Communication between microtubules and actin/myosin networks determine epithelial cell extrusion polarity.
Abstract
To preserve epithelial barrier function, dying cells are squeezed out of an epithelium by “apoptotic cell extrusion.” Specifically, a cell destined for apoptosis signals its live neighboring epithelial cells to form and contract a ring of actin and myosin II that squeezes the dying cell out of the epithelial sheet. Although most apoptotic cells extrude apically, we find that some exit basally. Localization of actin and myosin IIA contraction dictates the extrusion direction: basal extrusion requires circumferential contraction of neighboring cells at their apices, whereas apical extrusion also requires downward contraction along the basolateral surfaces. To activate actin/myosin basolaterally, microtubules in neighboring cells reorient and target p115 RhoGEF to this site. Preventing microtubule reorientation restricts contraction to the apex, driving extrusion basally. Extrusion polarity has important implications for tumors where apoptosis is blocked but extrusion is not, as basal extrusion could enable these cells to initiate metastasis.
Introduction
To preserve epithelial barrier function during cell turnover, dying cells are shoved out of the epithelium by a process we have termed “apoptotic cell extrusion” (Rosenblatt et al., 2001). Here, the cell destined for apoptosis signals its live neighboring epithelial cells to form a ring of actin and myosin II that contracts to squeeze the dying cell out of the epithelial sheet. This contraction not only ejects the dying cell out of the layer but also closes any gaps that may have resulted from the dying cell's exit.
Importantly, cells can be extruded either apically into the lumen or basally back into the tissue the epithelia encases. Previously, we only detected apical extrusion of apoptotic cells from the epithelia encasing vertebrate embryos and adult organs. However, we find that basal extrusion occurs occasionally in vertebrates and commonly in Drosophila embryonic epithelia (also see Gibson and Perrimon, 2005; Vidal et al., 2006; Ninov et al., 2007). The direction a cell is extruded is important for its fate, as corpses extruded basally must be engulfed, whereas those extruded apically may be expelled or trapped in the lumen. Extrusion direction is likely more important in cases where cells are blocked from death but still extrude. Apical extrusion would still eliminate these cells, whereas basal extrusion could allow live cells to be retained in the tissue to potentially migrate elsewhere. Because this decision is critical for the extruded cell's fate, we sought to identify the molecular mechanism that determines the direction a cell will extrude.
Although our previous work had established the fact that apoptotic cell extrusion requires Rho-mediated contraction of an intercellular actin/myosin II ring in the cells surrounding the dying cell (Rosenblatt et al., 2001), we did not examine how actin/myosin contraction is organized to squeeze the dying cell out apically into the lumen. Further, we did not notice the minor populations of apoptotic cells that extruded basally in tissue culture monolayers. Here, we analyze how actin and myosin contraction is regulated differently when cells extrude apically versus basally using cultured epithelial monolayers from canine kidney, MDCK, or human bronchia (16-HBE-14o), as well as the epidermis of live zebrafish. We find that apical extrusion requires reorientation of microtubules and associated p115 RhoGEF basolaterally, which activates Rho to contract actin/myosin basally and squeeze the dying cell out.
Results and discussion
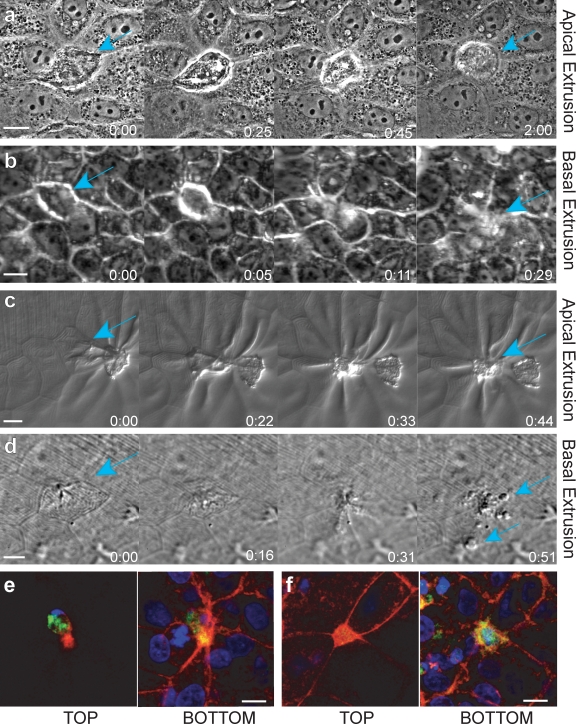
Using MDCK and 16-HBE-14o epithelial lines treated with UV254 to induce apoptosis, we found that although most apoptotic cells extrude apically, a small fraction extrudes basally. The direction of extrusion is apparent in the stills from phase videos (Fig. 1 and associated Videos 1–3). During apical extrusion, the cell body is pushed out of the monolayer into the medium (Fig. 1 a and Video 1), whereas during basal extrusion, the cell becomes trapped between the monolayer and the cover glass and migrates underneath the monolayer (Fig. 1 b and Videos 2 and 3). Because epithelial monolayers in culture may have artificially strong adhesion to glass, we wanted to assess if similar ratios of apical versus basal extrusion also occurred in vivo. We have found that zebrafish larval epidermises provide an excellent model system to follow extrusion live, which can also be manipulated with inhibitors. G-418 treatment of 4-d-old zebrafish larvae induces dying cells that predominantly extrude apically from the epidermis (Fig. 1 c and Video 4). Apically extruded cells exit into the fish water, whereas basally extruded cells become trapped beneath the epidermis (Fig. 1 d and Video 5).
