Human cytomegalovirus uses an E3 ubiquitin ligase to divert MHC I molecules into the ER-associated degradation pathway for destruction.
Abstract
The US2 and US11 gene products of human cytomegalovirus promote viral evasion by hijacking the endoplasmic reticulum (ER)–associated degradation (ERAD) pathway. US2 and US11 initiate dislocation of newly translocated major histocompatibility complex class I (MHC I) from the ER to the cytosol for proteasome-mediated degradation, thereby decreasing cell surface MHC I. Despite being instrumental in elucidating the mammalian ERAD pathway, the responsible E3 ligase or ligases remain unknown. Using a functional small interfering RNA library screen, we now identify TRC8 (translocation in renal carcinoma, chromosome 8 gene), an ER-resident E3 ligase previously implicated as a hereditary kidney cancer gene, as required for US2-mediated MHC I ubiquitination. Depletion of TRC8 prevents MHC I ubiquitination and dislocation by US2 and restores cell surface MHC I. TRC8 forms an integral part of a novel multiprotein ER complex that contains MHC I, US2, and signal peptide peptidase. Our data show that the TRC8 E3 ligase is required for MHC I dislocation from the ER and identify a new complex associated with mammalian ERAD.
Introduction
The ER quality control system ensures that nonnative or misfolded proteins are targeted for dislocation from the ER to the cytosol and degraded by the proteasome as part of the ER-associated degradation (ERAD) pathway (Vembar and Brodsky, 2008; Hirsch et al., 2009). ERAD is implicated in a wide variety of cellular processes, including cystic fibrosis, the serpinopathies such as α1-antitrypsin deficiency, and aggregation disorders, as seen with neurodegenerative diseases. An essential step in ERAD is ubiquitination of the protein substrate, a process which precedes or accompanies retrotranslocation of integral membrane proteins. The E3 ligase is the key enzyme in the dislocation pathway and crucial for substrate extraction, recognition, and degradation by the proteasome. The generation of a polyubiquitinated substrate requires three enzymes. The E1 ubiquitin–activating enzyme covalently activates and adds ubiquitin to an E2 ubiquitin–conjugating enzyme. Ubiquitin is then transferred from the ubiquitin-charged E2 to either the substrate or the growing ubiquitin chain by the action of the E3 ubiquitin ligase, which therefore confers the main element of specificity to the reaction. Current estimates suggest that there are two E1 enzymes, around 40 E2 enzymes, and several hundred E3 ligases, including the HECT (homologous to E6-AP C terminus) domain E3s, the RING (really interesting new gene) E3s, and the U box proteins (Li et al., 2008).
In yeast, the multimembrane-spanning, ER-resident Doa10 and Hrd1 (hydroxymethylglutaryl-CoA reductase degradation 1) E3 ligases are implicated in the degradation of every studied ubiquitinated ERAD substrate (Vembar and Brodsky, 2008). This contrasts with the greatly expanded mammalian repertoire of ERAD E3 ligases (Kostova et al., 2007), which include yeast Hrd1 homologues HRD1 and gp78, whereas MARCHVI (TEB4) is the Doa10 orthologue. Additional E3 ligases of the mammalian ERAD pathway include RMA1, which cooperates with gp78 to target mutant cystic fibrosis transport regulator for ERAD (Younger et al., 2006). Some of the additional 40–50 uncharacterized RING E3 ligases with predicted transmembrane segments are also likely to be involved in ERAD.
US2 and US11 are two well-characterized viral genes from human cytomegalovirus whose expression causes down-regulation of cell surface major histocompatibility complex class I (MHC I; Wiertz et al., 1996a,b). The US2 and US11 viral gene products have been instrumental in both defining the mammalian ERAD pathway and identifying novel components of the ERAD system. MHC I molecules play an essential role in the adaptive immune system, specifically in the defense against intracellular infections. They bind small peptides in the lumen of the ER and traffic them to the cell surface for presentation to cytotoxic T lymphocytes. Peptides derived from viruses are recognized as foreign, ensuring infected cells are killed by circulating cytotoxic T lymphocytes, thus preventing viral dissemination. The importance of the MHC I pathway is highlighted by the multiple strategies used particularly by the large double-stranded DNA viruses to prevent the presentation of viral peptides by disrupting the assembly and trafficking of MHC I, leading to decreased cell surface MHC I expression.
The US2 and US11 genes encode ER-resident membrane glycoproteins, which bind newly synthesized MHC I in the lumen of the ER. They initiate the dislocation of MHC I from the ER back to the cytosol before proteasome-mediated degradation (Wiertz et al., 1996a,b). Despite the apparent similarities in US2- and US11-mediated MHC I degradation, these two viral proteins use distinct ERAD pathways for MHC I dislocation. The US11 ERAD pathway is the better characterized and helped identify the Derlin-1–SEL1L complex (Lilley and Ploegh, 2004; Ye et al., 2004; Mueller et al., 2006), emphasizing the utility of these viral genes in defining ERAD components (Lilley and Ploegh, 2004; Mueller et al., 2006). Derlin-1 associates with the two ERAD E3 ligases HRD1 and gp78 (Lilley and Ploegh, 2005), although their involvement in US11-mediated MHC I dislocation has not been elucidated (Kikkert et al., 2004). SEL1L, the mammalian homologue of yeast Hrd3p, is required for US11-mediated MHC I dislocation (Mueller et al., 2006) and also helped identify additional components of the degradation complex (Mueller et al., 2008). Less progress has been made on the Derlin-independent US2 dislocation pathway. Signal peptide peptidase (SPP), a presenilin-like aspartyl protease which cleaves polypeptides within the membrane, is essential for US2-mediated MHC I degradation (Loureiro et al., 2006), although its function is unclear.
