A meiosis-conditional pds5 allele in yeast provides a more detailed understanding of homologue pairing and synaptonemal complex formation.
Abstract
During meiosis, homologues become juxtaposed and synapsed along their entire length. Mutations in the cohesin complex disrupt not only sister chromatid cohesion but also homologue pairing and synaptonemal complex formation. In this study, we report that Pds5, a cohesin-associated protein known to regulate sister chromatid cohesion, is required for homologue pairing and synapsis in budding yeast. Pds5 colocalizes with cohesin along the length of meiotic chromosomes. In the absence of Pds5, the meiotic cohesin subunit Rec8 remains bound to chromosomes with only minor defects in sister chromatid cohesion, but sister chromatids synapse instead of homologues. Double-strand breaks (DSBs) are formed but are not repaired efficiently. In addition, meiotic chromosomes undergo hypercondensation. When the mitotic cohesin subunit Mcd1 is substituted for Rec8 in Pds5-depleted cells, chromosomes still hypercondense, but synapsis of sister chromatids is abolished. These data suggest that Pds5 modulates the Rec8 activity to facilitate chromosome morphological changes required for homologue synapsis, DSB repair, and meiotic chromosome segregation.
Introduction
Eukaryotic chromosomes undergo precisely timed morphological changes during the cell cycle. Duplicated sister chromatids are associated along their length from S phase through metaphase, a process called sister chromatid cohesion. In both mitosis and meiosis, sister chromatids are condensed into rod-shaped structures before cohesion dissolution at anaphase. In meiosis, homologue synapsis, which is a unique chromosome morphogenetic process whereby homologues become juxtaposed along their length, is required for homologue disjunction. Synapsis is mediated by a tripartite synaptonemal complex (SC) located between juxtaposed homologues. The SC is composed of two lateral elements (LEs), which form along the length of each homologue, and a central element (CE) that is between the LEs and appears to connect them. From budding yeast to humans, SC formation and disassembly are believed to play a pivotal role in meiotic recombination and genome integrity (for review see Zickler and Kleckner, 1999).
Sister chromatid cohesion is largely the result of the activity of the cohesin complex (Guacci et al., 1997; Michaelis et al., 1997; Losada et al., 1998). In the budding yeast Saccharomyces cerevisiae, the four subunits of cohesin are called Smc1, Smc3, Mcd1/Scc1, and Scc3/Irr1 (for review see Onn et al., 2008). A meiosis-specific cohesin subunit, Rec8, largely replaces Mcd1 to form the predominant form of cohesin during meiosis (Klein et al., 1999). Mutational analysis shows that sister chromatid cohesion is required for proper chromosome structure, including homologue SC assembly (Klein et al., 1999; Revenkova et al., 2004; Xu et al., 2005; Novak et al., 2008). Cohesin itself is a major component of the chromosome axis, which provides a platform for LE formation (Eijpe et al., 2000; Revenkova et al., 2001). Remarkably, despite the orders of magnitude difference in chromosome size between yeast and humans, the distance between two LEs of an SC is ∼100 nm in both, suggesting a fundamentally conserved mechanism of SC formation and structure (for review see Zickler and Kleckner, 1999). Consistent with this view, components comprising both the LE and CE are functionally conserved among eukaryotes (Page and Hawley, 2004). Together, these observations support the idea that cohesin-mediated sister chromatid cohesion establishes a chromosome foundation required for the formation of higher order chromosome structures, including SCs. These data also raise an interesting question: why does an SC not form between sister chromatids?
After cohesion is established, its maintenance is facilitated by cohesin-associated factors. One such factor is called Pds5/Spo76/BIMD (Denison et al., 1993; van Heemst et al., 1999; Hartman et al., 2000; Panizza et al., 2000; Sumara et al., 2000; Tanaka et al., 2001). Both genetic and biochemical studies have confirmed that Pds5 is required primarily for the maintenance of sister chromatid cohesion (Hartman et al., 2000; Tanaka et al., 2001; Stead et al., 2003). Pds5 coimmunoprecipitates with cohesin and colocalizes with cohesin on chromosomes at cohesin-associated regions (Hartman et al., 2000; Panizza et al., 2000; Sumara et al., 2000; Tanaka et al., 2001; Lengronne et al., 2004). Despite this colocalization, the distinct roles of Pds5 and cohesin are evident because they are not mutually dependent for chromosomal binding. Pds5 localization to chromosomes requires cohesin, whereas cohesin localization to chromosomes does not depend on Pds5 (Hartman et al., 2000; Tanaka et al., 2001; Losada and Hirano, 2005; Zhang et al., 2005).
In meiosis, Pds5 mediates homologue interactions that facilitate SC formation and meiotic recombination in a timely manner, demonstrating a role for chromosome structural dynamics in DNA metabolism (Storlazzi et al., 2003, 2008; Zhang et al., 2005). Because PDS5 is an essential gene in most organisms, studies have used thermosensitive alleles or partially functional alleles of PDS5 (van Heemst et al., 1999, 2001; Hartman et al., 2000; Panizza et al., 2000; Stead et al., 2003; Wang et al., 2003; Ren et al., 2005; Zhang et al., 2005). The only exception is the fission yeast Schizosaccharomyces pombe, in which pds5-null strains are viable, but vegetative cells exhibit defects in cohesion maintenance after arrest in G2/M phase and increased chromosome loss rates (Tanaka et al., 2001). More pronounced defects were seen during fission yeast meiosis in which chromosomes showed hypercompaction, a pds5 mutant phenotype not observed in any other experimental system with thermosensitive pds5 alleles (Ding et al., 2006). The peculiar features of fission yeast meiosis, such as the absence of SC formation, may explain why Pds5 has a unique role in chromosome compaction. Alternatively, previous work with pds5 thermosensitive alleles may not have completely abrogated Pds5 activity.
Using a molecular approach, we created a meiosis-conditional pds5 allele in which Pds5 is depleted completely and specifically during meiosis in budding yeast. This organism has well-defined meiotic processes similar to those of other eukaryotes and an abundance of characterized chromosomal markers, including LE components Red1 and Hop1 and the CE component Zip1 (Rockmill and Roeder, 1988; Hollingsworth and Byers, 1989; Sym et al., 1993). Like previous work in budding yeast (Zhang et al., 2005), this study reveals only minor defects in cohesion, indicating that sister chromatid cohesion is largely intact in the absence of Pds5. We also find that meiotic cells without Pds5 are largely blocked at a pachytene-like stage. In contrast to previous work with a thermosensitive pds5 allele, we find that homologues fail to synapse and become hypercondensed when Pds5 is depleted. In addition, an SC-like structure forms between sister chromatids in these mutant cells. Finally, our data indicate that Pds5 inhibits SC formation between sister chromatids by specifically modulating the activity of the meiotic cohesin Rec8.
Results
Pds5 colocalizes with Rec8 on meiotic chromosomes in a cell cycle–dependent manner
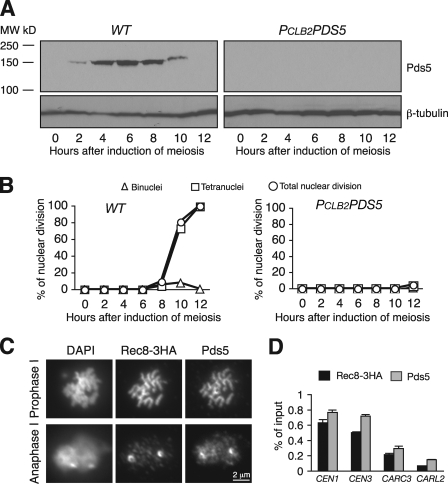
We investigated the role of Pds5 in meiotic chromosome morphological changes. First, we used an affinity-purified antibody against yeast Pds5 (Noble et al., 2006) to monitor Pds5 levels by conducting immunoblots in cells induced to undergo synchronous meiosis (Fig. 1). Pds5 is present in cells at all stages of the mitotic cell cycle (Stead et al., 2003) but is not detected in cells entering meiosis (Fig. 1 A, t = 0). Pds5 is detected at low levels 2 h after meiotic entry and reaches peak levels by 6 h (Fig. 1 A). This time frame corresponds to meiosis I, from premeiotic S phase through metaphase I (Fig. 1 B). Pds5 is no longer detected after 12 h of induction, as cells have exited meiosis (Fig. 1, A and B). These data show that Pds5 is degraded as a prelude to meiotic entry, is resynthesized during early meiosis when sister chromatid cohesion is established and homologue pairing occurs, and is degraded late in meiosis.
