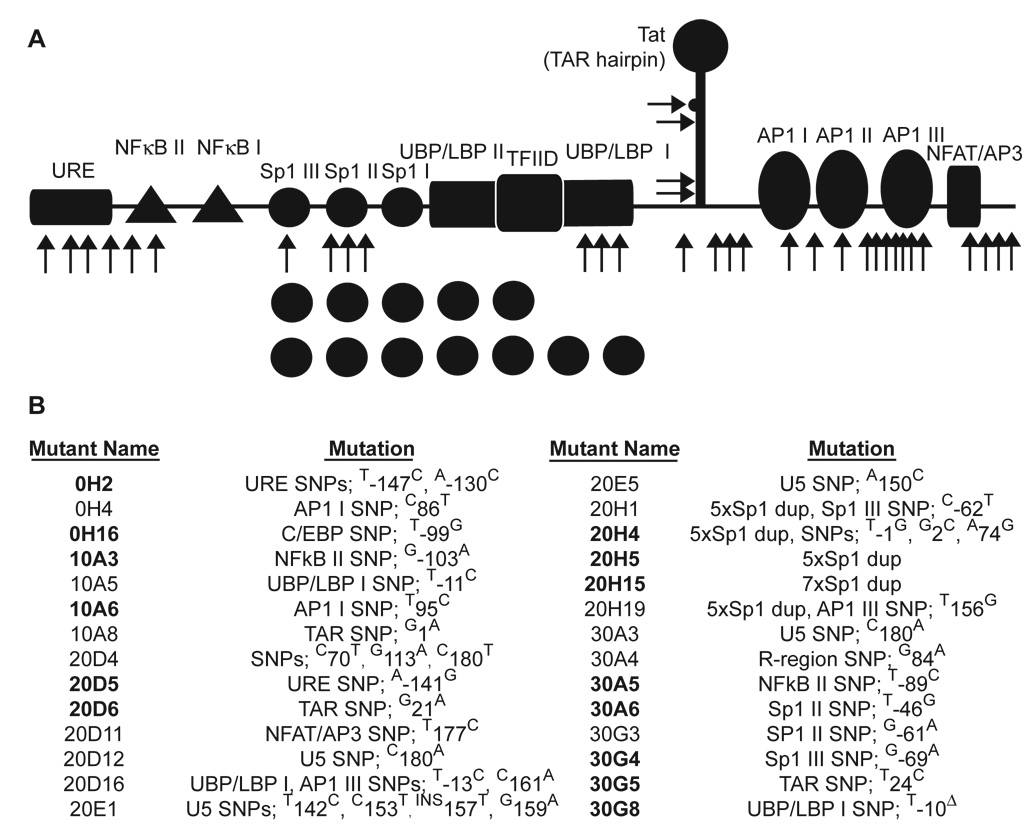
We next sought to determine whether viral populations had rebounded due to mutational adaptation. To date, viral escape from RNAi has been found to occur only by mutation in the targeted site, or in nearby regions that modulate RNA folding at the targeted sequence (Boden et al., 2003; Das et al., 2004; Gitlin et al., 2005; ter Brake et al., 2006; Westerhout et al., 2005). Intriguingly, sequence analysis of the LTRs of putative escape viral populations revealed no mutations in the RNAi target sequence within TAR that would be expected to alleviate RNAi-mediated suppression. Many mutations were instead present within or adjacent to transcription factor binding sites in the LTR, including NFκB, Sp1, UBP/LBP, AP-1, and NFAT (Figure 3). Although four separate point mutations were observed in TAR, they occurred outside of the RNAi target sequence. Two mutations (20D6 and 30G5) were adjacent to and within the 5’ side of the “bulge” responsible for Tat binding (nucleotides 23–25 in Figure 1A) (Roy et al., 1990), and two others (10A8 and 20H4) occurred at the base of the TAR hairpin. Only mutant 20D6 was predicted to change the secondary structure adjacent to the RNAi target sequence, according to the mFold algorithm (Zuker, 2003), but in a manner not likely to decrease accessibility of the target region to RISC, as was the case in a previously reported mechanism of HIV escape from RNAi (Westerhout et al., 2005) since the mutation in 20D6 creates a “window” of accessibility for target recognition (Ameres et al., 2007; Gredell et al., 2008). Importantly, no mutants were predicted to alter the structure and therefore accessibility of the target sequence itself (Supplementary Figure 5). Only one mutant expanded to dominate its viral population (culture 20H), and many viruses isolated from other cultures possessed the wild-type (WT) sequence in the region analyzed (Supplementary Table 2). Collectively, these data suggest that the virus did not directly escape anti-TAR RNAi but instead accumulated alterations in both positive and negative regulatory elements that potentially enhance viral replication in the presence of the RNAi inhibition. In addition, the sequencing results suggest that WT virus expansion in unprotected cells may have contributed to the recovery of viral replication.
Figure 3. Summary of potential escape mutants.