Figure 1.
Hallmarks of apical and basal apoptotic cell extrusion. Stills from phase videos of apoptotic cells extruding apically (a and c) and basally (b and d) in an MDCK cell culture epithelium (a and b) and from a zebrafish larval epidermis (c and d; teal arrows denote extruding cells). Time is given in hours and minutes. (e and f) An apoptotic cell extruding apically (e) and basally (f), immunostained for active caspase-3 (green), actin (red), and DNA (blue). Note that caspase and DNA are in the top z plane and a thick actin ring is in the bottom plane when a cell is extruded apically (e), whereas the actin ring is in the top plane and caspase and DNA are in bottom plane when extruding basally (f). Bars, 10 µm.
We visually scored apical versus basal extrusion in UV-treated, fixed monolayers (referred to as “extruding monolayers”) and zebrafish epidermises by assessing the location of the actin-extruding ring compared with the extruded body, marked by compacted DNA and activated caspase-3 staining (see Fig. 1, e and f for examples of apical and basal extrusion in MDCKs; and see Videos 6 and 7 and Fig. S2 for examples in zebrafish). During apical extrusion, the actin ring is thicker and extends to the base of the cell, whereas active caspase-3 and DNA (marking the extruded cell) are apical to the ring. In contrast, during basal extrusion, the actin ring is apical and similar in intensity to the cortical actin at adherens junctions in the live cells, and the caspase-3 and DNA are basal to the contracted apical ring. Using these hallmarks for examining the polarity of extrusion, we found that zebrafish and tissue culture epithelia predominantly extrude apoptotic cells apically. 89 ± 2.3% (SEM) from >500 extruding cells (n = 4 experiments) from zebrafish epidermises, 80 ± 6% (SEM) of 1,250 extruding cells (n = 5 experiments) from MDCK monolayers, and 79 ± 9% (SEM) of 700 extruding cells (n = 7 experiments) from 16-HBE-14o monolayers extruded apoptotic cells apically; the remainder extruded basally.
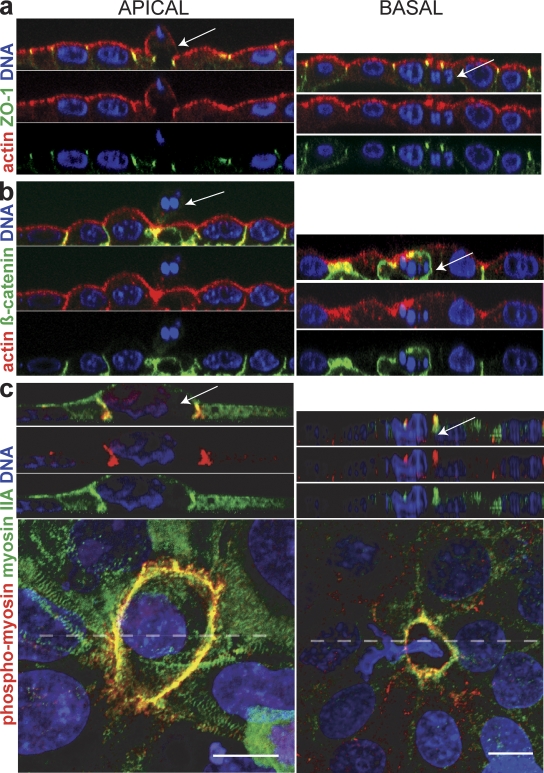
What determines whether cells are extruded apically or basally? Two models for differentially regulating the direction are: (1) the intrinsic polarity of neighboring cells may be altered or lost, positioning cortical actin differentially; or (2) actin and myosin may be independently localized apically or basally. To examine if polarity was altered when cells extruded apically versus basally, we immunostained extruding monolayers for the apical tight junction protein, ZO-1, and the basolateral adherens junction protein, β-catenin (Yu et al., 2005). ZO-1 remained apical and β-catenin basal during either apical or basal extrusion (Fig. 2, a and b, respectively). The preserved localization of polarized adhesion markers suggests that epithelial polarity is not markedly altered when cells extrude basally versus apically.
Figure 2.
Localization of active myosin IIA but not intrinsic polarity markers are altered when cells are extruded apically or basally. (a and b) Cross sections of tight junction protein Z0-1 (a), which remains apical, and β-catenin (b), which remains basal. (c) Myosin IIA and phospho-myosin II are basolateral during apical extrusion and apical during basal extrusion. Cross sections are taken from the broken line in pictures of 3D confocal reconstructions of MDCK cells. Arrows point to dying, extruded cells in each case. Bars, 10 µm.