Despite US2 showing a critical requirement for a functional ubiquitin system (Hassink et al., 2006) and ubiquitination being detected in US2-mediated MHC I degradation, the E3 ligase responsible for US2-mediated MHC I ubiquitination has remained elusive (Shamu et al., 2001; Furman et al., 2003; Hassink et al., 2006). US2 has no intrinsic E3 ligase activity itself but recruits newly synthesized MHC I molecules to the ERAD pathway. Therefore, the cellular ubiquitination machinery must participate in MHC I dislocation, but immunopurifications of the US2 complex have not identified candidate E3 ligases. We used a library of siRNAs that target ubiquitin E3 ligases in a genetic screen to identify siRNAs capable of rescuing US2-mediated MHC I degradation. The screen revealed that depletion of the TRC8 (translocation in renal carcinoma, chromosome 8 gene) E3 ligase leads to a loss of MHC I ubiquitination and dislocation and a dramatic rescue of MHC I to the cell surface in US2-expressing cells. These results implicate TRC8 as the E3 ligase required for MHC I ubiquitination and identify a novel ER complex involved in mammalian ERAD.
Results and discussion
An siRNA screen identifies TRC8 as the E3 ligase required for US2-mediated MHC I dislocation
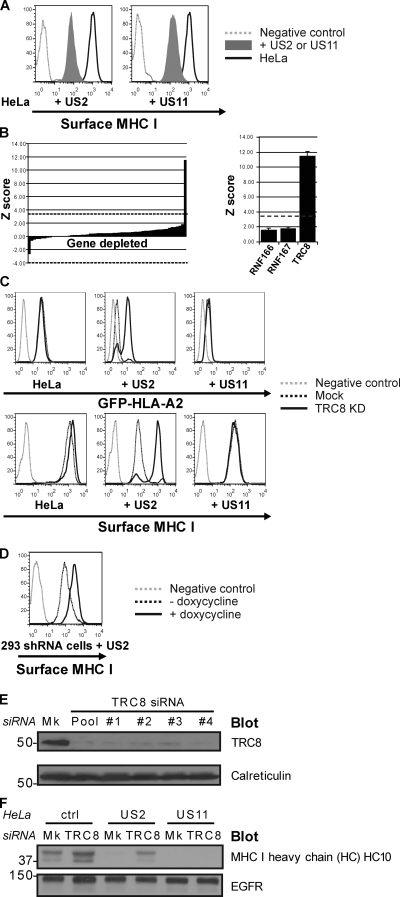
To identify the E3 ligase or ligases responsible for US2-mediated MHC I ubiquitination, we screened a custom siRNA library targeting putative members of the E3 ligase family (consisting of bioinformatically collated RING, HECT, and U box E3 ligases; Li et al., 2008) on HeLa cells stably expressing a lumenal GFP–MHC I heavy chain fusion protein (GFP-HLA-A2) and US2. Expression of US2 or US11 down-regulates cell surface MHC I (Fig. 1 A) as well as the GFP reporter (Fig. S1). We predicted that depletion of an E3 ligase responsible for US2-mediated MHC I ubiquitination would rescue GFP-HLA-A2 from proteasome-mediated degradation. Therefore, we used GFP-HLA-A2 fluorescence as an optical readout in a flow cytometry–based screen to identify the cellular E3 ligase recruited by US2. A related emerald GFP–MHC I chimera was previously shown to behave similarly to endogenous MHC I, including being targeted for retrograde translocation and proteasome-mediated degradation by US2 (Fiebiger et al., 2002). In control cells, GFP-HLA-A2 is expressed at the cell surface; expression of US2 causes proteasome-mediated degradation of GFP-HLA-A2 with a concomitant loss of fluorescence signal, which is rescued after proteasome inhibition (Fig. S1).
Figure 1.
An siRNA ubiquitin ligase screen identifies a requirement for TRC8 in MHC I dislocation. (A–F) siRNA-mediated TRC8 depletion rescues MHC I from US2-mediated dislocation. (A) Cytofluorometric analysis of cell surface MHC I in HeLa and US2- or US11-expressing HeLa cells demonstrating MHC I down-regulation. (B) Of the 373 E3 ligases screened, only TRC8 depletion gave a significant rescue in fluorescence signal with a Z score of 11.57. The bar chart shows the Z scores for the 60 genes on the TRC8-containing library plate. The dashed lines indicate the Bonferroni-corrected p-value threshold of 3.82 for the entire siRNA library. The graph on the right shows that the TRC8 siRNA pool caused the only significant increase in fluorescence signal. Error bars indicate standard error of the mean. (C) Cytofluorometric analysis of GFP (top) and MHC I (bottom) in GFP-HLA-A2–expressing HeLa, +US2, and +US11 cells after mock or TRC8 siRNA depletion. KD, knockdown. (D) US2-mediated MHC I down-regulation is also rescued by a doxycycline-inducible anti-TRC8 shRNA. Cytofluorometric analysis of MHC I in HEK-293–US2 cells (black dotted line) and doxycycline-induced TRC8 shRNA expression (black line) is shown. (E) Immunoblot analysis of endogenous TRC8 and control calreticulin levels in cells after siRNA mock (Mk) or TRC8 depletion with a pool of or individual (#1–4) TRC8-specific oligonucleotides. (F) Immunoblot analysis of MHC I heavy chain and control (ctrl; EGF receptor [EGFR]) in US2- and US11-expressing HeLa cells after mock or TRC8 siRNA depletion. MHC I heavy chain is rescued after TRC8 depletion. (E and F) Molecular mass is indicated in kilodaltons.