Figure 1.
Characterization of Pds5 protein level and localization to chromosomes during meiosis. (A) Immunoblot analysis of Pds5 in wild-type (WT) and PCLB2PDS5 cells during meiosis. Yeast cultures were induced to enter meiosis synchronously. Protein extracts were prepared at the indicated times. Pds5 was detected with a polyclonal anti-Pds5 antibody. β-Tubulin served as a loading control. (B) Meiotic nuclear division in wild-type and PCLB2PDS5 cells. Cell aliquots were withdrawn at the indicated times, fixed with 4% paraformaldehyde, stained by DAPI, and visualized by fluorescence microscopy. Only minimal nuclear division was observed in PCLB2PDS5 cells after 12 h of induction. (C) Pds5 and Rec8 colocalization to meiotic chromosomes is shown. Yeast nuclear spreads were prepared from synchronous meiotic cultures. Pds5 and Rec8-3HA were detected with anti-Pds5 and anti-HA antibodies (12CA5), respectively. Representative images from prophase I and anaphase I are shown. (D) ChIP assay of Pds5 and Rec8 binding to cohesin-associated regions at centromere 1 (CEN1), centromere 3 (CEN3), a cohesin site at the MAT locus (CARC3), and a cohesin site on chromosome XII (CARL2). Cells were arrested at pachytene by ndt80Δ after an 8-h induction. ChIP was performed as described previously by Yu and Koshland (2005) with anti-Pds5 and anti-HA antibodies. Note that Rec8 and Pds5 are not enriched at CARL2, which serves as a negative control. Error bars indicate SD.
We next investigated the association of Pds5 with meiotic chromosomes using anti-Pds5 antibodies for indirect immunofluorescence on spreads of yeast nuclei (Fig. 1 C). Early in meiosis I, Pds5 colocalizes with the cohesin subunit Rec8 along the entire length of chromosomes (Fig. 1 C, top). A similar pattern was previously seen with epitope-tagged Pds5 (Zhang et al., 2005). During anaphase I, Pds5 is predominantly localized to the centromeric region of the chromosome, as indicated by the chromosome colocalization with Rec8 (Fig. 1 C, bottom). This observation contrasts with a previous one that epitope-tagged Pds5 is not detectable during anaphase I (Zhang et al., 2005). We also find that Pds5 is not detectable on chromosomes in cells that have exited meiosis (unpublished data). This pattern is identical to that observed for Rec8 (Klein et al., 1999). The dynamic pattern of Pds5 localization to meiotic chromosomes is directly correlated with the times when Pds5 is present in cells as assayed by immunoblots (Fig. 1 A). Next, we used chromatin immunoprecipitation (ChIP) to perform a high resolution analysis of Pds5 association with chromosomes in staged pachytene cells by deleting the NDT80 gene (Xu et al., 1995). We assayed four representative chromosomal regions: two centromeric loci (CEN1 and CEN3) and two chromosome arm loci (CARC3 and CARL2). Cohesin binds robustly at these loci during mitosis (Glynn et al., 2004). Rec8 cohesin also robustly binds these loci during meiosis, except for CARL2, which has a very low level of binding (Glynn et al., 2004). During meiosis, we found that both Pds5 and Rec8 were highly enriched at loci near centromeres and present at lower levels at arm loci (Fig. 1 D). Therefore, Pds5 and Rec8 colocalize to chromosomes during meiosis.
Pds5 is required for meiotic cell progression beyond prophase I
Because Pds5 is degraded before meiotic entry, we were able to generate a meiosis-conditional null allele of PDS5, which depletes Pds5 only during meiosis. To construct this allele, we replaced the endogenous promoter of PDS5 with a mitosis-specific promoter from CLB2 to create PCLB2PDS5. This allele expresses PDS5 in vegetative cells but is completely repressed during meiosis. Consequently, Pds5 is absent from cells undergoing meiosis (Fig. 1 A, right). PCLB2PDS5 cells appear normal in cell viability and cell cycle progression during vegetative growth (unpublished data). To address whether Pds5 is required for meiotic nuclear division, we induced PCLB2PDS5 cells to undergo synchronous meiosis and monitored chromosome segregation by fluorescence microscopy. Meiotic nuclear division is largely absent in PCLB2PDS5 cells (Fig. 1 B). This observation is similar to that observed with the thermosensitive allele pds5-1 at a nonpermissive temperature (Zhang et al., 2005). About 90% of PCLB2PDS5 cells have only a single aster of microtubules, which is indicative of a prophase I arrest, whereas only 10% of mutant cells form a short bipolar spindle, which is indicative of metaphase I, a ratio that persisted even after 12 h of induction of meiosis (unpublished data). Therefore, when Pds5 is absent, the vast majority of cells are arrested before metaphase I.
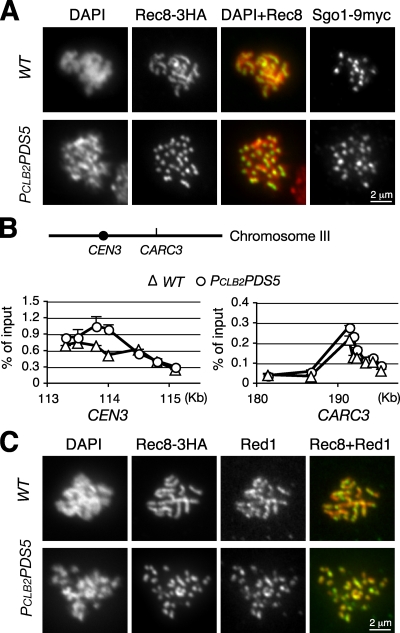
To further investigate the role of Pds5, we examined whether chromosome association of meiotic cohesin (shown with Rec8) depends on Pds5 using indirect immunofluorescence microscopy and ChIP. In wild-type cells at pachytene, Rec8 localizes along the entire length of meiotic chromosomes, forming linear rods (Fig. 2 A, top; and Fig. S1 A). In PCLB2PDS5 meiotic cells, chromosomes are strikingly compacted into short rods, as shown by both Rec8 and DAPI staining (Fig. 2, A and C). Our finding that Rec8 localizes to chromosomes when Pds5 is absent confirms previous data generated with the pds5-1 mutant allele (Zhang et al., 2005), but the highly compacted rods are a novel phenotype not previously observed in budding yeast. High resolution mapping by ChIP revealed that Rec8 associates with chromosomes at four representative chromosome regions in a similar manner in PCLB2PDS5 cells staged at pachytene (Fig. 2 B and not depicted). Consistent with the aforementioned result, similar levels of Rec8 are detected by immunoblotting in wild-type and PCLB2PDS5 cells during meiosis (Fig. S2). As in wild-type cells, the LE components Red1 and Hop1 were localized to meiotic chromosomes in PCLB2PDS5 cells (Fig. 2 C and Fig. S1 B), but in PCLB2PDS5 cells, Red1 and Hop1 formed short rods similar to those of Rec8. These results indicate that Pds5 is required for regulation of axial length but not for loading of meiotic cohesin Rec8, Red1, or Hop1 to chromosomes.
Figure 2.
Pds5 mediates chromosome morphological changes during meiosis. (A) Chromosome morphology at meiotic prophase I. Yeast cells were induced to enter synchronous meiosis for 5 (wild type [WT]) and 7 h (PCLB2PDS5), and nuclear spreads were prepared for immunofluorescence microscopy. Rec8-3HA and Sgo1-9MYC were detected with anti-HA (12CA5) and anti-MYC (9E10) antibodies, respectively. The chromosome number in PCLB2PDS5 cells appears to be twice that in wild-type cells. Red, DNA stained by DAPI; green, Rec8-3HA. (B) ChIP assay of Rec8 association with the chromosome. ChIP was performed as in Fig. 1 C. The locations of CEN3 and CARC3 are depicted on chromosome III. S. cerevisiae genome database coordinates are shown on the x axis. Error bars indicate SD. (C) Chromosome axis revealed by Red1 staining. Meiotic nuclear spreads were prepared as in A. Red1 (green) and Rec8-3HA (red) were detected with anti-Red1 and anti-HA antibodies, respectively.