(A) Locations of LTR mutations in isolates from “escaped” wells are indicated by black arrows. (B) A summary of individual mutants, with bold text indicating mutants that were selected for further analysis, is shown. Mutant names indicate where the mutant arose (“20D5” = isolate from a culture with 20% unprotected cells, culture replicate D, isolate #5 from this culture). Δ indicates a deletion, “ins” an insertion, and “dup” a duplication. Individual mutations are numbered using +1 for the transcriptional start site. Complete mutant LTR sequences are available in Supplementary Figure 4. Frequencies of each mutant are listed in Supplementary Table 2.
Fourteen mutants were selected for further analysis, based on several criteria. First, because several regions of documented importance were mutated in multiple variants, these variants were selected for analysis. These included mutations in the NFκB elements (variants 10A3 and 30A5), Sp1 elements (20D5, 20H4, 20H5, 20H15, 30A6 and 30G4), and adjacent to the TATA box (30G8). In addition, a set of variants whose mutation occurred within the R region of the LTR were chosen (10A6, 20D6 and 30G5), given their proximity to the RNAi-targeted region. Mutants (0H2 and 0H16) were also chosen to represent isolates from the various culture compositions (0, 10, 20 and 30% unprotected). To prepare genetically uniform stocks of each mutant, we developed and utilized a viral genomic plasmid containing a single LTR, analogous to a system developed previously (Leonard et al., 1989), to simplify genetic manipulation of the LTR and viral production (Supplementary Figure 6).
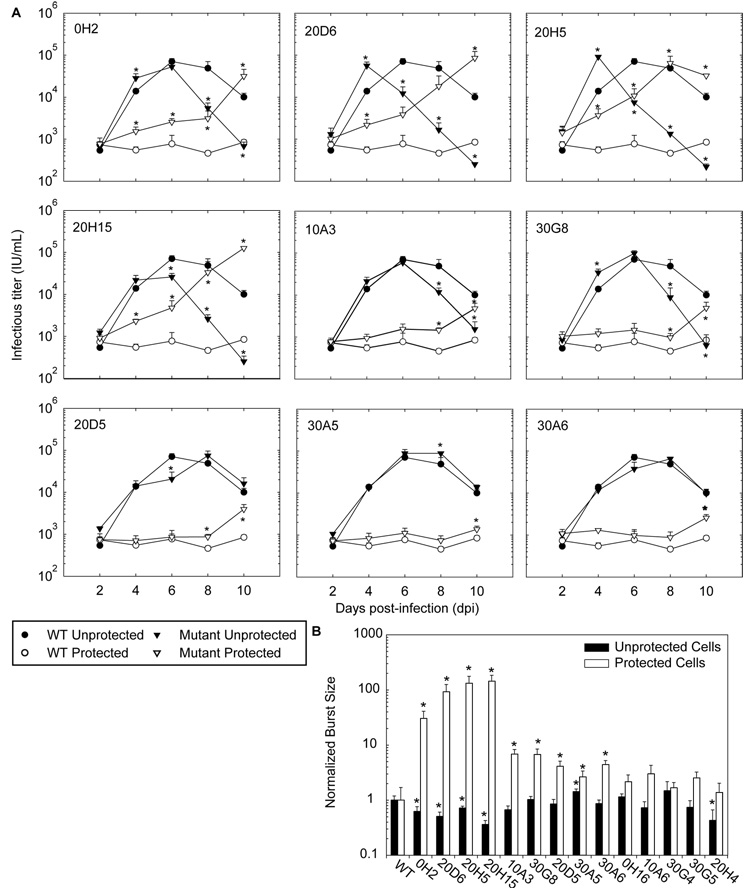
Using the resulting mutant viral stocks, homogeneous cultures of either protected or unprotected cells were infected at a MOI of 0.015. Over a 10-day period, nine of the fourteen mutants showed significantly enhanced replication in protected cells compared to WT virus (Figure 4A and Supplementary Figure 7). However, the replication of these mutants in protected cells was delayed compared to their expansion in unprotected cells, indicating that they are still susceptible to RNAi-mediated inhibition. Interestingly, six of the nine mutants with enhanced replication in protected cells (0H2, 20D6, 20H5, 20H15, 10A3 and 30G8) also exhibited faster than WT replication in unprotected cells. This resulted in an earlier peak in titer for three of these six mutants (relative to the WT virus peak) and a significantly accelerated decay in viral titer for all six mutants. This rapid decay can be explained by accelerated syncytia formation and cell death, as observed by light and fluorescence microscopy (Supplementary Figure 8). Since premature cell death may limit the number of progeny virions produced, it is likely that these mutants are less fit for replication in unprotected cells.
Figure 4. Enhanced replication of evading mutants.
(A) TAR4-protected or unprotected SupT1 cells were challenged with either WT virus or each potential evasion mutant at an MOI of 0.015. Mutants that did not show enhanced replication are included in Supplementary Figure 7. (B) Total viral yield (burst size) was calculated by integrating the titers from (A) and normalizing to WT virus (in identical cells). Experiments were performed in biological triplicate, error bars represent one standard deviation, and (*) indicates a significantly different titer (or burst size) as compared to WT virus replicating in the same cell type (p<0.05).