HeLa GFP-HLA-A2 cells stably expressing US2 were screened with a custom RNAi library using high throughput 96-well flow cytometry. Of the 373 ligases screened, only siRNA-mediated depletion of TRC8 (Z score 11.57) gave a significant rescue in fluorescence signal (Fig. 1, B and C, top) above the Bonferroni-corrected p-value threshold (P = 0.05) of 3.82. This rescue in fluorescence signal was achieved with four independent TRC8 siRNAs targeting oligonucleotides (Fig. S2 A). In the same US2-expressing HeLa cells, siRNA-mediated TRC8 depletion completely restored cell surface expression of endogenous MHC I (Fig. 1 C, bottom), an effect seen with all four TRC8-specific oligonucleotides (Fig. S2 B), as did doxycycline-induced TRC8 short hairpin RNA (shRNA) expression in HEK-293–US2 cells (Fig. 1 D). In contrast, in HeLa GFP-HLA-A2 US11-expressing cells, TRC8 depletion had no effect on either GFP-HLA-A2 fluorescence or MHC I cell surface expression (Fig. 1 C), implying that US2 utilizes a specific and distinct ERAD pathway.
Effective depletion of TRC8 was confirmed for all four individual siRNA oligonucleotides of the original pool by immunoblotting for endogenous TRC8, with no effect against unrelated calreticulin, confirming specificity (Fig. 1 E). Although depletion of TRC8 had little effect on endogenous MHC I heavy chain expression in control or US11-expressing HeLa cells, a rescue of MHC I heavy chain is seen by immunoblot in TRC8 siRNA–treated US2-expressing cells (Fig. 1 F).
TRC8 depletion rescues MHC I from the cytosol to the membrane fraction and is responsible for MHC I ubiquitination
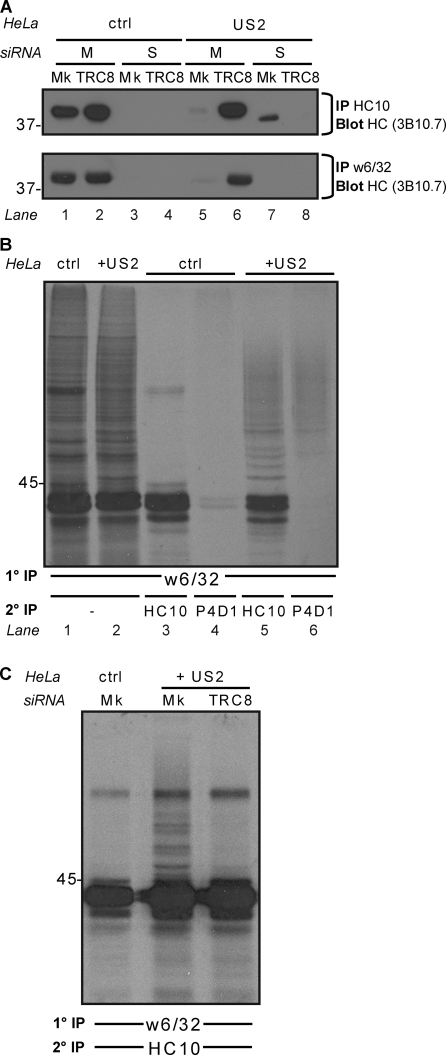
The signature of US2-mediated degradation of MHC I heavy chains is the appearance of a nonmembrane-bound, soluble, deglycosylated MHC I heavy chain, which is visualized as a faster migrating species in the presence of proteasome inhibitors (Wiertz et al., 1996b). The increased mobility results from cytosolic peptide: N-glycanase–mediated removal of the single N-linked MHC I glycan (Blom et al., 2004). To further determine the effect of TRC8 on MHC I degradation, we fractionated cells to separate membrane-bound and soluble cytosolic heavy chain (Hughes et al., 1997). After solubilization in Triton X-100, MHC I heavy chains were immunoprecipitated and visualized by immunoblotting. Immunoprecipitations with the HC10 antibody identified heavy chain in the membrane fraction of control cells (Fig. 2 A, lane 1). In contrast, in US2-expressing cells, heavy chain is predominantly isolated from the soluble cytosolic pool as the deglycosylated species (Fig. 2 A, lane 7). TRC8 depletion from US2-expressing cells shows a rescue of MHC I from the cytosolic to the membrane fraction as detected by the HC10 antibody, which recognizes unfolded MHC I (Fig. 2 A, lanes 6 and 8). This TRC8-mediated rescue was confirmed by immunoprecipitation with the MHC I–specific antibody (w6/32), which recognizes a conformation-dependent epitope on the lumenal domain of MHC I and showed a rescue of folded MHC I to the membrane fraction (Fig. 2 A, bottom, lane 5 vs. lane 6), which is consistent with the flow cytometry data (Fig. 1 C). Therefore, TRC8 is an ER ligase required for US2-mediated MHC I degradation.
Figure 2.