To examine where actin/myosin contraction occurred, we immunostained extruding monolayers with a phospho-myosin II antibody that recognizes only actively contracting myosin II (Matsumura et al., 1998). We find that myosin IIA contraction is active along the basolateral interface between the apoptotic cell and its neighboring cells during apical extrusion but is restricted to the apical interface during basal extrusion (Figs. 2 c and S1, control). Localization of phospho-myosin II and videos of extrusion suggest that all extrusion events require coordinated, circumferential contraction of the cortical actin and myosin II at the cell–cell contacts that interface with the dying cell. The difference in whether extrusion occurs apically rather than basally appears to be linked to whether basolateral contraction also occurs at this ring.
If extrusion direction corresponds to where myosin II assembles and contracts, what controls its localization? Because microtubules have been found to activate or inhibit myosin II contraction in cytokinesis and other contractile processes (Wittmann and Waterman-Storer, 2001; Matsumura, 2005), we examined whether microtubules regulate myosin II contraction during extrusion. Live 3D videos of mApple-actin and GFP-tubulin show that microtubules dynamically reorient toward the actin-extruding ring early and throughout extrusion process, but retract once extrusion is complete during both apical and basal extrusion (see Videos 8 and 9, respectively). Because microtubules grow dynamically toward the ring, we reasoned that the plus ends of microtubules reorient toward the ring and activate myosin II contraction. To confirm that microtubule plus ends abut where actin and myosin contract during both apical and basal extrusion, we immunostained fixed, extruding monolayers for EB1, a plus-end microtubule-binding protein (Carvalho et al., 2003), myosin II, and DNA. Many EB1 spots localized to the basolateral surface during apical extrusion (Fig. 3 A, 3–6 µm from base), whereas far fewer EB1 spots localize to the apex during basal extrusion (Fig. 3 B, ≥6 µm from the base). Quantification confirmed that more EB1 spots localized to the basolateral surface (0–4 µm from base) than the apical surface (4–8 µm from base) of the actin ring during apical extrusion compared with basal extrusion or to live cell–cell contacts, labeled “junction” (Fig. 3 C). These experiments suggest that microtubules in the cells surrounding an apoptotic cell dynamically reorient toward the basolateral surface that contacts the dying cell during apical extrusion.
Figure 3.
The growing ends of microtubules point toward the actin/myosin ring during extrusion. (a and b) EB1 (green), a plus-end microtubule-binding protein, colocalizes with the myosin II (red) ring during apical (a) and basal (b) extrusion. Arrows point to sites where EB1 abuts the myosin II ring. Note that most EB1 is from 0–4 µm from the base during apical extrusion, whereas during basal extrusion, only limited amounts of EB1 are seen at the apex (6 µm from the base). Bars, 10 µm. (c) Quantification of EB1 localization (>170 spots for each) from 12 extrusions each of apically extruding cells, basally extruding cells, and live junctions. Error bars indicate SEM and P < 0.0001, comparing spots from apical extrusion to basal extrusion.
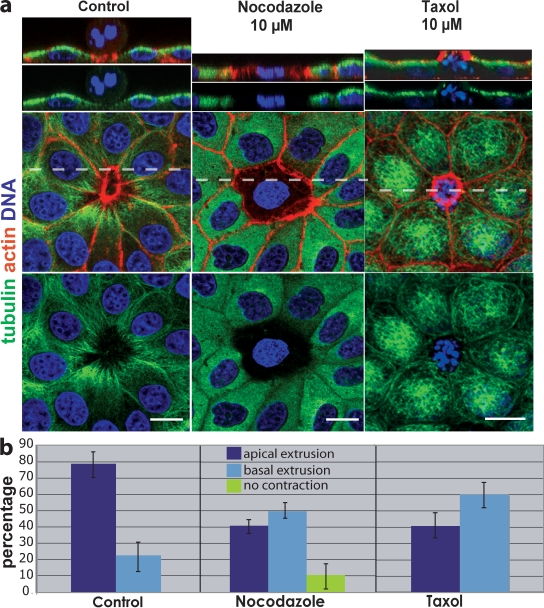
To test if microtubule reorientation toward the dying cell is required for basolateral contraction during apical extrusion, we used inhibitors to disrupt microtubules and analyzed fixed, immunostained extruding monolayers. Destabilizing microtubules with nocodazole reduced the number of apoptotic cells that extruded (no contraction) and increased the percentage that extruded basally (Fig. 4, a and b). To block the reorientation of microtubules toward the basal surface during apical extrusion, we treated extruding monolayers with the microtubule-stabilizing drug taxol. In contrast to control-treated cells, taxol treatment blocked microtubule reorientation toward the base of the dying cell (Fig. 4 a). Additionally, taxol treatment of monolayers shifted the direction of extrusion from predominantly apical to basal (Fig. 4 b). We found that disruption of microtubule dynamics shifted the balance of extrusions from apical to basal even more dramatically in zebrafish larval epidermises (from 89 ± 2.3% [SEM] to 27 ± 1.3% [SEM] in control vs. taxol treatments, from >900 extruding cells over four separate experiments, P < 0.0001). The drug treatments suggest that actin and myosin II contract at sites where microtubules contact the cortex. When microtubules are frozen at the apex, actin/myosin II contract at the apex and drive extrusion basally, but if microtubules are allowed to reorient toward the base, contraction occurs along the basolateral surface, driving extrusion apically.
Figure 4.