To quantitatively compare the ability of mutant and wild-type virus to produce progeny, we summed or integrated the infectious viral titers measured every 2 days over a 10-day period (the half life of infectious virus is ~8 hours (Perelson et al., 1996)), such that the result was a measure of viral “burst size” over the time course of the experiment (Figure 4B). The nine variants with enhanced replication kinetics also yielded significantly larger burst sizes in protected cells than did WT virus, indicating they are evading RNAi. Intriguingly, several mutants with the largest burst sizes in protected cells also had significantly decreased burst sizes in unprotected cells, compared to WT virus. These mutants thus apparently gained the ability to replicate in protected cells at the cost of reduced replication in unprotected cells, indicating that the acquired genetic changes can result in a fitness loss in some contexts.
Next, we tested whether the nine RNAi-evading mutants possessed altered transcriptional activity that may compensate for RNAi-mediated inhibition (Figure 5A). First, basal LTR-driven transcription was measured in unprotected cells by QPCR of GFP mRNA. All mutants except mutant 20D6 exhibited higher basal activities when measured as the RNA level. Since Tat-transactivated transcription is essential for viral replication, we also measured LTR activities in the presence of Tat. Mutants 0H2, 10A3, 20H5, 20H15, 30A6 and 30G8 had enhanced transcription in the presence of Tat compared to the WT LTR with Tat. Interestingly, mutant 20D6 appears to be virtually unresponsive to Tat. A luciferase assay confirmed these trends (Supplementary Figure 9). Collectively, these results suggest that HIV acquired the ability to replicate in RNAi-protected cells through indirect compensatory mutations that increase the basal and/or transactivated transcription rate, rather than by directly evading RISC-mediated downregulation of gene expression. This represents a novel mechanism of viral evasion of RNAi.
Figure 5. Transcriptional activity of mutant LTRs.
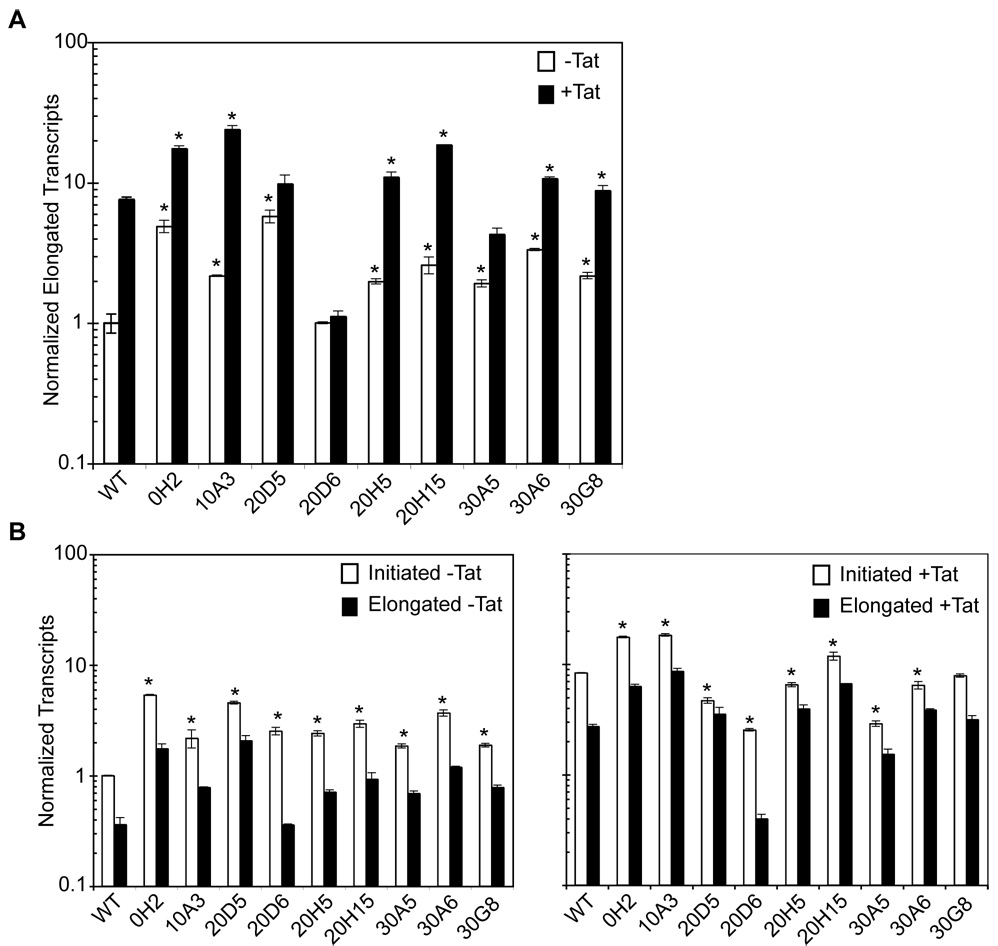
Tat-expressing or naïve SupT1 cells were transduced with vectors expressing GFP and luciferase from either WT or evading mutant HIV LTRs at MOIs of ~1.5. (A) Basal and transactivated transcription rates of each mutant were measured by QPCR of GFP mRNA using the ΔΔCT method (Livak and Schmittgen, 2001). All values were normalized for amplification efficiency. (B) The number of initiated and fully elongated transcripts were measured using QPCR as above. All values were normalized for amplification efficiency. The difference between these two values is considered the number of truncated transcripts (Williams et al., 2006). Experiments were performed in technical triplicate. Error bars represent one standard deviation and (*) indicates a statistically different value from the respective WT value (p<0.05).