US2-dependent MHC I dislocation and ubiquitination are abolished after TRC8 depletion. (A) Cellular fractionation shows that TRC8 depletion rescues MHC I from the soluble to the membrane fraction. siRNA mock (Mk) or TRC8-depleted cells ± US2 were separated into membrane (M) and soluble (S) fractions. MHC I was immunoprecipitated (mAb HC10 or w6/32) from each fraction and visualized by immunoblot analysis (mAb 3B10.7). After TRC8 depletion, conformational MHC I is found in the membrane fraction. (B and C) TRC8 depletion prevents MHC I ubiquitination. (B) Detergent lysates from 20-min radiolabeled control (ctrl) and US2 cells were immunoprecipitated with the MHC I (w6/32) mAb, denatured, and reprecipitated with heavy chain (HC10)– or polyubiquitin (P4D1)-specific mAb. HC10 precipitates were confirmed as ubiquitinated MHC I species. (C) siRNA mock or TRC8-depleted control and US2 cells were treated as in B. Ubiquitinated MHC I species were lost after TRC8 depletion. (A–C) Molecular mass is indicated in kilodaltons. 1°, primary; 2°, secondary; HC, heavy chain; IP, immunoprecipitation.
To determine the effect of TRC8 on MHC I ubiquitination, MHC I molecules were immunoprecipitated from radiolabeled cells with the w6/32 antibody (Furman et al., 2003). Samples were denatured in 1% SDS and reprecipitated with either heavy chain (HC10)– or polyubiquitin (P4D1)-specific antibodies. In US2-expressing but not control cells, reprecipitation with HC10 showed additional higher molecular mass bands above the 44-kD MHC I heavy chain, which were confirmed as ubiquitinated MHC I species by immunoprecipitation with the P4D1 antibody (Fig. 2 B, lane 6). TRC8 depletion of the US2-expressing cells resulted in a 76% loss of these ubiquitinated MHC I species (Fig. 2 C). Collectively, these results show that TRC8 is essential for MHC I dislocation and ubiquitination. After TRC8 depletion, the MHC I is neither ubiquitinated nor retained in the ER but traffics normally to the cell surface despite the presence of US2.
TRC8 RING mutants act as dominant negatives and rescue MHC I back to the cell surface
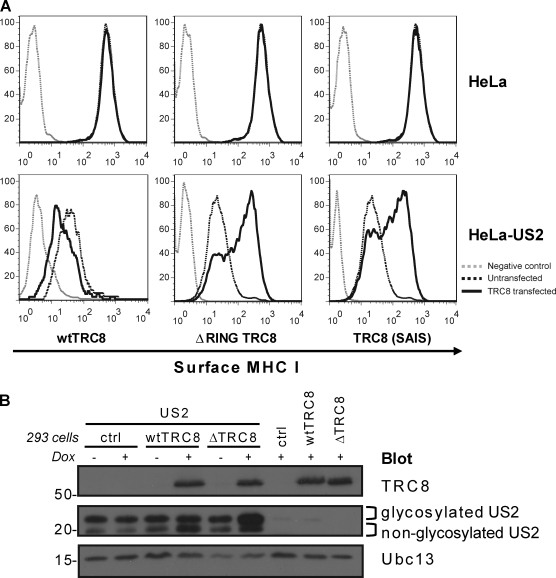
The TRC8 gene was originally identified in a family with hereditary kidney and thyroid cancer caused by a chromosomal translocation t(3;8)(3p14;q24.1); the disruption of TRC8 at 8q24 permitted its positional cloning and further analysis (Gemmill et al., 1998). TRC8 mutations were subsequently identified in sporadic cases of kidney cancer (Gemmill et al., 1998; Poland et al., 2007). TRC8 is a 66-kD ER-resident protein (Gemmill et al., 2002) predicted to span the membrane up to 10 times. Its C terminus has the defining feature of RING-type E3 ligases, a RING-H2 domain which coordinates zinc binding and was shown to catalyze in vitro ubiquitination (Brauweiler et al., 2007). To assess the requirement for a functional RING on TRC8 in vivo activity, we compared overexpression of wild-type and TRC8 RING mutants on cell surface MHC I levels in US2-expressing HeLa cells. Overexpression of wild-type TRC8 potentiated MHC I down-regulation (Fig. 3 A). The opposite effect was seen with both a TRC8 RING-less (ΔRING) and a TRC8 RING mutant (SAIS), in which two of the zinc-coordinating Cys residues are replaced with Ser residues, mutations which are known to destabilize the RING. Overexpression of either of these RING mutants increased cell surface MHC I to normal levels despite the presence of US2 (Fig. 3 A). Furthermore, doxycycline-induced expression of the RING-less TRC8 mutant in HEK-293 cells stabilized the more abundant glycosylated form of US2, whereas induction of wild-type TRC8 caused an increase in the faster migrating deglycosylated form of US2 (Fig. 3 B). Together, these results predict an interaction between TRC8 and US2 (as shown in Fig. 4), with the TRC8 RING mutants acting as dominant-negative forms competing with endogenous TRC8 for binding to US2.
Figure 3.