Pds5 is required for homologue pairing
A compelling feature of PCLB2PDS5 cells is that the number of Rec8-stained chromosome axes is approximately twice that of wild-type cells at a pachytene-like stage (Fig. 2, A and C). Consistent with this finding, we observed ∼32 kinetochores in PCLB2PDS5 cells by localizing the kinetochore-associated protein Sgo1 (Fig. 2 A). This situation contrasts with that in wild-type cells, which have only ∼16 Sgo1 foci, corresponding to 16 paired homologues in budding yeast at this stage (Fig. 2 A). In addition, the number of Red1-stained chromosome axes in PCLB2PDS5 cells appears to be roughly twice that in the wild type (Fig. 2 C). Together, these data suggest that in the absence of Pds5, homologues fail to pair, producing an increased number of observed chromosome axes.
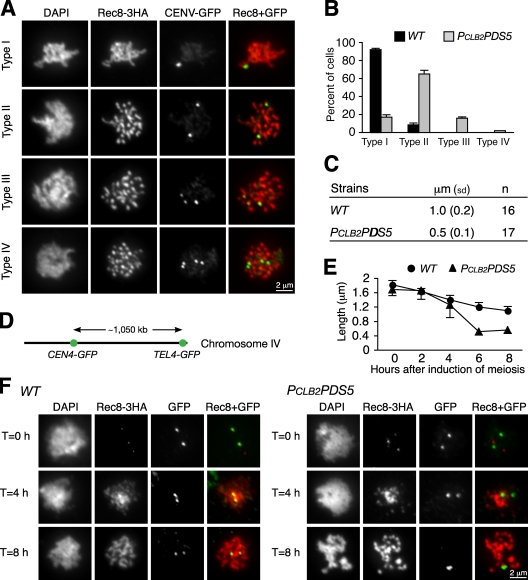
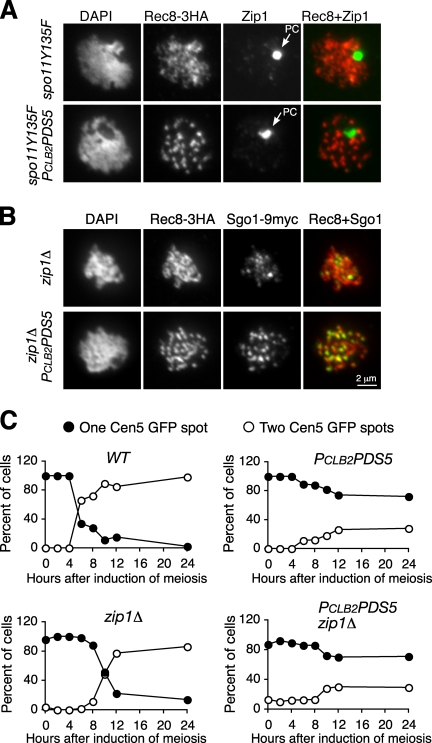
To confirm the role of Pds5 in homologue pairing, we integrated an array of tetO into both chromosome V homologues at the URA3 locus, which is ∼35 kb from centromere 5, and expressed tetR-GFP (Michaelis et al., 1997) so that we could track chromosome V by fluorescence microscopy (Fig. 3 A). In ndt80Δ cells arrested at pachytene, >90% of chromosome V homologues were paired, yielding a single GFP spot (Fig. 3, A and B). In contrast, in PCLB2PDS5 ndt80Δ cells, only 18% of cells had one GFP spot, suggesting a failure of homologue pairing.
Figure 3.
Pds5 is required for homologue pairing and limits chromosome compaction during meiosis. (A) Monitoring homologue pairing and sister chromatid cohesion during meiosis. CEN5 on both chromosome V homologues was marked by tetO/tetR-GFP and visualized by fluorescence microscopy. Wild-type and PCLB2PDS5 cells were arrested at pachytene with ndt80Δ. Four cell types were observed in PCLB2PDS5 cells: type I, a single GFP spot, indicating that homologues are paired; type II, two GFP spots, indicating that homologues fail to pair but sister chromatids stay together; type III, three GFP spots, indicating that homologues fail to pair and one pair of sister chromatids separate; and type IV, four GFP spots, indicating that homologues fail to pair and sister chromatid cohesion is lost on both homologues. (B) Quantitative measurement of homologue pairing and sister chromatid cohesion. At least 200 cells were scored for each strain. (C) Measurement of the axial length of chromosome V. Meiotic nuclear spreads were prepared. Chromosome V was identified by CEN5-GFP signal, and the entire length of the chromosome was determined by measurement of the Rec8-3HA staining (detected as in A). (D) The long arm of chromosome IV was marked by GFP at two loci (CEN4 and TEL4) with the lacO/lacI-GFP system. Only one homologue of chromosome IV was marked in these cells. (E) The length of chromosome IV arm was determined by measurement of the distance between two GFP spots. Cells were induced to enter synchronous meiosis, and aliquots were withdrawn at the indicated times for preparation of immunofluorescence. Error bars indicate SD. (F) Representative images from three time points are shown. WT, wild type.
The tetO/tetR-GFP chromosome-marking system also permitted us to evaluate sister chromatid cohesion (Fig. 3 A). If sister chromatids separated prematurely in meiotic cells, three or four GFP spots appeared as a result of sister separation on one or both homologues, respectively. In ndt80Δ cells, no cell had more than two GFP spots (Fig. 3 B). In PCLB2PDS5 ndt80Δ cells, ∼14% showed three GFP spots, and ∼2% showed four. This low percentage suggests that sister chromatin cohesion is weakly defective at a CEN-proximal locus in PCLB2PDS5 cells. Previous work monitoring cohesion at a telomeric locus on chromosome IV similarly revealed a low level of sister chromatid cohesion loss (Zhang et al., 2005). These data suggest that, although some cohesion defects occur, cohesion is relatively normal along most of the chromosomal length.
Pds5 limits chromosome condensation to prevent hypercompaction of the chromosome axis
Chromosome axes, as monitored by staining of axial components cohesin and Red1, appear much shorter in PCLB2PDS5 cells (Fig. 2 C and Fig. S1 A). From this result, we hypothesized that Pds5 limits chromosome axial compaction. We marked chromosome V using the tetO/tetR-GFP at URA3 and determined its entire axial length by staining with Rec8 (Fig. 3 A). Chromosome V is ∼570 kb of DNA and is 1.0 µm in axial length in wild-type cells at pachytene but only 0.5 µm in PCLB2PDS5 cells (Fig. 3 C). This result indicates that chromosome V is twice as condensed when Pds5 is absent.
We used a more direct method to assess chromosome axial compaction on chromosome IV, the second-longest yeast chromosome. Two loci on chromosome IV were marked with lacO arrays, one centromere proximal (CEN4-lacO/lacI-GFP) and one telomere proximal (TEL4-lacO/lacI-GFP), a distance spanning ∼1 Mb of DNA (Fig. 3 D). In wild-type cells, these loci were separated by ∼1.2 µm when the cells reach pachytene after 4–6 h of induction of meiosis (Fig. 3, E and F). In contrast, this distance is only half as great in PCLB2PDS5 cells (Fig. 3, E and F). By monitoring the kinetics of condensation, we found that chromosome hypercompaction started in PCLB2PDS5 cells after meiotic S phase and reached a maximum rate at the pachytene-like stage (Fig. 3 E and Fig. S3). Our observations are consistent with those from S. pombe, in which meiotic chromosomes become hypercondensed in pds5 mutants (Ding et al., 2006). We concluded that Pds5 serves to restrict the amount of axial chromosome condensation.