Since many of these mutants appeared to be evading RNAi at the transcriptional level, we next measured the production of initiated versus fully elongated transcripts for each mutant using a previously developed method (Williams et al., 2006) (Figure 5B). All mutants produced more initiated transcripts than WT virus in the absence of Tat. In addition, mutants 0H2, 10A3 and 20H15 exhibited enhanced initiation in the presence of Tat. Both of these scenarios suggest that elongation has become more efficient under for these mutants in the absence and perhaps even the presence of Tat. In contrast, mutant 20D6 showed an increase in initiated transcription in the absence of Tat, while fully elongated transcription was not enhanced compared to WT in either situation. This implies that a large number of transcripts are truncated for mutant 20D6.
Combinatorial Therapies for Enhanced Antiviral Activity
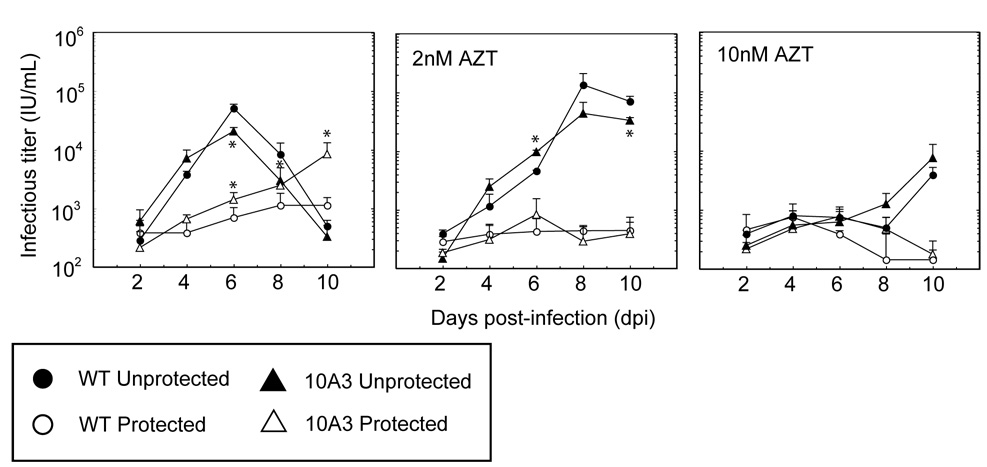
Since our mutants appear to evade RNAi by a general increase in gene expression, we investigated whether enhanced suppression of mutant viral replication could be achieved by using a combination of RNAi and a small molecule HIV inhibitor. One such combination strategy has demonstrated improved inhibition of both WT and drug-resistant HIV; however, in this study the siRNA and the small molecule antiviral targeted RT mRNA and protein, respectively (Huelsmann et al., 2006). We analyzed whether targeting different viral loci simultaneously, in a manner similar to HAART, may also enhance viral inhibition. In particular, we measured viral replication in TAR4-protected and unprotected SupT1 cells cultured with the nucleoside RT inhibitor (NRTI) zidovudine (AZT). Combinatorial inhibition enhanced the viral suppression exerted by either RNAi or the NRTI alone (Figure 6). When AZT was combined with TAR4 RNAi, complete suppression of WT viral replication was observed over 10 days, even at AZT concentrations that were unable to inhibit viral replication alone. Mutant 10A3, which was able to evade RNAi, was also able to replicate in unprotected cells in the presence of low concentrations AZT. However, when AZT was combined with RNAi, no viral replication was observed over 10 days. Similar trends were observed for mutants 0H2, 10A3, 20H5, and 20H15 at higher MOIs (data not shown).
Figure 6. Combinatorial inhibition of HIV with anti-TAR RNAi and antiviral drugs.
TAR4-protected or unprotected SupT1s were infected with WT or mutant 10A3 virus at an MOI of 0.015 in the presence of AZT. Titers were assayed every 2 days for 10 days. Experiments were performed in biological triplicate. Error bars represent one standard deviation, and (*) indicate statistically significant titer as compared to WT virus replicating in the same cell type (p<0.05).
Discussion
Here we describe a novel mechanism of HIV evolution when its replication is inhibited by RNAi. We identified a shRNA that targets a sequence in TAR that is highly conserved, even between HIV-1 subtypes, and that efficiently suppresses viral replication. However, mutants capable of evading RNAi emerged in long-term culture, particularly in mixtures of RNAi-protected and unprotected cells. Interestingly, no variants escaped the RNAi directly, but the virus instead upregulated its gene expression to compensate for this inhibition. Importantly, these results reveal that HIV can adaptively tune its gene regulation to enable viral evasion.