TRC8 RING mutants act as dominant negative to rescue US2-mediated MHC I down-regulation. (A) Cytofluorometric analysis of MHC I in US2-expressing HeLa cells (black dotted lines) transfected with wild-type (wt) TRC8, ΔRING TRC8, or a RING Cys-Ser mutant TRC8 (SAIS; black lines). In the absence of a functional RING, MHC I is rescued to the cell surface. (B) Overexpression of wild-type TRC8 increases deglycosylated US2, whereas ΔRING TRC8 stabilizes glycosylated US2. Immunoblot analysis of TRC8 and US2 in wild-type TRC8 and HA–ΔRING TRC8–inducible HEK-293 cells ± doxycycline (Dox). Ubc13 is the loading control (ctrl). Molecular mass is indicated in kilodaltons.
Figure 4.
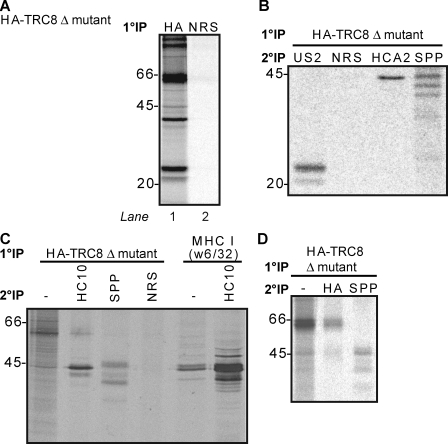
TRC8 forms a multimolecular ER complex with MHC I, US2, and SPP. (A) Doxycycline-induced, HA–ΔRING TRC8 HEK-293 cells transfected with US2 were radiolabeled for 30 min, and lysates were immunoprecipitated with HA or normal rabbit serum (NRS) control. (B) Doxycycline-induced HA–ΔRING TRC8 HEK-293 cells transfected with US2 and SPP were radiolabeled and immunoprecipitated with HA as in A, and samples were denatured and reprecipitated with normal rabbit serum, US2, SPP, or MHC I heavy chain (HCA2). (C) US2-expressing HeLa cells transfected with HA–ΔRING TRC8 were radiolabeled, and immunoprecipitations were performed as in B, or the cells were reprecipitated with MHC I heavy chain (HC10). (D) HA–ΔRING TRC8–overexpressing HEK-293 cells were radiolabeled, and immunoprecipitations were performed as in B. (A–D) Molecular mass is indicated in kilodaltons. 1°, primary; 2°, secondary; IP, immunoprecipitation.
TRC8 associates in the ER with US2, MHC I, and SSP
The identification of proteins binding to E3 ligases is often facilitated by the use of RING mutants. To identify potential TRC8-binding partners, doxycycline-induced, HA-tagged TRC8 ΔRING mutant HEK-293 cells (Brauweiler et al., 2007) transfected with US2 were radiolabeled and immunoprecipitated with anti-HA antibody (Fig. 4 A, lane 1). Together with the TRC8 ΔRING mutant (66 kD), prominent additional bands are detected at 20–23, 40–45, and 97 kD (Fig. 4 A, lane 1). The bands in the 40–45-kD region could represent either MHC I (44 kD) or SPP, which also runs at a similar molecular mass and is required for US2-mediated MHC I dislocation. To further delineate these bands, we repeated the same HA-specific primary immunoprecipitation on radiolabeled cells cotransfected with SPP and, after dissociating the complex in SDS, reprecipitated with US2-, MHC I (HCA2)–, and SPP-specific antibodies (Fig. 4 B). Glycosylated and deglycosylated US2 (20–23 kD) and MHC I heavy chain (44 kD) as well as monomeric (40–45 kD) forms of SPP were identified in association with TRC8. Identical reprecipitations from the same cells expressing wild-type TRC8 confirmed the association of MHC I and SPP with TRC8 (Fig. S3 A), although the MHC I band is weaker. The association of SPP and MHC I with TRC8 was also visualized in radiolabeled TRC8- and SPP-transfected HeLa-US2 cells (Fig. 4 C). Furthermore, SPP and TRC8 interact in the absence of US2 (Fig. 4 D), an interaction which was also confirmed with endogenous SPP (Fig. S3 B).
Our work identifies TRC8 as a novel ERAD ubiquitin E3 ligase recruited by US2 for the ubiquitination and dislocation of MHC I. The rescue of folded, mature MHC I back to the cell surface after TRC8 depletion suggests that for MHC I molecules, ubiquitination is an early, reversible event that precedes dislocation. Depletion of the mammalian E3 ligase gp78 resulted in the accumulation of its substrate CD3-δ in the ER membrane, also suggesting that ubiquitination was an early event that preceded dislocation (Chen et al., 2006). Any residual degradation likely reflects incomplete depletion of TRC8, but a subsidiary role of an additional ligase cannot be excluded.
Both the cytosolic tail and the transmembrane domain of US2 are critical for function (Furman et al., 2002; Chevalier and Johnson, 2003). Its tail interacts with SPP (Loureiro et al., 2006) and the p97 ATPase (Chevalier and Johnson, 2003), and engagement of TRC8 is likely to occur via the transmembrane domain. The role of SPP in the TRC8 complex remains undefined, in particular whether this multimembrane-spanning protein utilizes its aspartic protease activity, as no requirement for SPP catalytic activity in the dislocation process has been reported. It is tempting to speculate that after the SPP-mediated intramembrane cleavage of substrate polypeptides, there is an additional requirement for ubiquitination by TRC8 to target cleaved substrates for dislocation and degradation. However, SPP-mediated intramembrane cleavage has not been seen for either MHC I or US2. An alternative possibility is that the transmembrane segments of the multimembrane-spanning E3 ligases together with SPP either form or contribute to the retrotranslocation channel, as has been suggested for the yeast E3 ligases (Kreft et al., 2006; Kostova et al., 2007).
