Abstract
Paraspeckles are ribonucleoprotein bodies found in the interchromatin space of mammalian cell nuclei. These structures play a role in regulating the expression of certain genes in differentiated cells by nuclear retention of RNA. The core paraspeckle proteins (PSF/SFPQ, P54NRB/NONO, and PSPC1 [paraspeckle protein 1]) are members of the DBHS (Drosophila melanogaster behavior, human splicing) family. These proteins, together with the long nonprotein-coding RNA NEAT1 (MEN-ϵ/β), associate to form paraspeckles and maintain their integrity. Given the large numbers of long noncoding transcripts currently being discovered through whole transcriptome analysis, paraspeckles may be a paradigm for a class of subnuclear bodies formed around long noncoding RNA.
Introduction
The cell nucleus, especially in complex eukaryotes, is a highly organized structure. Individual chromosomes occupy discrete territories, and specific proteins and nucleic acids are enriched in subnuclear structures such as nucleoli, Cajal bodies, paraspeckles, and nuclear speckles (Platani and Lamond, 2004). Nuclear organization is linked to genome maintenance and to the control of gene expression and thus influences growth, development, and cellular proliferation. Moreover, disruption of nuclear organization is often correlated with disease states such as the loss of subnuclear promyelocytic leukemia bodies in acute promyelocytic leukemia (Weis et al., 1994).
In this review, we discuss the composition, formation, and function of paraspeckles, one of the most recent subnuclear bodies identified. We describe studies that demonstrate the role of paraspeckles in controlling gene expression by trapping adenosine to inosine (A to I) hyperedited RNA within the nucleus. New evidence suggests that this mechanism may be widely used to coordinate gene expression within a variety of different cellular contexts. We also review the recent findings by several groups that paraspeckles are formed around a long nuclear noncoding RNA (ncRNA), NEAT1 (Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). This finding has increased our knowledge of the functional capabilities of long ncRNAs and has opened up the possibility of more nuclear bodies being formed in this way.
Paraspeckles
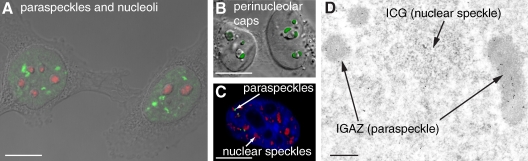
Paraspeckles are a relatively newly identified subnuclear body. They were discovered when a putative nucleolar protein was found to localize to nucleoplasmic foci that did not directly overlap with markers for any known subnuclear structure (Andersen et al., 2002; Fox et al., 2002). These foci were named paraspeckles because they were observed in the interchromatin space near to, yet distinct from, the nuclear speckles that are enriched in splicing factors (Fig. 1; Fox et al., 2002). The novel protein was named PSPC1 (paraspeckle protein 1) and has become the standard marker used to identify paraspeckles.
Figure 1.
Visualizing paraspeckles. (A) Combined differential interference contrast and fluorescence micrograph of HeLa cells stained with anti-PSPC1 to show paraspeckles (green) as nucleoplasmic foci distinct from nucleoli (stained with B23 antibody; red). (B) HeLa cells showing reorganization of the DBHS protein PSPC1 (green) to perinucleolar caps after treatment with actinomycin D to inhibit RNA Pol II transcription. (C) HeLa cell stained with anti-PSPC1 (green), anti-SC35 (red), and DAPI (blue) to show the relationship between paraspeckles abutting nuclear speckles in the interchromatin space. (D) TEM image of a HeLa cell section immunogold labeled with anti-PSPC1. The labeled IGAZs are usually found in close proximity to the interchromatin granules (ICGs; nuclear speckles). This image was provided by S. Souquere and G. Pierron (Institut André Lwoff, Villejuif, France). Panels B and C are adapted from Fox et al. (2002) with permission from Elsevier. Bars: (A–C) 10 µm; (D) 0.5 µm.