Existing HAART drugs interfere with viral replication via binding directly to target HIV proteins and inhibiting their function by either competing for catalytic sites or blocking conformational changes required for activity. Consequently, mutations in the targeted proteins may alter the physical interactions with antiviral drugs and thereby allow these proteins to function in the presence of the associated inhibitors. While viral resistance mutations may initially impair the function of these proteins, additional compensatory mutations in the targeted protein (or in other viral proteins) can restore overall viral fitness. For example, mutations in HIV Protease results in virus that is resistant to protease inhibitors, and compensatory mutations in Protease’s viral substrate, Gag, restores viral fitness (reviewed in (Clavel and Hance, 2004)). However, in all such cases of resistance to existing HIV therapies, the underlying mutations occur exclusively within the protein-coding sequences affected by the therapy. Likewise, in all cases of evolved resistance to RNAi described to date, viruses have acquired point mutations or deletions in the target sequence (Boden et al., 2003; Das et al., 2004; Gitlin et al., 2005; ter Brake et al., 2006; Wilson and Richardson, 2005; Wu et al., 2005) or have structurally arranged the target sequence to render it inaccessible to RISC (Westerhout et al., 2005). In each case, resistance was conferred by a direct alleviation of RNAi-mediated inhibition.
We describe a distinct mechanism of viral evolution in response to RNAi. No direct escape from RNAi occurred, presumably because TAR is essential for replication, and neither the primary RNA sequence nor secondary structure can be modified without severely compromising viral function (Selby et al., 1989). Instead, we identified variants harboring mutations within the HIV LTR promoter that confer the ability to replicate in the presence of the anti-TAR shRNA. However, all mutants continue to be partially inhibited by this RNAi. Several mutants (0H2, 10A3, 20D5, 20H5, and 20H15, 30A5, 30A6 and 30G8) acquired increased basal or Tat-transactivated transcription rates relative to the WT promoter, suggesting they persisted by overwhelming the RNAi machinery with viral transcripts. Thus, when RNAi is directed against a highly conserved viral target, such as TAR, the virus adapts by modulating genetic elements that regulate overall levels of viral gene expression. Interestingly, the concept that viruses can overwhelm endogenous pathways has some parallels in protein translation in adenovirus biology (Mathews and Shenk, 1991); however, to our knowledge, this type of viral evolution to evade a therapeutic inhibitor has not been previously described, and more broadly M. tuberculosis is the only pathogen reported to acquire drug resistance through a promoter mutation (Rinder et al., 1998).
An examination of the mutations that conferred enhanced viral replication suggests several possible mechanisms by which HIV may modulate its genetic regulation. One class of mutations occurred in sites that can either enhance or suppress expression. The HIV LTR contains two NFκB binding sites. In mutants 10A3 and 30A5, NFκB site II was mutated to sequences shown to impair binding of the repressive homodimer (Wang et al., 2003), which may tip the regulatory scales in the favor of transcription. The LTR also contains binding sites for Sp1, which again recruits both positive (p300) and negative (HDACs) regulators (Doetzlhofer et al., 1999; Suzuki et al., 2000). Mutants 20H5 and 20H15 contain tandem Sp1 site duplications, apparently acquired through sequential recombination events during viral replication. While p300-bound Sp1 is known to have a reduced affinity for DNA relative to Sp1 or HDAC-bound Sp1 (Suzuki et al., 2000), the cooperative nature of Sp1 binding to DNA (Mastrangelo et al., 1991) may again push the balance towards activation of transcription.
It should be noted that this type of duplication of Sp1 binding sites has been observed during the long-term passage of the triple-deletion HIV strain used in this study (Berkhout et al., 1999). We therefore sought to address the possibility that this mutation is a consequence of in vitro culturing of attenuated HIV. We confirmed that in the full-length HIV-1 strain NL4-3, the Sp1 duplication mutant (20H5) confers a significant replication advantage compared to WT virus in protected cells. Furthermore, the inclusion of this mutation in the full-length strain results in accelerated replication kinetics in unprotected cells and a lower total burst size over 10 days compared to the WT virus (Supplementary Figure 10). While the differences in replication are less pronounced in the full-length strain, the phenotypes are consistent with results using the triple-deletion strain. This further supports the interpretation that the observed Sp1 duplications are an adaptation that helps HIV to overcome both RNAi-mediated pressure and attenuation by gene deletion, since this adaptive advantage can be observed in both the wild type and attenuated HIV strains used in this study.
The observed increase in initiated transcription in a number of mutants in the absence of Tat, coupled with enhanced basal elongation rates, suggests that some mutants may “jump-start” viral gene expression by producing a large number of initiated transcripts. Notably, none of the observed mutations were predicted to alter the structure of the TAR hairpin in a substantial manner, suggesting that target accessibility to RISC should not be decreased. However, mutant 20D6 contains a mutation below the Tat-binding “bulge”, which may increase the size of the bulge, as predicted by Mfold. This mutant exhibited reduced initiation and basal transcription rates, and it remains unresponsive to Tat, suggesting that the larger bulge may affect Tat binding and transactivation. Additionally, the large difference in initiated vs. elongated transcripts produced by this mutant suggests that truncated transcripts may also serve as decoys that saturate TAR4-loaded RISC machinery. Finally, seemingly non-adaptive mutants (and apparently WT virus) may have propagated through coupled replication with adaptive variants, and it remains possible that some variants possessed adaptive mutations outside of the analyzed region.
We have shown that RNAi directed against TAR can inhibit HIV replication; however, viral evasion occurred via novel mechanisms involving compensatory upregulation of gene expression. It remains to be seen whether similar behavior exists in vivo, but our results combining inhibitory RNAi with antiviral drugs suggest a strategy for suppressing replication, and correspondingly, viral escape. In summary, evolution of viral gene regulation may represent a general mechanism by which viruses adapts to evolutionary pressure and escapes antiviral therapy.