In addition to its RING domain, TRC8 also possesses a putative sterol-sensing domain, which is identified in a diverse range of membrane proteins, including hydroxymethylglutaryl-CoA reductase and Patched (Gemmill et al., 1998), but not seen in other E3 ligases. Overexpression of TRC8 represses genes involved in cholesterol and fatty acid biosynthesis that are transcriptionally regulated by the sterol response element-binding proteins, although how TRC8 regulates these proteins is unclear (Brauweiler et al., 2007). Sterol-sensing domains play a role in cholesterol homeostasis as well as vesicle trafficking and could therefore provide a link between the proposed role for lipids and lipid droplet formation with the dislocation of proteins from the ER (Ploegh, 2007). However, effective sterol depletion with lipoprotein-depleted serum, mevastatin, mevalonate, and hydroxypropyl-β-cyclodextrin did not affect down-regulation of MHC I by US2 (unpublished data).
The involvement of TRC8 in both hereditary and sporadic cases of clear cell renal carcinoma is intriguing, particularly as TRC8 is the second E3 ligase, after the von Hippel-Lindau gene product, to be implicated in the etiology of renal tumors. Because disruption of TRC8 predisposes to renal cell carcinoma, increasing levels of proteins usually disposed of by TRC8 may be involved in tumor development, making the identification of physiological substrates of this novel ERAD E3 ligase all the more imperative.
Materials and methods
RNAi screening
Cells were reverse transfected with a custom E3 ubiquitin ligase siRNA library (Thermo Fisher Scientific) using Oligofectamine (Invitrogen). Each gene was targeted with an siRNA pool of four individual oligonucleotides. The 373 candidate E3 ligases were determined bioinformatically using InterPro family accessions (http://www.ebi.ac.uk/interpro), the Human Protein Reference Database (http://www.hprd.org), and National Center for Biotechnology Information Entrez Gene RefSeq accessions (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene). The TRC8 siRNA pool was subsequently deconvoluted into four individual oligonucleotides (oligonucleotide #1 [J-006942-05], 5′-GGGAAAAGCTTGACGATTA-3′; oligonucleotide #2 [J-006942-06], 5′-AGAGAGACTTTACTGTTTA-3′; oligonucleotide #3 [J-006942-07], 5′-GGGAGCCGCTTACAAGAAA-3′; and oligonucleotide #4 [J-006942-08], 5′-TGACAGGCGTCTTGGCTTT-3′; Thermo Fisher Scientific). A custom US2-specific siRNA was transfected as a positive control sequence, 5′-CGATCCGAAGGCCGATTATGG-3′. Cells were cultured for 60 h and then assayed by FACSCalibur (BD). Data were analyzed with FlowJo (Tree Star, Inc.). Z scores were used to standardize our data, which had a normal distribution (99% of data had Z scores between −2 and 2), to allow comparisons both across siRNA plates and between US2 depletion controls. Plates were screened n = 1 or n = 2 when it appeared a hit might be present. Data were normalized to mock-transfected controls (five on each plate), and the mean was taken of repeats where present. Z scores were calculated from the mean and standard deviation of the entire dataset. Using a Bonferroni-corrected p-value (0.05), a Z score of >3.82 is statistically significant. TRC8 depletion yielded a Z score of 11.57.
Cell culture
HeLa cells were stably transduced with pK1 GFP-HLA-A2 (a gift from L. Boyle, University of Cambridge, Cambridge, England, UK; Boyle et al., 2006). GFP-HLA-A2–expressing HeLa cells were subsequently transduced with the retroviral constructs pLZRS US2-IRES-ΔNGFR or pLZRS US11-IRES-ΔNGFR (Hassink et al., 2006). Doxycycline-inducible HEK-293 TRC8 cells were previously described (Brauweiler et al., 2007). In brief, TRC8-HA in GFPC1 (Gemmill et al., 2002) was transferred to pcDNA5/FRT/TO using KpnI and verified by sequence analysis. Site-directed mutagenesis generated all RING mutations. HEK-293 FlpIn lines carrying doxycycline-inducible constructs were generated by cotransfection of pcDNA5/FRT/TO with pOG44 encoding flippase-recombinase. After 2 d, recombinant cells were selected in 50 µg/ml hygromycin B (Invitrogen) and 5 µg/ml blasticidin (Invitrogen).
Antibodies
The following antibodies were used: rabbit anti-HA (Sigma-Aldrich), rabbit anti-SPP (Abcam), mouse anti-TRC8 (Abnova), mouse anti–ubiquitin P4D1 (Santa Cruz Biotechnology, Inc.), rabbit anti-Ubc13 (AbD Serotec), mouse anticalreticulin (Thermo Fisher Scientific), rabbit anti–EGF receptor (Santa Cruz Biotechnology, Inc.), mouse mAb HC10 (anti–MHC I heavy chain), mouse mAb HCA2 (anti–MHC I heavy chain), mouse w6/32 (anti–MHC I), rat 3B10.7 (anti–MHC I heavy chain), and rabbit anti-US2 (177-5).