Disruption of microtubules alters the direction a cell extrudes. (a) In control (DMSO-treated) MDCK monolayers, microtubules dip down toward the basolateral surface of the actin ring, whereas nocodazole either blocks contraction of the ring (as shown) or drives extrusion basally. Stabilizing microtubules with taxol freezes microtubules at the apex compared with control and drives extrusion basally. Broken lines indicate the site of cross sections from 3D confocal reconstructions above. Bars, 10 µm. (b) Quantification of drug-treatments from six experiments, n = 1,350 extruding cells per treatment. P-values for apical and basal extrusions, respectively, were 0.0011 and 0.0066 for nocodazole and 0.0023 and 0.0023 for taxol. Error bars indicate SEM.
To determine if basolateral microtubule reorientation localizes myosin II or its activators to this surface, we immunostained extruding monolayers treated with nocodazole or taxol for myosin II and phospho-myosin II. Both myosin II and phospho-myosin II localized to the extruding ring in control-treated monolayers but were substantially reduced in the presence of nocodazole, and restricted to the apex in the presence of taxol (Fig. S1). Disruption of both myosin II localization and activation with microtubule inhibitors suggests that microtubules promote both assembly and activation of the actin/myosin ring.
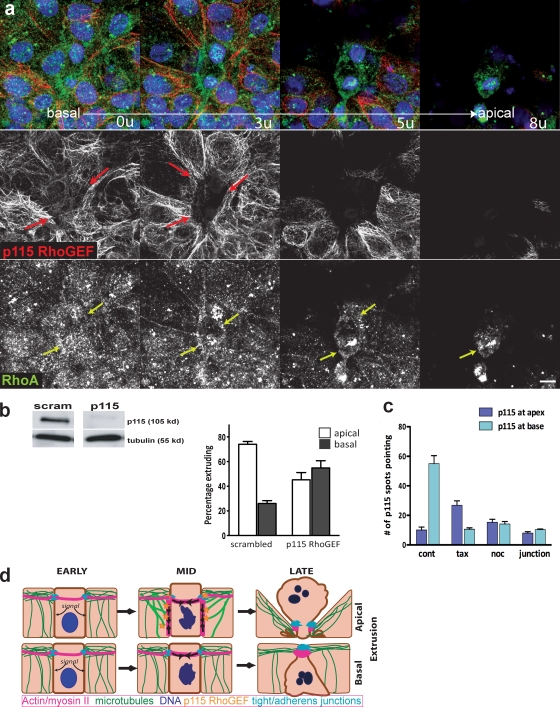
Because Rho activation in neighboring cells is required for extrusion contraction (Rosenblatt et al., 2001) and because microtubules can regulate several RhoGEFs (Krendel et al., 2002; Rogers et al., 2004), we hypothesized that microtubules could regulate extrusion direction by locally activating Rho. Total RhoA protein localized throughout the live–dead cell interface, although myosin II contraction occurred only at the bottom of the actin/myosin ring during apical extrusion (Fig. 5 a, bottom), suggesting that Rho activation is restricted to the basolateral surface. Because the Drosophila RhoGEF2 associates with microtubule plus ends to drive Rho-mediated actin/myosin contraction during gastrulation (Rogers et al., 2004), we investigated whether members of the regulator of G protein signaling (RGS) family of RhoGEFs, mammalian homologues of RhoGEF2 (Fukuhara et al., 2001), were required for apical extrusion. We treated 16-HBE-14o monolayers with siRNAs directed against PDZ RhoGEF, p115 RhoGEF, and leukemia-associated RhoGEF (LARG), and assayed the direction of extrusion in the resulting monolayers. Knockdown of PDZ and LARG RhoGEFs had no obvious effect on extrusion direction (Fig. S3, a and b). However, knockdown of p115 RhoGEF dramatically reduced the percentage of apically extruding cells, whereas scrambled p115 RhoGEF siRNA had no effect on extrusion direction (Fig. 5 b). The percentage of cells that shifted from predominantly apical to basal was similar to that seen with taxol treatment, which is consistent with p115 RhoGEF and microtubules acting in the same pathway.
Figure 5.
p115 RhoGEF is required for apical extrusion. (a) p115 RhoGEF (red; separate middle panel) form filaments that point toward the basolateral surface of the extruding ring during apical extrusion, whereas RhoA (green; bottom panel) is present through the apical and basal regions of the ring. Corresponding colored arrows indicate sites of interaction with the actin/myosin ring. Bar, 10 µm. (b) siRNA-mediated knockdown of p115 RhoGEF (above, lanes from one immunoblot) disrupts apical extrusion, whereas scrambled siRNA does not. The graph shows the percentage of apical versus basal extrusion from >1,000 cells from four siRNAs experiments, where P < 0.0001. (c) p115 RhoGEF localizes to the base of the extruding ring during apical extrusion, whereas it does not when microtubules are disrupted with nocodazole or taxol, or in control live junctions, where P-values are <0.0001 for control versus each treatment; n = 10 extrusions per treatment. (d) Model for how microtubules might control extrusion direction. An early apoptotic cell sends signals to its live neighboring epithelial cells, which react by reorienting microtubules toward the basolateral surfaces of the cells that contact the dying cell. p115 RhoGEF, by association with microtubules, assembles and activates Rho-mediated actin and myosin II contraction along the basolateral surface, extruding the cell apically (top). Basal extrusion occurs if the microtubules do not reorient or if p115 RhoGEF is absent (bottom).