Experimental Procedures
Cell culture
HEK 293T cells (ATCC) were cultured in IMDM (Mediatech) with 10% FBS (Gibco), and 100 U/mL penicillin + 100 µg/mL streptomycin (Gibco). SupT1 cells and CEM GFP cells, obtained via the NIH AIDS Research and Reference Reagent Program, were cultured in RPMI (Mediatech) containing 10% FBS, and antibiotics as above.
RNAi expression constructs
RNAi-inducing cassettes using the human U6 promoter from pTZU6+1 (Scherer et al., 2004) were constructed by PCR (Supplementary Table 1). Targets in GFP (Leirdal and Sioud, 2002) and LacZ (Qin et al., 2003) were previously described. U6-shRNA cassettes were inserted into pBS SK+ (Stratagene) for transient expression. The U6-TAR4 cassette was subcloned into the self-inactivating lentiviral vector pHIV CS (Miyoshi et al., 1998), and a CMV-NeoR selection cassette from pcDNA3.1 (Invitrogen) was inserted upstream of the 3' LTR to generate a vector for inducing sustained TAR4 shRNA expression.
Cell line generation
For stable GFP expression, the lentiviral vector LGIT and the murine retroviral vector CLPIT-GFP were packaged as previously described (Yu and Schaffer, 2006). HEK293 cells were infected with LGIT at a MOI of 1, and with with CLPIT-GFP at a MOI = 0.1 followed by selection in 1 µg/mL puromycin (Sigma-Aldrich). SupT1 cells were infected with TAR4 shRNA lentiviral vectors (produced as above) at an MOI of 0.1, and cells were selected with 500 µg/mL G418 sulfate (Invitrogen).
HIV stock preparation and titering
WT HIV stocks were generated from the hemigenomic plasmids p210-19 and p210-8 (NIH AIDS Program) as described (Gibbs et al., 1994). Individual mutant viruses were produced using a single-LTR packaging platform (psLTR HIV) (Supplementary Figure 6), which was generated by combining p210-19 and p210-8. Mutations were introduced into psLTR HIV by QuikChange PCR (Stratagene) (primer sequences available upon request), and each mutant LTR was sequenced and subcloned back into the parental plasmid to avoid unintended mutations. Full-length HIV was generated by combining hemigenomic plasmids p83-2 and p83-10 (NIH AIDS Program). To generate mutant and full-length HIV stocks, HEK 293Ts were transfected with psLTR templates and helper plasmids (pcDNA3 IVS VSV-G, pMDLg/pRRE, pRSV Rev, and pCLPIT-tat mCherry) to increase packaging efficiency. The resulting virus was amplified on SupT1s and titered on CEM GFP indicator cells (Gervaix et al., 1997). To titer virus, 100–300 µL of viral supernatant was used to infect 1×105 CEM GFP cells using 2 µg/mL polybrene (American Bioanalytical) and 0.1 µM saquinivir (NIH AIDS Program). After 3 days, CEM GFP cells were fixed in 2% paraformaldehyde, and transduction efficiency was assessed by flow cytometry (Supplementary Figure 3) and used to calculate viral titer.
HIV propagation experiments
4 × 105 TAR4-protected and unprotected SupT1s were mixed as indicated in 1.5 mL of medium and infected with HIV at a MOI of 0.015. At 2-day intervals, 700 µL of medium were removed for titering and replaced with fresh medium. For long-term experiments, every 8th day 250 µL of supernatant was transferred to a new culture of 4 × 105 cells (matching the initial composition of the cell population). Potentially resistant cultures were isolated at 32 dpi, enriched on RNAi-protected cells for up to 18 days, and titered on CEM GFP cells. Cellular genomic DNA was also harvested from these cells as previously described (Delassus et al., 1991; Meyerhans et al., 1989), and 5’ LTR sequences were recovered by PCR (primer sequences available upon request) and inserted in pBS SK+ for sequencing.
LTR transcription assays
A lentiviral vector (LLIG) was created by modifying LGIT to place a luciferase-IRES-GFP cassette under the transcriptional control of the HIV LTR. Mutants were produced as above and packaged as previously described (Weinberger et al., 2005). SupT1s were infected with LLIG variants, and Tat expression was induced by transduction with a lentiviral vector expressing Tat from a ubiquitin promoter. Cells were cultured for 3 weeks to allow transcription to reach steady-state, and transcriptional activity was measured by detection of GFP mRNA using QPCR. Values were normalized for LLIG infection efficiency was quantified by QPCR of viral stocks using GFP primers.