SCs form between sister chromatids in the absence of Pds5
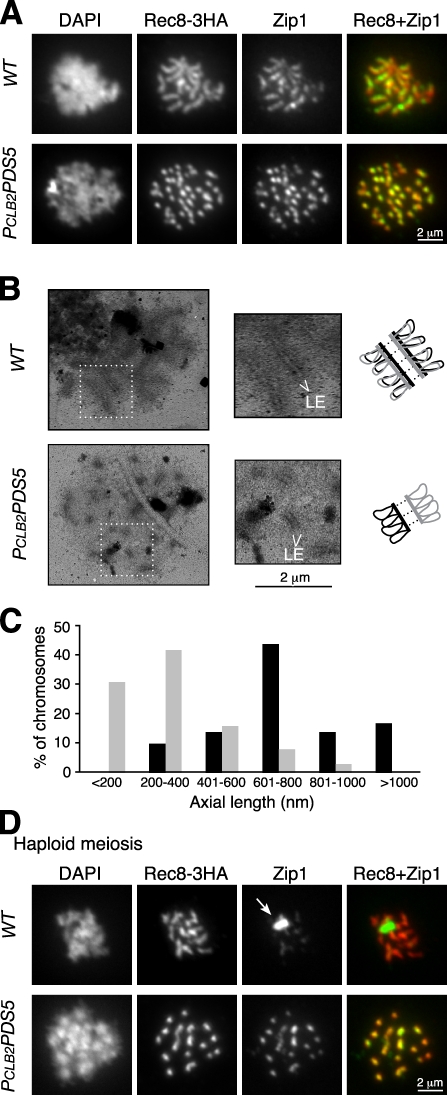
Two LE components, Red1 and Hop1, are still localized to chromosomes in PCLB2PDS5 cells (Fig. 2 C and Fig. S1 B). To determine whether this localization represents formation of SCs, we first assayed binding of the CE component Zip1 to chromosomes. In wild-type cells at pachytene, Zip1 localizes along the lengths of the 16 synapsed homologues (Fig. 4 A). In PCLB2PDS5 cells, Zip1 still localizes to chromosomes but forms ∼32 short rods (Fig. 4 A). Zip1 staining was also found on chromosomes in thermosensitive pds5-1 mutant cells (Zhang et al., 2005). The robust chromosome staining of Zip1 mirrors that of Red1 and Rec8 (Fig. 2 C and Fig. 4 A). Because a diploid yeast cell contains 16 pairs of homologues but 32 pairs of sister chromatids, our data are consistent with the idea that, in PCLB2PDS5 cells, an SC forms on each chromosome consisting of two sister chromatids.
Figure 4.
SC formation during meiosis. (A) Immunofluorescence analysis of SC formation in wild-type (WT) and PCLB2PDS5 cells. Yeast cells were induced to undergo synchronous meiosis, and nuclear spreads were prepared for immunofluorescence microscopy as in Fig. 2 A. Zip1 (green) and Rec8-3HA (red) were detected with anti-Zip1 and anti-HA antibodies, respectively. Note that Zip1 still localizes to chromosomes in PCLB2PDS5 cells despite the absence of homologue synapsis. (B, left) EM of SC formation in wild-type and PCLB2PDS5 cells. Yeast nuclear spreads were stained with silver nitrate and visualized by EM. (middle) Magnified views of boxed regions are shown. (top right) A twofold enlargement of regions of interest is shown. (bottom right) Diagrams of sister chromatids are shown in black and gray. Note that short stretches of LEs are formed in PCLB2PDS5 cells despite the absence of homologue synapsis. (C) Distribution of chromosome axial length from wild type (black bars) and PCLB2PDS5 (gray bars). Eight cells from each stain were scored. (D) Immunofluorescence analysis of SC formation in haploid yeast cells. Haploids were induced to undergo synchronous meiosis for 8 h and processed as described in A. These haploid strains could enter meiosis because the SIR2 gene had been deleted, resulting in activation of both mating types. Rec8-3HA (red) and Zip1 (green) were detected as described in A. The arrow shows the polycomplex formed by aggregation of SC components.
To assay more definitively for intersister SC formation, we used a silver staining method to visualize spreads of yeast nuclei with EM (Dresser and Giroux, 1988). In wild-type cells at pachytene, a pair of homologues forms two prominent parallel lines heavily stained by silver, which correspond to the two LEs of an SC (Fig. 4 B). Each LE is formed by one chromosome consisting of sister chromatids. Therefore, the width of the SC is judged by the distance between the two LEs, which in wild-type cells is ∼103 ± 9 nm (n = 24). In PCLB2PDS5 cells, we observed that 8 out of 15 cells have 16 or more identifiable short stretches of SC-like structures in spread nuclei (Fig. 4 B). LEs were symmetrical, and their ends appeared open and even. The observed SC-like structures were therefore less likely to be formed by either nonhomologous chromosome pairing or the folding back of the same chromosome. The width of the SC-like structure on each chromosome in PCLB2PDS5 cells is ∼107 ± 13 nm (n = 26), which is not statistically different from the width of the SC formed between homologues in the wild-type cells. The lengths of SC structures in PCLB2PDS5 cells are on average 334 ± 176 nm (n = 8 cells; Fig. 4 C), which is less than half that in wild-type cells (mean, 768 ± 327 nm; n = 8 cells; Fig. 4 C). The shorter SC is consistent with the increased axial compaction found in PCLB2PDS5 cells (Fig. 3). These results suggest that SCs can form between sister chromatids in PCLB2PDS5 cells, which implies that Pds5 normally functions to inhibit this intersister SC formation.
To provide further evidence for intersister SC formation, we examined meiosis in haploid yeast cells. The use of haploids eliminates homologues and halves the chromosome number, simplifying the assessment of intersister SC formation. We forced haploids to enter meiosis and assayed for SC formation. In wild-type haploids, only minimal SCs form (Fig. 4 D), which are believed to be between nonhomologous chromosomes (Loidl et al., 1991). The failure of SC formation in wild-type haploids leads to accumulation of SC components in a polycomplex (Fig. 4 D, top). In contrast, large polycomplexes rarely accumulate in PCLB2PDS5 haploids, and most cells show ∼16 short Zip1-staining rods, which is the haploid chromosome number (Fig. 4 D, bottom). If SC formation in PCLB2PDS5 haploids were a consequence of nonhomologous chromosome pairing, the number of Zip1 rods would have been reduced to as few as eight. Our data unequivocally show that sister chromatids can form SC-like structures when Pds5 is absent.
Intersister SC formation depends on Spo11 and Zip3, but it does not contribute to sister chromatid cohesion
Our observations did not preclude the possibility that SC formation between sister chromatids was actually centromere coupling, a phenomenon in spo11 mutants in which SCs are initiated at centromeres but fail to elongate (Tsubouchi and Roeder, 2005). To determine whether this phenomenon could occur in PCLB2PDS5 cells, we constructed PCLB2PDS5 spo11-Y135F double mutants and monitored Zip1 staining. In the absence of Spo11 activity, SC components, shown with Zip1 staining, are concentrated in the polycomplex whether Pds5 is present or not (Fig. 5 A). This result indicates that meiotic recombination, promoted by Spo11, is necessary for intersister SC formation. In addition, in PCLB2PDS5 cells lacking Zip3, a protein that promotes SC assembly, polycomplexes form, but SCs fail to assemble onto chromosomes (Fig. S4, A and B). As previously demonstrated, besides the polycomplex, Zip1 forms weak foci on meiotic chromosomes (Fig. 5 A and Fig. S5). A similar 61% of Zip1 foci overlap with the staining of the kinetochore protein Sgo1 in both spo11-Y135F and PCLB2PDS5 spo11-Y135F cells (Fig. S5). Therefore, our data suggest that intersister SC formation depends on Spo11 and Zip3 and that some SCs are initiated at the centromeres.
Figure 5.
SC formation depends on SPO11 but SC does not contribute to cohesion in PCLB2PDS5 cells. (A) SC formation requires Spo11 activity. Yeast cells were induced to undergo synchronous meiosis, and nuclear spreads were prepared for immunofluorescence microscopy as in Fig. 2 A. Zip1 (green) and Rec8-3HA (red) were detected with anti-Zip1 and anti-HA antibodies, respectively. In the absence of Spo11 activity, Zip1 localizes to the polycomplex (PC; arrows). (B) Chromosome morphology in the absence of Zip1. Yeast cells were induced to undergo synchronous meiosis, and nuclear spreads were prepared for immunofluorescence microscopy as in Fig. 2 A. Red, Rec8-3HA; green, Sgo1-9Myc. (C) Sister chromatid cohesion assayed with CEN5-GFP in wild-type (WT), PCLB2PDS5, zip1Δ, and PCLB2PDS5 zip1Δ cells. Only one homologue of chromosome V was marked by tetO/tetR-GFP. Yeast cells were induced to undergo synchronous meiosis, aliquots were withdrawn at the indicated times, fixed with 4% formaldehyde, and visualized by fluorescence microscopy. At least 100 cells were counted at each time point.