Constructs
Tetracycline-inducible shRNA expression constructs were designed based on TRC8 siRNA (Applied Biosystems) and inserted into pSuperior.puro. Reverse complementary oligonucleotides were synthesized with the following sequences: forward, 5′-GATCCGCTTGACGATTATGTCTACTTCAAGAGAGTAGACATAATCGTCAAGCTTTTTTGGCCC-3′; and reverse, 5′-TCGAGGGCCAAAAAAGCTTGACGATTATGTCTACTCTCTTGAAGTAGACATAATCGTCAAGCG-3′. Stable transfectants in HEK-293 FlpIn TRex cells were selected with 3 µg/ml puromycin and 5 µg/ml blasticidin. Human SPP cDNA (Strausberg et al., 2002) was cloned into pCMV SPORT6.0 (a gift from P. St. George-Hyslop, University of Cambridge, Cambridge, England, UK). US2 transfections were performed using pCIneo US2.
Flow cytometry
HeLa or HEK-293 cells were stained with the mAb w6/32 in PBS and visualized with goat anti–mouse Cy5-conjugated secondary antibody (Jackson ImmunoResearch Laboratories). Cells were fixed in PBS with 1% PFA (AnalaR BDH), read on a FACSCalibur (BD), and analyzed in FlowJo.
Radiolabeling and immunoprecipitation
In brief, cells were treated with 20 µM MG132 (EMD) in starve and pulse. Cells were starved for 30 min in Met- and Cys-free medium and labeled with [35S]Met and [35S]Cys for the indicated time periods. N-ethylmaleimide (Sigma-Aldrich) was added to cells 2 min before lysis at 0.5 mM and solubilized in 1% Triton X-100 or 1% digitonin in 10 mM Tris and 150 mM NaCl (TBS), pH 7.4, containing 1 mM iodoacetamide (Sigma-Aldrich), 0.5 mM PMSF (Sigma-Aldrich), and protease inhibitor cocktail tablets (Roche). After a protein A–Sepharose preclear, samples were immunoprecipitated with the indicated antibody and protein A–Sepharose for 1–3 h. For reimmunoprecipitations, samples were dissociated with 1% SDS-TBS and, after SDS sequestration, with 1% Triton X-100 and reimmunoprecipitated for 12 h. After washing in 0.1% lysis buffer, immunoprecipitates were dissociated from the beads at 70°C in SDS-reducing sample buffer, separated by SDS-PAGE, processed for autoradiography with a STORM scanner (GE Healthcare), and analyzed with ImageQuant TL (GE Healthcare). Visualization of ubiquitin bands required autoradiography for 5–14 d.
Cell fractionation and immunoblotting
After a 3-h incubation with MG132 at 37°C, cells were fractionated by freeze–thaw permeabilization (Hughes et al., 1997), and the nuclear fraction was removed. Membranes were pelleted by ultracentrifugation at 100,000 g for 1 h, and both membrane and cytosolic fractions were solubilized in 1% Triton X-100. Samples were immunoprecipitated, separated by SDS-PAGE (8, 9, or 10% Tris-Gly gels), and after transfer onto polyvinylidene fluoride membranes, blocked with 5% dried milk (Marvel; Premier International Foods Ltd) in 0.2% PBS Tween (NBS Biologicals) for 30 min. Membranes were probed with specific primary antibodies for 1 h at room temperature or overnight at 4°C, washed, and then incubated with 1:10,000 dilutions of HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). Reactive bands were visualized by ECL (GE Healthcare) with SuperSignal West Pico or West Dura reagent (Thermo Fisher Scientific).
Online supplemental material
Fig. S1 shows that GFP-HLA-A2 is degraded in a proteasome-dependent fashion in the presence of US2. Fig. S2 shows that four independent TRC8 siRNAs rescue US2-mediated MHC I degradation. Fig. S3 shows that TRC8 associates with US2, MHC I, and SPP. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200906110/DC1.
Acknowledgments
We thank all members of the Lehner Laboratory for helpful discussions and Tim Rayner for statistical assistance.
This work was supported by the Medical Research Council, Wellcome Trust, Newton Trust, and Cambridge Biomedical Research Centre. P.J. Lehner is a Lister Prize Fellow.