Paraspeckles are restricted to mammalian nuclei and are observed in transformed and primary cell lines, embryonic fibroblasts, tissues, and tumorigenic biopsies (Fox et al., 2002; Prasanth et al., 2005; Clemson et al., 2009; Sunwoo et al., 2009; unpublished data). They are also dynamic structures; for instance, paraspeckles are not present in human embryonic stem cells but only appear upon differentiation (Chen and Carmichael, 2009). These bodies are ∼0.5–1.0 µm in size, and their numbers vary both within cell populations and depending on cell type. For example, HeLa have 13–17 paraspeckles per nucleus, whereas NIH3T3 have 5–10 foci per nucleus (Fig. 1; Fox et al., 2002; Cardinale et al., 2007; Clemson et al., 2009). At the EM level, paraspeckle markers label distinct nuclear structures that are electron dense and rich in RNA (Fig. 1 D; Prasanth et al., 2005; Cardinale et al., 2007). These transmission EM (TEM) paraspeckles correspond to the interchromatin granule–associated zones (IGAZs; Visa et al., 1993). IGAZs are electron-dense fibrillar structures found in close proximity to interchromatin granules/nuclear speckles, whose function has remained unknown since their identification in the early 1990s.
At present, paraspeckles are known to contain a small number of proteins with reported roles in transcription and/or RNA processing (Table I and next section). However, paraspeckles do not directly overlap with sites of active transcription, as measured by bromo-UTP incorporation (Fox et al., 2002; Xie et al., 2006), although they may still form in association with some active genes (discussed in Role of paraspeckles in nuclear retention of RNA and Paraspeckle formation). Nevertheless, paraspeckles are intimately linked with transcription because of the presence of active RNA polymerase II (Pol II) and newly made RNA at their periphery (Xie et al., 2006).
Table I.
Paraspeckle proteins
| Protein | Synonyms | Features | Referencea |
| Core paraspeckle proteins | |||
| P54NRB | NONO, NMT55, NRB54 | DBHS; required for paraspeckle integrity in HeLa cells | Fox et al., 2002 |
| PSF | SFPQ | DBHS; required for paraspeckle integrity in HeLa cells | Prasanth et al., 2005 |
| PSPC1 | PSP1 | DBHS | Fox et al., 2002 |
| Other paraspeckle proteins | |||
| CoAAb | PSP2, RBM14, SIP, SYTIP1 | Transcriptional/splicing coregulator | Fox et al., 2002 |
| CFIm68 | CPSF6, HPBRII-4 | RNA 3′ end cleavage factor; also found in nuclear speckles | Dettwiler et al., 2004 |
| SOX9bc | SRA1 | Developmental transcription factor | Hata et al., 2008 |
| WTXb | NA | Wilms tumor protein, tumor suppressor | Rivera et al., 2009 |
| WT1(+KTS)b | WAGR | Wilms tumor transcription factor, partial colocalization with paraspeckles | Dutton et al., 2006 |
| BCL11Abc | CTIP1, ZNF856 | Zinc finger transcription factor | Liu et al., 2006 |
| RNA Pol II | NA | Also found associated with chromatin and nuclear speckles | Xie et al., 2006 |
NA, not applicable.
Localization to paraspeckles was first shown in these studies.
Only overexpressed proteins have been assessed for localization in paraspeckles.
Further studies need to address whether these proteins are genuine paraspeckle components or are retargeting DBHS proteins into different subnuclear locations.
Paraspeckle proteins
Currently, paraspeckle proteins are defined by their colocalization in subnuclear foci with a member of the mammalian DBHS (Drosophila melanogaster behavior, human splicing) protein family, consisting of PSPC1, P54NRB/NONO, or PSF/SFPQ. These three members of the DBHS protein family are the most well-studied intrinsic protein components of paraspeckles. Reported interactions between all members of this family suggest that they exist as either homo- or heterodimers in vivo (Myojin et al., 2004; Fox et al., 2005). They share >50% sequence identity within two N-terminal RNP-type RNA recognition motifs and a C-terminal coiled-coil domain. One of these RNP-type RNA recognition motifs and the coiled-coil domain (which mediates dimerization) are required for PSPC1 to be targeted to paraspeckles (Fox et al., 2005). DBHS proteins are dynamic within the nucleus: they cycle between the nucleoplasm, paraspeckles, and the nucleolus under normal conditions and accumulate within perinucleolar cap structures when RNA Pol II transcription is inhibited (Fig. 1 B; Fox et al., 2002; Shav-Tal et al., 2005). This latter finding explains the discovery of PSPC1 in the nucleolar proteome. Knockdown of either of the two highly expressed DBHS proteins (P54NRB/NONO and PSF/SFPQ) in HeLa cells results in the loss of paraspeckles (Sasaki et al., 2009). In contrast, knockdown of the less abundant DBHS protein PSPC1 in HeLa cells has no effect on paraspeckles (Sasaki et al., 2009). Thus, highly expressed DBHS protein dimers are at the core of paraspeckle structural integrity.