We next asked whether SCs formed between sister chromatids help promote sister chromatid cohesion. Previous work showed that defects in SC formation caused by the absence of Zip1 lead to a mild defect in sister chromatid cohesion (Sym and Roeder, 1994). We deleted the ZIP1 gene to disrupt SC formation (Fig. 5 B). In PCLB2PDS5 cells lacking Zip1, chromosomes still hypercondensed, but no increase in precocious sister separation was seen (Fig. 5 C). Similarly, no changes were seen when HOP1 was deleted (unpublished data). Therefore, SCs formed between sister chromatids do not contribute to sister chromatid cohesion in PCLB2PDS5 cells.
Rec8 is required for intersister SC formation
How does Pds5 inhibit SC formation between sister chromatids? Because Pds5 localization to chromosomes depends on Rec8 (Zhang et al., 2005; Ding et al., 2006), we hypothesized that Pds5 interacts with meiotic cohesin Rec8 to ensure that sister chromatid axes are held together to form a single platform for LE formation. To test this idea, we first asked whether cohesin activity is required for the phenotypes we observed in PCLB2PDS5 cells. In the absence of meiotic cohesin Rec8, meiotic chromosomes fail to establish a linear chromosome axis regardless of whether Pds5 is present or absent (Fig. S4 C). These data further confirm that Rec8 is epistatic to Pds5 in chromosome axial formation.
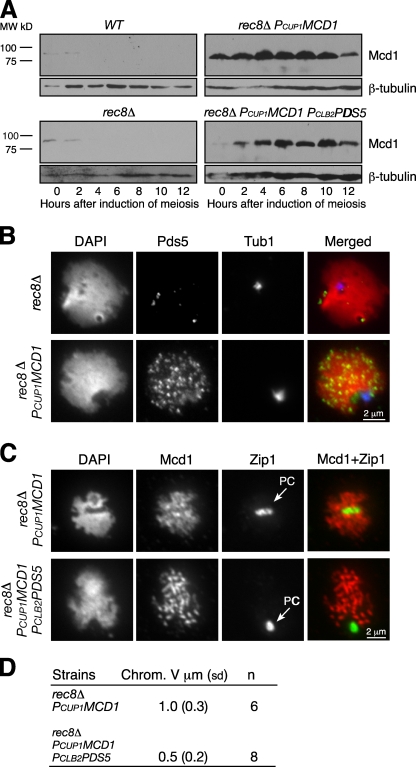
Next, we asked whether meiotic cohesin Rec8 is specifically required for Pds5-mediated chromosome morphogenesis, including SC formation and chromosome compaction. Only small amounts of the mitotic cohesin subunit Mcd1 are present as cells enter meiosis, and this protein is no longer detected 4 h after induction of meiosis (Fig. 6 A). Moreover, the MCD1 promoter is repressed during meiosis (Chu et al., 1998). Therefore, we placed MCD1 under control of the inducible CUP1 promoter to permit meiotic expression of MCD1 at high levels in both rec8Δ and rec8Δ PCLB2PDS5 cells (Fig. 6 A). Mcd1 can replace Rec8 for generation of sister chromatid cohesion during meiosis (Buonomo et al., 2000; unpublished data). Ectopic expression of MCD1 in rec8Δ cells results in a dramatic increase in the amount of Pds5 loaded onto meiotic chromosomes (Fig. 6 B). More importantly, SCs fail to form in either rec8Δ PCUP1MCD1 or rec8Δ PCUP1MCD1 PCLB2PDS5 cells, as revealed by formation of only a large polycomplex by Zip1 staining (Fig. 6 C). Interestingly, the chromosome axes, as measured by Mcd1 staining, also hypercondense in rec8Δ PCUP1MCD1 PCLB2PDS5 cells to a level identical to that in PCLB2PDS5 cells (Fig. 3, C and D). Therefore, Pds5 specifically modulates Rec8 to facilitate SC formation. In contrast, Pds5 can exert its role in regulating chromosome axial compaction through either the mitotic or the meiotic form of cohesin.
Figure 6.
Pds5 interacts with cohesin in meiotic chromosome morphogenesis. (A–D) Yeast cultures were induced to undergo synchronous meiosis. Protein extracts were prepared at the indicated times for immunoblotting (A) and meiotic nuclear spread for immunofluorescence (B–D). (A) Ectopic production of Mcd1 during meiosis. To induce PCUP1MCD1 expression, we added 60 µM (final concentration) CuSO4 to the sporulation medium after induction of meiosis. Mcd1 was detected with a polyclonal anti-Mcd1 antibody. β-Tubulin served as a loading control. WT, wild type. (B) Cohesin is required for Pds5 localization to chromosomes. Pds5 (green) was detected with anti-Pds5 antibody, microtubules (blue) by a monoclonal anti–α-tubulin antibody, and DNA (red) with DAPI. (C) Localization of ectopically expressed Mcd1 during meiosis. Mcd1 bound to meiotic chromosomes, but the SC component Zip1 (green) was present in the polycomplex (PC; arrows). (D) Quantitative analysis of chromosome V compaction in rec8Δ PCUP1MCD1 and rec8Δ PCUP1MCD1 PCLB2PDS5 cells. SD is shown in parentheses.
Pds5 is not necessary for double-strand break (DSB) formation but is required for DSB repair
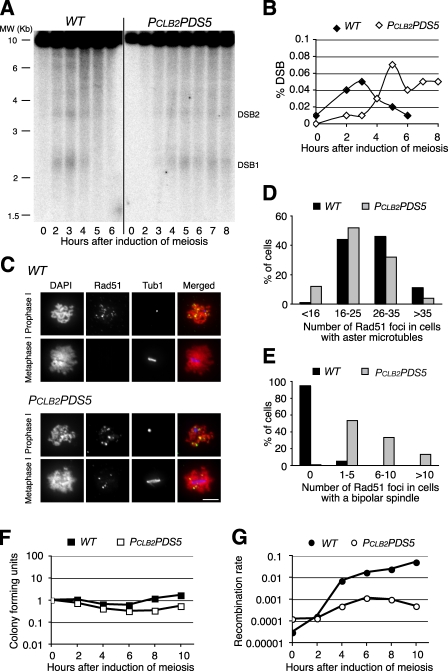
Having shown that Pds5 was required for proper SC formation, we asked whether Pds5 is required for meiotic recombination. First, we used a molecular approach to monitor DSB formation directly at the recombination hotspots at the YCR047c/YCR048w locus (Smith et al., 2001). DSB accumulation peaks 3 h after induction of meiosis in the wild type (Fig. 7 A). The peak accumulation of DSB is delayed in PCLB2PDS5 cells by 2 h, but the level of DSBs reaches that observed in the wild type (Fig. 7 A). This delay is probably the result of PCLB2PDS5 cells exhibiting a 2-h delay in meiotic S phase (Fig. S3). DSBs are formed in the absence of Pds5 but persist, suggesting that DSBs are not fully repaired (Fig. 7, A and B). We next used a cytological approach to assess DSB focus formation by monitoring Rad51 in spread meiotic nuclei. In wild-type cells, Rad51 (DSB) foci are seen in prophase I, as defined by the presence of a monopolar spindle (aster microtubules), then disappear by metaphase I, as defined by the presence of a short bipolar spindle (Fig. 7, C–E). In PCLB2PDS5 cells, 90% of which are arrested in a pachytene-like stage (monopolar spindles), Rad51 foci are also visible (Fig. 7, C and D), but in the remaining 10% of PCLB2PDS5 cells that do reach metaphase I (short bipolar spindle), Rad51 foci persist, indicating that DSBs remain (Fig. 7, C and E). These data show that Pds5 is not required for DSB formation but is required for DSB repair.
Figure 7.