Footnotes
Abbreviations used in this paper:
- ERAD
- ER-associated degradation
- MHC I
- major histocompatibility complex class I
- RING
- really interesting new gene
- shRNA
- short hairpin RNA
- SPP
- signal peptide peptidase
References
- Blom D., Hirsch C., Stern P., Tortorella D., Ploegh H.L. 2004. A glycosylated type I membrane protein becomes cytosolic when peptide: N-glycanase is compromised.EMBO J. 23:650–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle L.H., Gillingham A.K., Munro S., Trowsdale J. 2006. Selective export of HLA-F by its cytoplasmic tail.J. Immunol. 176:6464–6472 [DOI] [PubMed] [Google Scholar]
- Brauweiler A., Lorick K.L., Lee J.P., Tsai Y.C., Chan D., Weissman A.M., Drabkin H.A., Gemmill R.M. 2007. RING-dependent tumor suppression and G2/M arrest induced by the TRC8 hereditary kidney cancer gene.Oncogene. 26:2263–2271 [DOI] [PubMed] [Google Scholar]
- Chen B., Mariano J., Tsai Y.C., Chan A.H., Cohen M., Weissman A.M. 2006. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site.Proc. Natl. Acad. Sci. USA. 103:341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier M.S., Johnson D.C. 2003. Human cytomegalovirus US3 chimeras containing US2 cytosolic residues acquire major histocompatibility class I and II protein degradation properties.J. Virol. 77:4731–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebiger E., Story C., Ploegh H.L., Tortorella D. 2002. Visualization of the ER-to-cytosol dislocation reaction of a type I membrane protein.EMBO J. 21:1041–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman M.H., Ploegh H.L., Tortorella D. 2002. Membrane-specific, host-derived factors are required for US2- and US11-mediated degradation of major histocompatibility complex class I molecules.J. Biol. Chem. 277:3258–3267 [DOI] [PubMed] [Google Scholar]
- Furman M.H., Loureiro J., Ploegh H.L., Tortorella D. 2003. Ubiquitinylation of the cytosolic domain of a type I membrane protein is not required to initiate its dislocation from the endoplasmic reticulum.J. Biol. Chem. 278:34804–34811 [DOI] [PubMed] [Google Scholar]
- Gemmill R.M., West J.D., Boldog F., Tanaka N., Robinson L.J., Smith D.I., Li F., Drabkin H.A. 1998. The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8.Proc. Natl. Acad. Sci. USA. 95:9572–9577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmill R.M., Bemis L.T., Lee J.P., Sozen M.A., Baron A., Zeng C., Erickson P.F., Hooper J.E., Drabkin H.A. 2002. The TRC8 hereditary kidney cancer gene suppresses growth and functions with VHL in a common pathway.Oncogene. 21:3507–3516 [DOI] [PubMed] [Google Scholar]
- Hassink G.C., Barel M.T., Van Voorden S.B., Kikkert M., Wiertz E.J. 2006. Ubiquitination of MHC class I heavy chains is essential for dislocation by human cytomegalovirus-encoded US2 but not US11.J. Biol. Chem. 281:30063–30071 [DOI] [PubMed] [Google Scholar]
- Hirsch C., Gauss R., Horn S.C., Neuber O., Sommer T. 2009. The ubiquitylation machinery of the endoplasmic reticulum.Nature. 458:453–460 [DOI] [PubMed] [Google Scholar]
- Hughes E.A., Hammond C., Cresswell P. 1997. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome.Proc. Natl. Acad. Sci. USA. 94:1896–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. 2004. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum.J. Biol. Chem. 279:3525–3534 [DOI] [PubMed] [Google Scholar]
- Kostova Z., Tsai Y.C., Weissman A.M. 2007. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation.Semin. Cell Dev. Biol. 18:770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft S.G., Wang L., Hochstrasser M. 2006. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI).J. Biol. Chem. 281:4646–4653 [DOI] [PubMed] [Google Scholar]
- Li W., Bengtson M.H., Ulbrich A., Matsuda A., Reddy V.A., Orth A., Chanda S.K., Batalov S., Joazeiro C.A. 2008. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling.PLoS One. 3:e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley B.N., Ploegh H.L. 2004. A membrane protein required for dislocation of misfolded proteins from the ER.Nature. 429:834–840 [DOI] [PubMed] [Google Scholar]
- Lilley B.N., Ploegh H.L. 2005. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane.Proc. Natl. Acad. Sci. USA. 102:14296–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro J., Lilley B.N., Spooner E., Noriega V., Tortorella D., Ploegh H.L. 2006. Signal peptide peptidase is required for dislocation from the endoplasmic reticulum.Nature. 441:894–897 [DOI] [PubMed] [Google Scholar]
- Mueller B., Lilley B.N., Ploegh H.L. 2006. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER.J. Cell Biol. 175:261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B., Klemm E.J., Spooner E., Claessen J.H., Ploegh H.L. 2008. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins.Proc. Natl. Acad. Sci. USA. 105:12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploegh H.L. 2007. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum.Nature. 448:435–438 [DOI] [PubMed] [Google Scholar]
- Poland K.S., Azim M., Folsom M., Goldfarb R., Naeem R., Korch C., Drabkin H.A., Gemmill R.M., Plon S.E. 2007. A constitutional balanced t(3;8)(p14;q24.1) translocation results in disruption of the TRC8 gene and predisposition to clear cell renal cell carcinoma.Genes Chromosomes Cancer. 46:805–812 [DOI] [PubMed] [Google Scholar]
- Shamu C.E., Flierman D., Ploegh H.L., Rapoport T.A., Chau V. 2001. Polyubiquitination is required for US11-dependent movement of MHC class I heavy chain from endoplasmic reticulum into cytosol.Mol. Biol. Cell. 12:2546–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg R.L., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences.Proc. Natl. Acad. Sci. USA. 99:16899–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar S.S., Brodsky J.L. 2008. One step at a time: endoplasmic reticulum-associated degradation.Nat. Rev. Mol. Cell Biol. 9:944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz E.J., Jones T.R., Sun L., Bogyo M., Geuze H.J., Ploegh H.L. 1996a. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol.Cell. 84:769–779 [DOI] [PubMed] [Google Scholar]
- Wiertz E.J., Tortorella D., Bogyo M., Yu J., Mothes W., Jones T.R., Rapoport T.A., Ploegh H.L. 1996b. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction.Nature. 384:432–438 [DOI] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Yun C., Ron D., Rapoport T.A. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol.Nature. 429:841–847 [DOI] [PubMed] [Google Scholar]
- Younger J.M., Chen L., Ren H.Y., Rosser M.F., Turnbull E.L., Fan C.Y., Patterson C., Cyr D.M. 2006. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator.Cell. 126:571–582 [DOI] [PubMed] [Google Scholar]