Moreover, p115 RhoGEF, unlike RhoA, localizes only to sites of active myosin II contraction during both apical and basal extrusion. p115 RhoGEF appears in fibers that point to the base of the extruding ring during apical extrusion (Fig. 5 a, top; and Fig. 5 c). Unlike RhoGEF2 that localizes to microtubule tips (Rogers et al., 2004), we found that p115 RhoGEF colocalizes along microtubules, suggesting that it either associates with microtubules or with a microtubule-associated matrix (Fig. S3 c). To test if p115 RhoGEF localization was dependent on microtubules, we quantified the number of p115 RhoGEF fibers in control-extruding monolayers compared with nocodazole or taxol-treated monolayers. Compared with control-treated monolayers, taxol treatment restricted p115 RhoGEF to the apex (Figs. 5 c and S3 d, for a representative picture), whereas nocodazole treatment eliminated p115 RhoGEF at the dying cell, similar to levels seen at live cell–cell junctions (Figs. 5 c and S3 e). These results suggest that microtubules localize p115 RhoGEF to activate basolateral contraction during apical extrusion. Although Rho exists throughout the ring, and microtubules and p115 span the extruding live cells, Rho is activated only at sites where it intersects with p115 RhoGEF.
Based on the effects of disrupting microtubules and p115 RhoGEF on the direction of extrusion, we propose a model for how microtubules control the direction of extrusion (Fig. 5 d). When cells are extruded apically (Fig. 5 d, top), the cells surrounding a cell destined to die reorient their microtubules toward the basolateral surfaces that interface with the dying cell. Microtubules target p115 RhoGEF to the basolateral surface, where it activates Rho, present at the live–dead cell interface, to assemble and activate myosin IIA contraction. The extrusion ring contracts circumferentially around the dying cell as well as basolaterally to squeeze the apoptotic cell out apically. Failure of p115 RhoGEF to target to the basolateral surface, either by its knockdown or by disruption of microtubule reorientation, results in basal extrusion (Fig. 5 d, bottom). Here, actin and myosin IIA already present at apical adherens junctions could readily contract circumferentially around the dying cell. However, because basolateral actin/myosin contraction is absent, contraction is restricted to the apex, forcing the apoptotic cell out basally. During both types of extrusion, adherens and tight junctions are maintained above the sites of contraction, which preserves the barrier function of the epithelium throughout the extrusion process (as seen in Fig. 2, a and b).
Although we have found that microtubule reorientation is important for apical extrusion, microtubule disruption did not reverse the extrusion direction in 100% of cells, which suggests that other factors may also contribute to extrusion polarity. The extrusion polarity effects from taxol treatment were more dramatic in zebrafish than in culture, which suggests that this decision is regulated more tightly in vivo. Other factors affecting extrusion polarity may include matrix, intrinsic polarity, or cell height. A more adhesive surface may encourage apical extrusion, whereas basal extrusion might dominate in monolayers on a less adhesive substratum. Taller epithelia, such as in the gut, would likely show more dramatic effects in extrusion polarity than would flatter epithelia.
Several observations suggest that extrusion may not be limited to apoptotic cells, but could be a common mechanism for living cells to exit an epithelium without disrupting epithelial function. Apoptotic extrusion looks morphologically similar to developmental events where single cells exit the epithelium, such as neuroblast delamination in Drosophila melanogaster (Hartenstein et al., 1994). Here, cells exit the neuroepithelium and, instead of dying, divide asymmetrically before differentiating into neurons. We have previously found that blocking cell death with a caspase inhibitor does not block cell extrusion (Rosenblatt et al., 2001). Further, many high-grade epithelial tumors have mutations in apoptosis (Yang et al., 2003; Schulze-Bergkamen and Krammer, 2004; Meulmeester and Jochemsen, 2008) that would block apoptosis at steps we predict would not block extrusion.
In situations where cells can extrude without dying, the direction a cell extrudes is pivotal for its fate. In cancer, apical extrusion could act as a tumor suppressor, eliminating tumor cells through the lumen. Even in cases where tumors cells accumulate in a closed lumen, such as in breast ductal carcinoma in situ, they rarely invade and metastasize (Kopans et al., 2003). Basal extrusion, however, could provide an exit path for these tumor cells to initiate metastasis and migrate to other sites in the body. Evidence for basal extrusion promoting invasion exists in Drosophila larvae and pupae when cell death is blocked, as extrusion in these tissues occurs almost exclusively basally (Pagliarini and Xu, 2003; Gibson and Perrimon, 2005; Thompson et al., 2005; Vidal et al., 2006; Ninov et al., 2007; Hogan et al., 2009). Further work will test if basal extrusion of tumor cells could enable their invasion and metastasis through the matrix and into the bloodstream in vertebrates. Our results identifying a role for microtubules in regulating the direction of extrusion in zebrafish will allow us to follow the fate of basally extruded live cells in a vertebrate model where single cells can be readily imaged and tracked (Redd et al., 2006).