Pds5 is necessary for meiotic recombination. (A) A physical assay of DSB formation and processing at the YCR047c/YCR048w locus. Yeast cells were induced to undergo synchronous meiosis, and DNA samples were extracted at the indicated time points. DSBs were detected by Southern blotting. Two prominent DSB sites at this location are depicted (DSB1 and DSB2). (B) Quantitative measurement of DSB formation in wild-type (WT) and PCLB2PDS5 cells. The ratio of the intensity of DSB1 to that of the parental band is shown on the y axis. (C) Rad51 focus formation during meiosis. Yeast cells were induced to enter meiosis as in A. Yeast nuclear spreads were prepared for immunofluorescence. Rad51 was detected with an anti-Rad51 antibody and microtubules with an anti–α-tubulin antibody. Red, DNA stained by DAPI; green, Rad51; blue, microtubules. Bar, 4 µm. (D and E) Quantification of Rad51 foci in cells with aster microtubules and short bipolar spindles is shown. Note that Rad51 foci persist in PCLB2PDS5 cells with a bipolar spindle. 20 cells from each strain were scored. (F) Cell viability was assayed by return to growth. (E) Yeast cells were induced to enter meiosis, and aliquots were withdrawn at the indicated times and plated on both YPD and Arg minus plates. Colony-forming units are defined as 1 at time 0. (G) Meiotic recombination at the ARG4 locus. Two heteroalleles of ARG4 (arg4-Bgl and arg4-Nsp) are present in the strains assayed. Recombination between the heteroalleles generates a wild-type ARG4 allele, which is detected as an Arg-positive colony. The ratio of colonies formed on Arg minus plates to those on YPD plates determines the recombination rate.
To assess DSB repair genetically in PCLB2PDS5 cells, we used a return to growth assay to determine meiotic recombination at a representative hotspot at ARG4 (Fig. 7, F and G). Yeast cells were induced to undergo synchronous meiosis, and aliquots were withdrawn at the specified times and plated on both YPD (1% yeast extract, 2% peptone, and 2% dextrose) and arginine dropout media. The number of colony-forming units on YPD was reduced less than twofold in the mutant after 10 h of sporulation (Fig. 7 F), but heteroallelic recombination at ARG4 was reduced by almost two orders of magnitude (Fig. 7 G). These data further suggest that Pds5 is required for DSB repair for completion of meiotic recombination.
Discussion
We report that Pds5 is required for homologue synapsis and cell progression through meiosis I. When Pds5 is absent, most cells are arrested at prophase I with highly compacted chromosomes, and an SC-like structure forms between sister chromatids. Cohesion between sisters is largely intact, as only small amounts of precocious sister separation are observed. DSBs form but are not fully resolved, indicating that DSB repair is defective. Our work significantly extends previous observations of the role of Pds5 in formation of meiotic chromosome structure. These novel observations may arise because our experiments involved a meiotic pds5-null allele, whereas previous studies involved thermosensitive pds5 alleles. These thermosensitive alleles make mutated proteins that still bind to chromosomes and therefore retain residual Pds5 activity (Storlazzi et al., 2008; unpublished data).
Previous work in budding yeast indicated that Rec8 promotes SC formation and homologue synapsis because neither of these events occurs when Mcd1 replaces Rec8 (Klein et al., 1999; Buonomo et al., 2000). We show that Pds5 serves an important inhibitory role because when Pds5 is absent, SCs form between sisters rather than homologues. This intersister SC formation requires Rec8 because when Mcd1 replaces Rec8 in Pds5-depleted cells, no SCs form. These data are consistent with the idea that Pds5 restrains the SC-promoting activity of Rec8. This reasoning seems at odds with the situation in vertebrates, where intersister SCs form when REC8 is deleted (Xu et al., 2005). This result suggests that vertebrate Rec8 serves as an inhibitor, much as Pds5 does in yeast. One possibility is that yeast and vertebrate cells prevent SC formation between sisters by distinct mechanisms. Alternatively, vertebrates also contain other meiosis-specific cohesins, such as SMC1β and STAG3 (Prieto et al., 2001; Revenkova et al., 2001). Therefore, deleting vertebrate REC8 could generate meiotic cohesin with a reduced affinity for Pds5 to relieve the inhibition of intersister SC formation, but these other meiosis-specific subunits could also provide cues for SC formation or have acquired the roles entirely. Further experiments assessing the chromosomal binding of one or both Pds5 orthologues in vertebrates and the consequence of their absence can distinguish between these possibilities.
A remarkable feature of meiotic recombination is its preference for using homologues as repair templates during DSB repair (Haber, 2000). This distinction must be made to ensure that DSBs promote synapsis between homologues rather than the more proximal sister chromatid. One model posits that fusion of sister chromatid axes removes the choice so that only the homologue presents a target for exchange and SC formation (for review see Zickler and Kleckner, 1999). In such a model, Pds5 could be directly required for axial fusion or could promote it by modulating the activity of other chromosomal factors, such as cohesin. Indeed, our EM analysis of the chromosome axis in Pds5-depleted cells suggests that sister axes are apart. Similarly, sister axes are often split in Sordaria macrospora cells bearing a thermosensitive allele of spo76-1/pds5 (van Heemst et al., 1999; Storlazzi et al., 2003). This splitting provides two sister axes in close proximity and, therefore, a potential substrate on which DSBs can nucleate intersister SC formation. During homologue pairing, discrete loci initiate SC formation simultaneously along the chromosome (Zickler, 2006). Localized sister chromatid and axial separation is thought to be part of the process used for homologue exchange (for review see Zickler and Kleckner, 1999). Paradoxically, localized sister separation would seem to provide a sister template to compete with the homologue for pairing rather than promoting it. One solution to this paradox is that axial separation is a consequence of localized dissolution of cohesion, but the separated sisters are then blocked for cohesion reestablishment. We propose that Pds5 exerts its inhibitory function at this step. Cohesin may remain bound but in a form that is inhibited for cohesion reestablishment, as was previously proposed for the regulation of S phase cohesion (Guacci, 2007). Work from both budding and fission yeast shows that Pds5 serves as an inhibitor to cohesion establishment by opposing the action of the conserved establishment factor Eco1 on cohesin (Tanaka et al., 2001; Rowland et al., 2009; Sutani et al., 2009). This result could explain how Pds5 acts as an inhibitor of cohesion and, as such, potentially of intersister SC formation even after sister axial splitting occurred as part of homologue exchange.
One might expect that sister chromatid exchange would increase in pds5-null cells at the expense of homologue exchange because SCs form between sisters rather than homologues. Previous work on pds5 thermosensitive alleles suggested that Pds5 serves to bias recombination toward interhomologue exchange rather than intersister exchange (van Heemst et al., 1999; Storlazzi et al., 2003; Kateneva and Dresser, 2006), which is also consistent with such an expectation, but we do not see evidence for such a shift because the level of unequal sister chromatid exchange remains low in Pds5-depleted cells compared with that in the wild type (unpublished data). Moreover, we find that DSBs are not completely repaired in Pds5-depleted cells in budding yeast, suggesting that intersister SC formation alone is insufficient to promote sister chromatid exchange/repair. Cohesin is essential for efficient repair of DSBs in vegetative cells (for review see Onn et al., 2008). Given that Pds5 interacts with cohesin to modulate cohesin function, the impairment of DSB repair in the absence of Pds5 during meiosis is not surprising. In addition, only part of the machinery necessary for sister exchange may be set up for intersister SC formation in pds5-null cells, possibly leading to the persistence of DSBs.
A model for chromosome structure in which chromosome condensation is regulated by cohesin was first proposed more than a decade ago (Guacci et al., 1997). It posits that cohesin binds at intervals along the chromosome arms, creating DNA loops between cohesin-binding sites that can be compacted. In this model, higher cohesin density forms smaller loops, whereas lower cohesin density forms larger loops, which would result in lesser and greater axial compaction, respectively (Guacci et al., 1997). Experimental data consistent with this view came from studies of meiotic chromosomes (Ding et al., 2006; Novak et al., 2008). In both budding and fission yeast cells lacking Pds5, meiotic chromosomes hypercondense (Ding et al., 2006; this study). In fission yeast pds5 mutants, Rec8 still binds chromosomes, but the binding sites are more widely spaced, a result that is consistent with the idea that loop size is inversely proportional to axial condensation (Ding et al., 2006). In budding yeast, cohesin binding to chromosomes appears to be normal, but sister chromatid cohesion is partially lost (this study; unpublished data). Importantly, chromosomes still hypercondense in Pds5-depleted cells when ectopically expressed Mcd1 replaces Rec8. These data suggest that cohesin is required to provide an axial template to permit proper chromosome compaction. In vertebrate cells, chromosomes become hypercondensed when the meiosis-specific cohesin subunit SMC1β has been deleted (Revenkova et al., 2004; Novak et al., 2008). Unlike budding yeast, vertebrate cells have high levels of mitotic cohesin, and REC8-containing meiotic cohesin is still present in cells lacking SMC1β (Novak et al., 2008). Because these complexes bind chromosomes, one or both types of cohesin are probably able to serve as axis templates and permit loop formation (Novak et al., 2008). The reduced cohesin activity in vertebrates that leads to a partial loss of sister chromatid cohesion could mimic a yeast Pds5 depletion phenotype, resulting in axial hypercompaction and the formation of heterogeneous chromatin loops.