RNA extraction and quantification by QPCR
Total RNA from SupT1 cells was isolated using Trizol (Invitrogen) and transcripts were quantified using the QuantiTect SYBR Green RT-PCR kit (Qiagen) on the Bio-Rad iCycler with the ΔΔCT method (Livak and Schmittgen, 2001). Initiated transcripts were detected with TAR primers HIVTAR5 (5′-GTTAGACCAGATCTGAGCCT-3′) and HIVTAR3 (5′-GTGGGTTCCCTAGTTAGCCA-3′) (Williams et al., 2006). Elongated transcripts were detected with GFP primers GFP5 (5′- AGCAAAGACCCCAACGAGAA-3′) and GFP3 (5′-CGTCCATGCCGAGAGTGAT-3′) (Weinberger et al., 2005). For each sample, measurements were normalized by the corresponding levels of β-Actin mRNA, which were quantified with primers β-Actin5 (5′-ACCTGACTGACTACCTCATGAAGATCCTCACCGA-3′) and β-Actin3 (5′-GGAGCTGGAAGCAGCCGTGGCCATCTCTTGCTCGAA-3′) (Weinberger et al., 2005). Triplicate RT-QPCR measurements were performed for all samples for each primer set and melt curves were performed on the Bio-Rad iCycler for all samples to confirm the specificity of QPCR reaction.
Statistical Analysis
All statistical analysis was conducted using the Student’s t-test with a threshold p-value of 0.05.
Supplementary Material
Acknowledgements
We thank John Rossi (Beckman Research Institute of the City of Hope, Duarte, CA) for pTZU6+1 and Albert Keung, Siddharth Dey and Randolph Ashton (Department of Chemical Engineering, U.C. Berkeley) for their assistance in data collection. This work was supported by NIH R01 GM73058 and NDSEG and a NSF Graduate Fellowship (to PSS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Berkhout B, Verhoef K, van Wamel JL, Back NK. Genetic instability of live, attenuated human immunodeficiency virus type 1 vaccine strains. J Virol. 1999;73:1138–1145. doi: 10.1128/jvi.73.2.1138-1145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoux L, Pechoux C, Darlix JL. Multiple effects of an anti-human immunodeficiency virus nucleocapsid inhibitor on virus morphology and replication. J Virol. 1999;73:10000–10009. doi: 10.1128/jvi.73.12.10000-10009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr A. Toxicity of antiretroviral therapy and implications for drug development. Nat Rev Drug Discov. 2003;2:624–634. doi: 10.1038/nrd1151. [DOI] [PubMed] [Google Scholar]
- Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, Soifer H, Gatignol A, Riggs A, Rossi JJ. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Tommasi S, Li M, Li H, Yanow S, Pfeifer GP, Rossi JJ. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol Ther. 2005;12:179–183. doi: 10.1016/j.ymthe.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Clavel F, Hance AJ. HIV drug resistance. The New England journal of medicine. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science (New York, NY. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- Delassus S, Cheynier R, Wain-Hobson S. Evolution of human immunodeficiency virus type 1 nef and long terminal repeat sequences over 4 years in vivo and in vitro. J Virol. 1991;65:225–231. doi: 10.1128/jvi.65.1.225-231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G, Wintersberger E, Seiser C. Histone deacetylase 1 can repress transcription by binding to Sp1. Molecular and cellular biology. 1999;19:5504–5511. doi: 10.1128/mcb.19.8.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du B, Wolf A, Lee S, Terwilliger E. Changes in the host range and growth potential of an HIV-1 clone are conferred by the vpu gene. Virology. 1993;195:260–264. doi: 10.1006/viro.1993.1370. [DOI] [PubMed] [Google Scholar]
- Feinberg MB, Baltimore D, Frankel AD. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci U S A. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervaix A, West D, Leoni LM, Richman DD, Wong-Staal F, Corbeil J. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc Natl Acad Sci U S A. 1997;94:4653–4658. doi: 10.1073/pnas.94.9.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JS, Regier DA, Desrosiers RC. Construction and in vitro properties of HIV-1 mutants with deletions in "nonessential" genes. AIDS research and human retroviruses. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Stone JK, Andino R. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J Virol. 2005;79:1027–1035. doi: 10.1128/JVI.79.2.1027-1035.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredell JA, Berger AK, Walton SP. Impact of Target mRNA Structure on siRNA Silencing Efficiency: A Large-Scale Study. Biotechnology and bioengineering. 2008 doi: 10.1002/bit.21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nature biotechnology. 2007;25:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsmann PM, Rauch P, Allers K, John MJ, Metzner KJ. Inhibition of drug-resistant HIV-1 by RNA interference. Antiviral research. 2006;69:1–8. doi: 10.1016/j.antiviral.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science (New York, NY. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Imai K, Okamoto T. Transcriptional repression of human immunodeficiency virus type 1 by AP-4. The Journal of biological chemistry. 2006;281:12495–12505. doi: 10.1074/jbc.M511773200. [DOI] [PubMed] [Google Scholar]
- Jacque JM, Triques K, Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Horikoshi M, Roeder RG. Repression of HIV-1 transcription by a cellular protein. Science (New York, NY. 1991;251:1476–1479. doi: 10.1126/science.2006421. [DOI] [PubMed] [Google Scholar]
- Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Molecular Biology. 2007;8 doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Dykxhoorn DM, Kumar P, Ranjbar S, Song E, Maliszewski LE, Francois-Bongarcon V, Goldfeld A, Swamy NM, Lieberman J, et al. Lentiviral delivery of short hairpin RNAs protects CD4 T cells from multiple clades and primary isolates of HIV. Blood. 2005;106:818–826. doi: 10.1182/blood-2004-10-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leirdal M, Sioud M. Gene silencing in mammalian cells by preformed small RNA duplexes. Biochemical and biophysical research communications. 2002;295:744–748. doi: 10.1016/s0006-291x(02)00736-2. [DOI] [PubMed] [Google Scholar]