Materials and methods
Cell culture
MDCK II cells (gift from K. Matlin, University of Chicago, Chicago, IL) were cultured in Dulbecco's minimum essential medium (DMEM) high glucose with 5% FBS and 100 µg/ml penicillin/streptomycin (all from Invitrogen) at 5% CO2, 37°C. 16-HBE-14o (provided by D. Gruenert [California Pacific Medical Center, San Francisco, CA] and J. Porter [University College London, London, England, UK]) were cultured in MEM containing Earle's salts, 1 g/liter glucose, 2.2 g/liter NaHCO3, l-glutamine, 10% FBS, and 100 µg/ml penicillin-streptomycin (Invitrogen) in a flask coated with human fibronectin type I (BD), bovine collagen I (Purecol; Inamed biomaterials), and BSA (Invitrogen) at 5% CO2, 37°C.
Transfections
siRNAs were synthesized at the University of Utah oligo and peptide synthesis core to the following sequences: p115 RhoGEF (5′-GCAGCUCUGAGAACGGCAA-3′), LARG RhoGEF (5′-GAAACUCGUCGCAUCUUCC-3′), PDZ RhoGEF (5′-ACUGAAGUCUCGGCCAGCU-3′), and scrambled control siRNA (5′-UUCUCCGAACGUGUCACGU-3′). Antisense and sense duplexes LARG RhoGEF and P115 RhoGEF were annealed in 100 mM potassium acetate, 30 mM Hepes-KOH, 2 mM magnesium acetate, pH 7.4, and PDZ RhoGEF duplexes in 50 mM Tris, pH 8.0, and 100 mM NaCl at 90°C for 1 min and 37°C for 1 h. 50 pMol (LARG and P115) or 30 pMol (PDZ) siRNAs were transfected by nucleofection using the solution V-program G016 (Amaxa) or by RNAimax Reverse Transfection according to the manufacturer's instructions (Invitrogen). We also confirmed our results using two independent shRNAs directed against p115, PDZ, and LARG RhoGEFs (gift from A. Jaffe [Novartis, Boston, MA] and A. Hall [Memorial Sloan Kettering Cancer Center, New York, NY]).
Cell staining
For EB1 and myosin IIA staining, cells were fixed with 100% methanol at −20°C for 45 s. For RhoGEF p115 staining, monolayers were prepermeablized with PBS with 0.1% Triton X-100 containing 1 µg/ml phalloidin for 60 s before fixation with 4% formaldehyde in PBS at 37°C for 20 min. For all other immunostainings, cells were fixed with 4% formaldehyde in PBS at 37°C for 20 min. Fixed cells were rinsed three times in PBS, permeabilized for 5 min with PBS containing 0.5% Triton X-100, and blocked in AbDil (PBS with 0.1% Triton X-100 and 2% BSA), then incubated in primary antibody (in AbDil) for 1 h, washed three times with PBS with 0.1% Triton X-100, and incubated in secondary antibodies. Antibody concentrations used for immunostaining were: 1:200 anti-tubulin ascites (DM1α; Sigma-Aldrich); 1:50 anti-EB1 monoclonal clone 5 (BD); 1:50 nonmuscle myosin IIA polyclonal antibody (Sigma-Aldrich); 1:50 phosphomyosin II antibody (Cell Signaling Technology); 1:100 mouse β-catenin antibody (BD), 1:25 anti-Rho A (BD), or 1:500 rabbit anti–ZO-1 (Zymed); and 1:100 rabbit anti–active caspase-3 (Cell Signaling Technology). Actin was detected using 0.1 µg/ml Alexa Fluor 568–phalloidin (Invitrogen) or with 1:200 actin polyclonal antibody (A2066 [Sigma-Aldrich] when fixed with methanol). Alexa Fluor 488 goat anti–mouse and Alexa Fluor 568 goat anti–rabbit IgG were used as secondary antibodies (Invitrogen). DNA was detected with 1 µg/ml Hoechst 33342 (Sigma-Aldrich) or 5 µm DRAQ5 (Axxora) in all fixed cell experiments.
Zebrafish staining
To score the percentage of epidermal cells extruding apically versus basally, we treated 4-d zebrafish larvae with 450 µg/ml G-418 (Invitrogen) and 20 µM taxol or 0.2% DMSO in fish water (60 mg Instant Ocean mix per liter of distilled H2O) for 1 or 2 h. Larvae were then fixed in 4% formaldehyde, 0.15% glutaraldehyde, 1 mM MgCl2, 0.2% Triton X-100, and 25 mM Pipes, pH 6.9, for 2 h, blocked in 0.25% casein overnight, and stained with active caspase-3 antibody and Alexa Fluor 594–phalloidin + DAPI or DRAQ5 overnight with three washes of 0.5% Triton X-100 in PBS between each incubation. Tails were cut and mounted between a coverslip and slide in ProLong Gold (Invitrogen).
Drug and UV treatment
Cells were treated with 10 µM nocodazole, 10 µM taxol, or 0.1% DMSO (for control) for 10 min before UV treatment. To induce apoptosis, monolayers plated on glass coverslips were exposed to 1,200 μJ/cm2 UV254 irradiation in a UV series II (Spectroline) and incubated for 2 h before fixation.