Materials and methods
Yeast strains
Yeast strains used in this study are diploids isogenic to SK1, except the sir2Δ strains, which are haploids (Table S1). To create the PCLB2PDS5 allele, we used a PCR-based approach (Yu and Koshland, 2005) with the primers 5′-TTTAGCCGCCAAGGGAAAATATGCACTACCCGGAATGATGGCGTTAAAGCATAGGCCACTAGTGGATCTG-3′ and 5′-GATATTATAGGTGAGTTAAACTTCAGTTTAGTAACAGCACCTTTAGCCATAGCGTAATCTGGAACGTCATA-3′. We used the same method to create PCUP1MCD1 using plasmid pHG40 as a template in PCR reactions. The primers used were 5′-TGATGGTGATGGAACTCCAATTACCTAATCAAAAATATGAATGTTTATGCGCATAGGCCACTAGTGGATCTG-3′ and 5′-TTGGTGGCAAGTCTTAAAACAGTAAGACGTTGAGGATTTTCTGTAACAGCAGCGTAATCTGGAACGTC-3′. A 1-Kb DNA sequence upstream of the CUP1 open reading frame was used to replace the endogenous MCD1 promoter. All strains were confirmed by yeast colony PCR and were backcrossed to the wild type. To mark CEN5 with GFP, we incorporated tandem arrays of tetO at the URA3 locus and expressed tetR-GFP (Michaelis et al., 1997). To mark CEN4 and TEL4 with GFP, we incorporated tandem arrays of lacO at the TRP1 locus and a telomere IV locus simultaneously and expressed lacI-GFP (Milutinovich et al., 2007). To induce haploid cells to enter meiosis, we deleted the SIR2 gene with pClonatMX4 using the aforementioned PCR-based method. Primers are available upon request.
Yeast culture methods
Before yeast cells were induced to enter meiosis in 2% KOAC, they were grown in the YPA medium with vigorous shaking for ∼12 h to an optical density (λ = 600) of 1.5. All yeast cultures were incubated at 30°C. To induce PCUP1MCD1 expression during meiosis, 60 µM (final concentration) CuSO4 was added to the sporulation medium after induction of meiosis.
Meiotic nuclear spread and immunofluorescence
To prepare surface spreads of yeast nuclei, aliquots were withdrawn from meiotic cultures and subjected to spheroplasting by lyticase (Sigma-Aldrich). Isolated nuclei were spread on a clean microscope slide and fixed immediately with 4% paraformaldehyde for 20 min (Yu and Koshland, 2005). Fixed nuclear spreads were incubated with primary rabbit polyclonal antibodies, including anti-Pds5 (Noble et al., 2006), anti-Red1 and anti-Zip1 (provided by G.S. Roeder, Yale University, New Haven, CT), anti-Rad51 (provided by D.K. Bishop, University of Chicago, Chicago, IL), and anti-Hop1 (provided by N. Hollingsworth, Stony Brook University, Stony Brook, NY), as well as mouse monoclonal antibodies, including anti–α-tubulin (YOL1/34; AbD Serotec), anti-HA (12CA5; Roche), and anti-Myc (9E10; Roche). Secondary antibodies (FITC-conjugated goat anti–rabbit, rhodamine-conjugated goat anti–mouse, and Cy3-conjugated goat anti–rat) were purchased from Jackson ImmunoResearch Laboratories. Chromosomal DNA was stained by DAPI. All fluorescence images were acquired with a Plan Apochromat 100× 1.40 NA objective lens mounted on a motorized epifluorescence microscope (AxioImager; Carl Zeiss, Inc.) by AxioVision software (Carl Zeiss, Inc.) at room temperature. Chromosome V length in Figs. 3 C and 6D was determined using AxioVision measurement tools by tracking Rec8- and Mcd1-stained chromosome axes, respectively. Chromosome V length was measured in spread nuclei only when its ends were clearly separated from other chromosomes. The length of the chromosome IV fragment in Fig. 3 E was determined by the distance between two marked GFP spots. Displayed images were processed with AxioVision for pseudo coloring.
EM
For EM analysis of meiotic chromosomes, yeast nuclei were spread on formvar (0.3% wt/vol)-coated slides and stained with AgNO3 (Dresser and Giroux, 1988). Silver-stained nuclear spreads were transferred to 75-mesh copper grids and visualized under a transmission electron microscope (CM120; Phillips) at 80 kV. A 12-bit charge-coupled device camera (Tem-Cam F224; Tietz) was used to acquire EM images. To determine the width of SCs, we used measurement tools provided by the IPLab software (BD). Lengths of SC structures that could be individualized in 10 representative cells were measured. We used the IPLab software to adjust the contrast of displayed EM images.
Protein extraction and immunoblotting
To prepare yeast protein extracts, aliquots were withdrawn from meiotic cultures and incubated with equal volumes of trichloroacetic acid for 10 min on ice. Cell pellets were resuspended in acetone and air dried. Standard SDS-PAGE and Western blotting were performed. Polyclonal antibodies against Pds5 and Mcd1 (1:20,000) were used to detect those proteins in protein extracts (Noble et al., 2006). For HA-tagged proteins, we used a monoclonal anti-HA antibody (12CA5; Roche) at 1:5,000. A β-tubulin antibody (1:10,000) was used to detect β-tubulin for a loading control.
ChIP
Yeast cells were fixed with 1% formaldehyde for 2 h and followed by protein extraction and immunoprecipitation. We used a Pds5-specific antibody and an anti-HA (12CA5; Roche) antibody for immunoprecipitation of Pds5 and Rec8-3HA, respectively. Chromosomal DNA cross-linked to Pds5 and Rec8-3HA was purified by phenol chloroform extraction. A semiquantitative PCR method was used to detect Pds5 and Rec8 binding at the following four cohesin-associated regions: centromere 1 (CEN1), centromere 3 (CEN3), a cohesin site at the MAT locus (CARC3), and a cohesin site on chromosome XII (CARL2; Glynn et al., 2004). Primers used in the ChIP assay are available upon request.
Physical analysis of DSBs
Yeast DNA extracts were prepared from synchronous meiotic cultures, digested with AseI, and separated on a 0.8% agarose gel. A 1.6-kb probe was used to detect the hotspots at the YCR047c/YCR048w locus by Southern blotting (Yu and Koshland, 2003). Images were scanned and quantified by the Typhoon (GE Healthcare).
Online supplemental material
Fig. S1 shows chromosome morphology in ndt80Δ and hop1Δ cells during meiosis. Fig. S2 shows immunoblot analysis of Rec8 protein levels in wild-type and PCLB2PDS5 cells. Fig. S3 shows S phase progression during meiosis by FACS analysis. Fig. S4 shows Zip1 localization in zip3Δ and Red1 in rec8Δ cells during meiosis. Fig. S5 shows localization of Zip1 and Sgo1 in spo11Δ cells. Table S1 shows the yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200810107/DC1.
Acknowledgments
We thank H.W. Bass and D.M. Gilbert for discussions and comments. We are grateful to G.S. Roeder for Red1 and Zip1 antibodies and D.K. Bishop for the Rad51 antibody. F. Contreras and K. Riddle provided technical assistance. A.B. Thistle assisted in text editing.
This work was supported in part by the March of Dimes Foundation (grant #5-FY08-111) and the Florida Biomedical Research Program (grant 08BN-08).