- Leonard J, Parrott C, Buckler-White AJ, Turner W, Ross EK, Martin MA, Rabson AB. The NF-kappa B binding sites in the human immunodeficiency virus type 1 long terminal repeat are not required for virus infectivity. J Virol. 1989;63:4919–4924. doi: 10.1128/jvi.63.11.4919-4924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JN, Schaffer DV. Computational design of antiviral RNA interference strategies that resist human immunodeficiency virus escape. J Virol. 2005;79:1645–1654. doi: 10.1128/JVI.79.3.1645-1654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li H, Rossi JJ. RNAi in combination with a ribozyme and TAR decoy for treatment of HIV infection in hematopoietic cell gene therapy. Annals of the New York Academy of Sciences. 2006;1082:172–179. doi: 10.1196/annals.1348.006. [DOI] [PubMed] [Google Scholar]
- Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J, Akkina R, Rossi JJ. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Margolis DM, Somasundaran M, Green MR. Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J Virol. 1994;68:905–910. doi: 10.1128/jvi.68.2.905-910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo IA, Courey AJ, Wall JS, Jackson SP, Hough PV. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci U S A. 1991;88:5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Shenk T. Adenovirus virus-associated RNA and translation control. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- Meyerhans A, Cheynier R, Albert J, Seth M, Kwok S, Sninsky J, Morfeldt-Manson L, Asjo B, Wain-Hobson S. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell. 1989;58:901–910. doi: 10.1016/0092-8674(89)90942-2. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science (New York, NY. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Posada D, Crandall KA, Holmes EC. The causes and consequences of HIV evolution. Nat Rev Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Anderson EM, Vermeulen A, Fedorov Y, Robinson K, Leake D, Karpilow J, Marshall WS, Khvorova A. Induction of the interferon response by siRNA is cell type-and duplex length-dependent. RNA (New York, NY. 2006;12:988–993. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinder H, Thomschke A, Rusch-Gerdes S, Bretzel G, Feldmann K, Rifai M, Loscher T. Significance of ahpC promoter mutations for the prediction of isoniazid resistance in Mycobacterium tuberculosis. Eur J Clin Microbiol Infect Dis. 1998;17:508–511. doi: 10.1007/BF01691135. [DOI] [PubMed] [Google Scholar]
- Rossi JJ, June CH, Kohn DB. Genetic therapies against HIV. Nature biotechnology. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Delling U, Chen CH, Rosen CA, Sonenberg N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes & development. 1990;4:1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- Scherer LJ, Yildiz Y, Kim J, Cagnon L, Heale B, Rossi JJ. Rapid assessment of anti-HIV siRNA efficacy using PCR-derived Pol III shRNA cassettes. Mol Ther. 2004;10:597–603. doi: 10.1016/j.ymthe.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Schumacher AJ, Hache G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby MJ, Bain ES, Luciw PA, Peterlin BM. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes & development. 1989;3:547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- Sodroski J, Goh WC, Rosen C, Dayton A, Terwilliger E, Haseltine W. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature. 1986;321:412–417. doi: 10.1038/321412a0. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kimura A, Nagai R, Horikoshi M. Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells. 2000;5:29–41. doi: 10.1046/j.1365-2443.2000.00302.x. [DOI] [PubMed] [Google Scholar]
- ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- von Eije KJ, ter Brake O, Berkhout B. Human immunodeficiency virus type 1 escape is restricted when conserved genome sequences are targeted by RNA interference. J Virol. 2008;82:2895–2903. doi: 10.1128/JVI.02035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JK, Li TX, Bai YF, Lu ZH. Evaluating the binding affinities of NF-kappaB p50 homodimer to the wild-type and single-nucleotide mutant Ig-kappaB sites by the unimolecular dsDNA microarray. Analytical biochemistry. 2003;316:192–201. doi: 10.1016/s0003-2697(03)00049-6. [DOI] [PubMed] [Google Scholar]
- Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. The EMBO journal. 2006;25:139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JA, Richardson CD. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J Virol. 2005;79:7050–7058. doi: 10.1128/JVI.79.11.7050-7058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HL, Huang LR, Huang CC, Lai HL, Liu CJ, Huang YT, Hsu YW, Lu CY, Chen DS, Chen PJ. RNA interference-mediated control of hepatitis B virus and emergence of resistant mutant. Gastroenterology. 2005;128:708–716. doi: 10.1053/j.gastro.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari K, Miyagishi M, Taira K. Effects on RNAi of the tight structure, sequence and position of the targeted region. Nucleic Acids Res. 2004;32:691–699. doi: 10.1093/nar/gkh221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Schaffer DV. Selection of novel vesicular stomatitis virus glycoprotein variants from a peptide insertion library for enhanced purification of retroviral and lentiviral vectors. J Virol. 2006;80:3285–3292. doi: 10.1128/JVI.80.7.3285-3292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. The EMBO journal. 1998;17:3681–3691. doi: 10.1093/emboj/17.13.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.