Image and video acquisition
For live fluorescent imaging, MDCK cells transfected with GFP-tubulin (Clontech Laboratories, Inc.) and mApple-actin-7 (a gift from Michael W. Davidson, Florida State University, Tallahassee, FL) were grown to confluence on glass-bottom delta T culture dishes (Bioptechs) in DMEM. After 48 h, DMEM was replaced with DMEM/F12 without phenol red or Hepes buffer (Invitrogen) + 5% FBS. Medium was maintained at 37°C, 5% CO2, and 3% humidity using a Ludin Cube and Brick (Life Imaging Services) fitted to the microscope and stage. Cells were treated with 800–1,000 μJ/cm2 UV254 irradiation, then imaged with a spinning disc confocal system (Andor Technologies) mounted on an inverted microscope (TE300; Nikon) with a 60× 1.4 NA Plan-Apochromat lens (Nikon). Solid-state 488 nm and 568 nm lasers (Melles Griot) were used to excite GFP and mApple fluorophores, respectively. Images were acquired with an electron-multiplied cooled charge-coupled device (CCD) camera (DV887 1004X1002; Andor Technologies) driven by Andor IQ imaging software, and 4D movies were assembled with IQ software (Andor Technologies) and processed further with MetaMorph (GE Healthcare) and QuickTime Pro (Apple) software. A sample thickness of 9 µm in 4 z steps of 2.25 µm each was imaged every 2 min for 44 min.
For live phase video microscopy, cultured monolayers were imaged using a microscope (90i; Nikon) with a 40× phase lens. Images were acquired with a cooled CCD camera (Roper Scientific) driven by MetaMorph software. For filming zebrafish extrusion, larvae were treated with 450 µg/ml G-418 (Invitrogen) and 20 µM taxol or 0.2% DMSO for 15 min and mounted in 1% low melt agarose in the same treatment mix with 0.2% tricaine covered with fish water and a coverslip. Zebrafish videos were filmed using a 40× water immersion fluor differential interference contrast (DIC) lens on an upright microscope (Eclipse 90i). One frame per minute was captured for 2 h for all videos. MetaMorph software was used to analyze data.
Confocal sections were taken using a TCS SP5 microscope (Leica) using a 63× oil lens at 1,024 × 1,024 resolution. For montage pictures of EB1/myosin II and p115 RhoGEF/Rho, we made stacks from four (p115) or five (EB1) projections of five (EB1) or six (p115) consecutive 0.35-µm z slices from LSM confocal scans of each extrusion event. We displayed consecutive projections using the “montage” function on MetaMorph software. All images were processed further using Image J, Photoshop (Adobe), and Illustrator (Adobe).
Quantification of EB1 and p115 RhoGEF
To assess quantitatively the distribution of EB1 in 16-HBE-14o cells, we analyzed the abundance of EB1 fluorescent spots at actin/myosin rings in 12 randomly selected cell extrusions (>170 z sections). The number of EB1 spots at the base was averaged from the bottom 12 of 24 z sections and at the apex from the top 12 of 24 z sections, where the error bars indicate SEM from the variation of 12 separate apical and basal extrusions, and 12 cell–cell contacts where p-values from a t test are <0.0001 for apical versus basal extrusion. For quantification of p115 RhoGEF fibers intersecting with the extruding ring, we created montages (as described in the image acquisition section) and manually counted the number of p115 RhoGEF spots abutting the extrusion ring in the first 0–4 µm (base) and the next 4–8 µm (apex) from the bottom of the cell. Statistical analysis was done on 10 separate extrusions from control, nocodazole, taxol, and junctions from each, where error bars represent the SEM and the p-values from a t test are <0.0001 for each compared with the control.
Online supplemental material
Fig. S1 shows that disruption of microtubules mislocalizes myosin IIA and phospho-myosin II in 16-HBE-14o cell monolayers. Fig. S2 shows that taxol treatment induces basal extrusion in zebrafish larval epidermis. Fig. S3 shows that siRNA-mediated knockdown of other RGS-RhoGEFs has no effect on the direction of extrusion, and that disruption of microtubules disrupts localization of RhoGEF p115. Videos 1–3 show typical basal and apical extrusion from MDCK cell monolayers. Videos 4–7 shows apical and basal extrusion from the epidermis of 4-d-old zebrafish. Videos 8–9 show apical and basal extrusion, respectively, in MDCK cells expressing mApple-actin and GFP-tubulin. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903079/DC1.
Acknowledgments
We thank Aron Jaffe and Alan Hall for shRNAs of several RhoGEFs, Karl Matlin for MDCK cells, Dieter Gruenert, and Jo Porter for 16-HBE-14o cells, Carl Thummel for use of his LSM confocal microscope, and Michael Davidson for mApple-actin 7. We also give thanks to Katie Ullman, Mark Metzstein, and Julie Kadrmas for helpful comments.
This work was supported by Cancer Research UK project grant No. C8836/A3275, Biotechnology and Biological Sciences Research project grant No. 31/C18722, and a National Institutes of Health Innovator Award No. DP2 OD002056-01 to J. Rosenblatt, and P30 CA042014 awarded to the Huntsman Cancer Institute for core facilities.