Footnotes
Abbreviations used in this paper:
- CE
- central element
- ChIP
- chromatin immunoprecipitation
- DSB
- double-strand break
- LE
- lateral element
- SC
- synaptonemal complex
References
- Buonomo S.B., Clyne R.K., Fuchs J., Loidl J., Uhlmann F., Nasmyth K. 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin.Cell. 103:387–398 [DOI] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P.O., Herskowitz I. 1998. The transcriptional program of sporulation in budding yeast.Science. 282:699–705 [DOI] [PubMed] [Google Scholar]
- Denison S.H., Käfer E., May G.S. 1993. Mutation in the bimD gene of Aspergillus nidulans confers a conditional mitotic block and sensitivity to DNA damaging agents.Genetics. 134:1085–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D.Q., Sakurai N., Katou Y., Itoh T., Shirahige K., Haraguchi T., Hiraoka Y. 2006. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast.J. Cell Biol. 174:499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser M.E., Giroux C.N. 1988. Meiotic chromosome behavior in spread preparations of yeast.J. Cell Biol. 106:567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijpe M., Heyting C., Gross B., Jessberger R. 2000. Association of mammalian SMC1 and SMC3 proteins with meiotic chromosomes and synaptonemal complexes.J. Cell Sci. 113:673–682 [DOI] [PubMed] [Google Scholar]
- Glynn E.F., Megee P.C., Yu H.-G., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L. 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae.PLoS Biol. 2:e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V. 2007. Sister chromatid cohesion: the cohesin cleavage model does not ring true.Genes Cells. 12:693–708 [DOI] [PubMed] [Google Scholar]
- Guacci V., Koshland D., Strunnikov A. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae.Cell. 91:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. 2000. Partners and pathwaysrepairing a double-strand break.Trends Genet. 16:259–264 [DOI] [PubMed] [Google Scholar]
- Hartman T., Stead K., Koshland D., Guacci V. 2000. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae.J. Cell Biol. 151:613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N.M., Byers B. 1989. HOP1: a yeast meiotic pairing gene.Genetics. 121:445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kateneva A.V., Dresser M.E. 2006. Sister chromatid cohesion remodeling and meiotic recombination.Cell Cycle. 5:467–471 [DOI] [PubMed] [Google Scholar]
- Klein F., Mahr P., Galova M., Buonomo S.B., Michaelis C., Nairz K., Nasmyth K. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis.Cell. 98:91–103 [DOI] [PubMed] [Google Scholar]
- Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F. 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription.Nature. 430:573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J., Nairz K., Klein F. 1991. Meiotic chromosome synapsis in a haploid yeast.Chromosoma. 100:221–228 [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano T. 2005. Dynamic molecular linkers of the genome: the first decade of SMC proteins.Genes Dev. 19:1269–1287 [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano M., Hirano T. 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion.Genes Dev. 12:1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids.Cell. 91:35–45 [DOI] [PubMed] [Google Scholar]
- Milutinovich M., Unal E., Ward C., Skibbens R.V., Koshland D. 2007. A multi-step pathway for the establishment of sister chromatid cohesion.PLoS Genet. 3:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble D., Kenna M.A., Dix M., Skibbens R.V., Unal E., Guacci V. 2006. Intersection between the regulators of sister chromatid cohesion establishment and maintenance in budding yeast indicates a multi-step mechanism.Cell Cycle. 5:2528–2536 [DOI] [PubMed] [Google Scholar]
- Novak I., Wang H., Revenkova E., Jessberger R., Scherthan H., Höög C. 2008. Cohesin Smc1β determines meiotic chromatin axis loop organization.J. Cell Biol. 180:83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I., Heidinger-Pauli J.M., Guacci V., Unal E., Koshland D.E. 2008. Sister chromatid cohesion: a simple concept with a complex reality.Annu. Rev. Cell Dev. Biol. 24:105–129 [DOI] [PubMed] [Google Scholar]
- Page S.L., Hawley R.S. 2004. The genetics and molecular biology of the synaptonemal complex.Annu. Rev. Cell Dev. Biol. 20:525–558 [DOI] [PubMed] [Google Scholar]
- Panizza S., Tanaka T., Hochwagen A., Eisenhaber F., Nasmyth K. 2000. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion.Curr. Biol. 10:1557–1564 [DOI] [PubMed] [Google Scholar]
- Prieto I., Suja J.A., Pezzi N., Kremer L., Martínez-A C., Rufas J.S., Barbero J.L. 2001. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I.Nat. Cell Biol. 3:761–766 [DOI] [PubMed] [Google Scholar]
- Ren Q., Yang H., Rosinski M., Conrad M.N., Dresser M.E., Guacci V., Zhang Z. 2005. Mutation of the cohesin related gene PDS5 causes cell death with predominant apoptotic features in Saccharomyces cerevisiae during early meiosis.Mutat. Res. 570:163–173 [DOI] [PubMed] [Google Scholar]
- Revenkova E., Eijpe M., Heyting C., Gross B., Jessberger R. 2001. Novel meiosis-specific isoform of mammalian SMC1.Mol. Cell. Biol. 21:6984–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E., Eijpe M., Heyting C., Hodges C.A., Hunt P.A., Liebe B., Scherthan H., Jessberger R. 2004. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination.Nat. Cell Biol. 6:555–562 [DOI] [PubMed] [Google Scholar]
- Rockmill B., Roeder G.S. 1988. RED1: a yeast gene required for the segregation of chromosomes during the reductional division of meiosis.Proc. Natl. Acad. Sci. USA. 85:6057–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland B.D., Roig M.B., Nishino T., Kurze A., Uluocak P., Mishra A., Beckouët F., Underwood P., Metson J., Imre R., et al. 2009. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity.Mol. Cell. 33:763–774 [DOI] [PubMed] [Google Scholar]
- Smith K.N., Penkner A., Ohta K., Klein F., Nicolas A. 2001. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis.Curr. Biol. 11:88–97 [DOI] [PubMed] [Google Scholar]
- Stead K., Aguilar C., Hartman T., Drexel M., Meluh P., Guacci V. 2003. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion.J. Cell Biol. 163:729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A., Tessé S., Gargano S., James F., Kleckner N., Zickler D. 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division.Genes Dev. 17:2675–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A., Tesse S., Ruprich-Robert G., Gargano S., Pöggeler S., Kleckner N., Zickler D. 2008. Coupling meiotic chromosome axis integrity to recombination.Genes Dev. 22:796–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I., Vorlaufer E., Gieffers C., Peters B.H., Peters J.M. 2000. Characterization of vertebrate cohesin complexes and their regulation in prophase.J. Cell Biol. 151:749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T., Kawaguchi T., Kanno R., Itoh T., Shirahige K. 2009. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction.Curr. Biol. 19:492–497 [DOI] [PubMed] [Google Scholar]
- Sym M., Roeder G.S. 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein.Cell. 79:283–292 [DOI] [PubMed] [Google Scholar]
- Sym M., Engebrecht J.A., Roeder G.S. 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis.Cell. 72:365–378 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Hao Z., Kai M., Okayama H. 2001. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism.EMBO J. 20:5779–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T., Roeder G.S. 2005. A synaptonemal complex protein promotes homology-independent centromere coupling.Science. 308:870–873 [DOI] [PubMed] [Google Scholar]
- van Heemst D., James F., Pöggeler S., Berteaux-Lecellier V., Zickler D. 1999. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs.Cell. 98:261–271 [DOI] [PubMed] [Google Scholar]
- van Heemst D., Kafer E., John T., Heyting C., van Aalderen M., Zickler D. 2001. BimD/SPO76 is at the interface of cell cycle progression, chromosome morphogenesis, and recombination.Proc. Natl. Acad. Sci. USA. 98:6267–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Yoder J., Antoshechkin I., Han M. 2003. Caenorhabditis elegans EVL-14/PDS-5 and SCC-3 are essential for sister chromatid cohesion in meiosis and mitosis.Mol. Cell. Biol. 23:7698–7707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Ajimura M., Padmore R., Klein C., Kleckner N. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae.Mol. Cell. Biol. 15:6572–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Beasley M.D., Warren W.D., van der Horst G.T., McKay M.J. 2005. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis.Dev. Cell. 8:949–961 [DOI] [PubMed] [Google Scholar]
- Yu H.-G., Koshland D.E. 2003. Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages.J. Cell Biol. 163:937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.-G., Koshland D.E. 2005. Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis.Cell. 123:397–407 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ren Q., Yang H., Conrad M.N., Guacci V., Kateneva A., Dresser M.E. 2005. Budding yeast PDS5 plays an important role in meiosis and is required for sister chromatid cohesion.Mol. Microbiol. 56:670–680 [DOI] [PubMed] [Google Scholar]
- Zickler D. 2006. From early homologue recognition to synaptonemal complex formation.Chromosoma. 115:158–174 [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. 1999. Meiotic chromosomes: integrating structure and function.Annu. Rev. Genet. 33:603–754 [DOI] [PubMed] [Google Scholar]