Footnotes
Abbreviations used in this paper:
- CCD
- charge-coupled device
- DIC
- differential interference contrast
- DMEM
- Dulbecco's minimum essential medium
- LARG
- leukemia-associated RhoGEF
- RGS
- regulator of G protein signaling
References
- Carvalho P., Tirnauer J.S., Pellman D. 2003. Surfing on microtubule ends.Trends Cell Biol. 13:229–237 [DOI] [PubMed] [Google Scholar]
- Fukuhara S., Chikumi H., Gutkind J.S. 2001. RGS-containing RhoGEFs: the missing link between transforming G proteins and Rho? Oncogene. 20:1661–1668 [DOI] [PubMed] [Google Scholar]
- Gibson M.C., Perrimon N. 2005. Extrusion and death of DPP/BMP-compromised epithelial cells in the developing Drosophila wing.Science. 307:1785–1789 [DOI] [PubMed] [Google Scholar]
- Hartenstein V., Younossi-Hartenstein A., Lekven A. 1994. Delamination and division in the Drosophila neurectoderm: spatiotemporal pattern, cytoskeletal dynamics, and common control by neurogenic and segment polarity genes.Dev. Biol. 165:480–499 [DOI] [PubMed] [Google Scholar]
- Hogan C., Dupré-Crochet S., Norman M., Kajita M., Zimmermann C., Pelling A.E., Piddini E., Baena-López L.A., Vincent J.P., Itoh Y., et al. 2009. Characterization of the interface between normal and transformed epithelial cells.Nat. Cell Biol. 11:460–467 [DOI] [PubMed] [Google Scholar]
- Kopans D.B., Rafferty E., Georgian-Smith D., Yeh E., D'Alessandro H., Moore R., Hughes K., Halpern E. 2003. A simple model of breast carcinoma growth may provide explanations for observations of apparently complex phenomena.Cancer. 97:2951–2959 [DOI] [PubMed] [Google Scholar]
- Krendel M., Zenke F.T., Bokoch G.M. 2002. Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton.Nat. Cell Biol. 4:294–301 [DOI] [PubMed] [Google Scholar]
- Matsumura F. 2005. Regulation of myosin II during cytokinesis in higher eukaryotes.Trends Cell Biol. 15:371–377 [DOI] [PubMed] [Google Scholar]
- Matsumura F., Ono S., Yamakita Y., Totsukawa G., Yamashiro S. 1998. Specific localization of serine 19 phosphorylated myosin II during cell locomotion and mitosis of cultured cells.J. Cell Biol. 140:119–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E., Jochemsen A.G. 2008. p53: a guide to apoptosis.Curr. Cancer Drug Targets. 8:87–97 [DOI] [PubMed] [Google Scholar]
- Ninov N., Chiarelli D.A., Martín-Blanco E. 2007. Extrinsic and intrinsic mechanisms directing epithelial cell sheet replacement during Drosophila metamorphosis.Development. 134:367–379 [DOI] [PubMed] [Google Scholar]
- Pagliarini R.A., Xu T. 2003. A genetic screen in Drosophila for metastatic behavior.Science. 302:1227–1231 [DOI] [PubMed] [Google Scholar]
- Redd M.J., Kelly G., Dunn G., Way M., Martin P. 2006. Imaging macrophage chemotaxis in vivo: studies of microtubule function in zebrafish wound inflammation.Cell Motil. Cytoskeleton. 63:415–422 [DOI] [PubMed] [Google Scholar]
- Rogers S.L., Wiedemann U., Häcker U., Turck C., Vale R.D. 2004. Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner.Curr. Biol. 14:1827–1833 [DOI] [PubMed] [Google Scholar]
- Rosenblatt J., Raff M.C., Cramer L.P. 2001. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism.Curr. Biol. 11:1847–1857 [DOI] [PubMed] [Google Scholar]
- Schulze-Bergkamen H., Krammer P.H. 2004. Apoptosis in cancer—implications for therapy.Semin. Oncol. 31:90–119 [DOI] [PubMed] [Google Scholar]
- Thompson B.J., Mathieu J., Sung H.H., Loeser E., Rørth P., Cohen S.M. 2005. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila.Dev. Cell. 9:711–720 [DOI] [PubMed] [Google Scholar]
- Vidal M., Larson D.E., Cagan R.L. 2006. Csk-deficient boundary cells are eliminated from normal Drosophila epithelia by exclusion, migration, and apoptosis.Dev. Cell. 10:33–44 [DOI] [PubMed] [Google Scholar]
- Wittmann T., Waterman-Storer C.M. 2001. Cell motility: can Rho GTPases and microtubules point the way? J. Cell Sci. 114:3795–3803 [DOI] [PubMed] [Google Scholar]
- Yang L., Cao Z., Yan H., Wood W.C. 2003. Coexistence of high levels of apoptotic signaling and inhibitor of apoptosis proteins in human tumor cells: implication for cancer specific therapy.Cancer Res. 63:6815–6824 [PubMed] [Google Scholar]
- Yu W., Datta A., Leroy P., O'Brien L.E., Mak G., Jou T.S., Matlin K.S., Mostov K.E., Zegers M.M. 2005. Beta1-integrin orients epithelial polarity via Rac1 and laminin.Mol. Biol. Cell. 16:433–445 [DOI] [PMC free article] [PubMed] [Google Scholar]