Proteins of the DBHS family have been implicated in a wide array of functions. They have been shown to bind to both double- and single-stranded DNA and RNA and have been copurified in numerous different complexes, leading to the catch-all label of “multifunctional nuclear proteins” (for review see Shav-Tal and Zipori, 2002). These functions encompass many aspects of transcription and RNA processing, including transcription initiation (Dong et al., 1993; Yang et al., 1993, 1997), coactivation (Kuwahara et al., 2006; Amelio et al., 2007), and corepression (Mathur et al., 2001; Dong et al., 2005), constitutive and alternative splicing (Patton et al., 1993; Peng et al., 2002; Kameoka et al., 2004; Ito et al., 2008), and transcriptional termination (Kaneko et al., 2007). A further function relevant to paraspeckles is the involvement of PSF/SFPQ and P54NRB/NONO in the nuclear retention of RNA, specifically preventing A to I hyperedited RNA from leaving the nucleus (Zhang and Carmichael, 2001). RNA hyperediting of long double-stranded RNA (optimally 100 bp) occurs in the nucleus and results in the conversion of up to half of all adenosines (A) in the RNA to inosines (I). A to I hyperediting mostly occurs on transcribed repeat elements (as discussed in more detail in Role of paraspeckles in nuclear retention of RNA).
With roles in constitutive processes such as splicing and transcription, the biological implications for the DBHS proteins are wide ranging. One interesting example indicates a conserved role for P54NRB/NONO in mammals and NonA, a DBHS orthologue in Drosophila, in the control of circadian rhythms. P54NRB/NONO is required for mammalian circadian rhythm maintenance via association with the PERIOD-1 protein (Brown et al., 2005), and NonA mutants are nearly arrhythmic. Brown et al. (2005) speculate that P54NRB/NONO serves to dampen the effects of transcriptional noise on circadian rhythms. DBHS proteins likely carry out their diverse functions by varying their binding partners, posttranslational modification, and subcellular and subnuclear localization (Proteau et al., 2005; for review see Shav-Tal and Zipori, 2002).
Paraspeckle RNAs
After the initial discovery of paraspeckles, several observations suggested that in addition to proteins, paraspeckles would contain RNA. First, paraspeckles are degraded after incubation with RNase A (which degrades single-stranded RNA); however, DNase I does not affect their structural integrity (Fox et al., 2005; Prasanth et al., 2005). Second, all of the major paraspeckle proteins contain RNA-binding motifs, and many have previously described functions in RNA processing (see previous section and Table I). Third, PSPC1 requires its RNA-binding domains for paraspeckle targeting (Fox et al., 2005). Finally, paraspeckles disassemble in the absence of active Pol II transcription and subsequently reassemble on its restoration, suggesting that their formation may be dependent on RNA production (Fox et al., 2002, 2005). In line with this evidence, two types of RNA have now been identified that specifically localize to paraspeckles, each providing clues to paraspeckle formation and function.
Role of paraspeckles in nuclear retention of RNA.
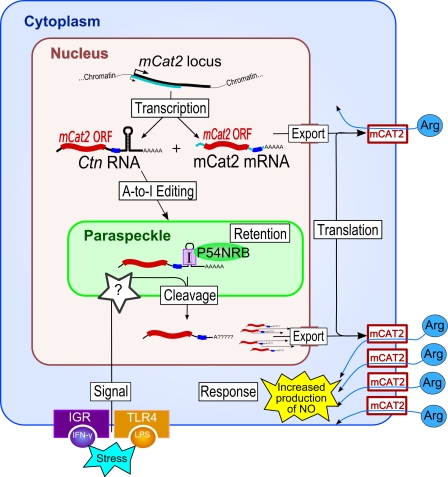
The discovery in mouse of the first paraspeckle RNA revealed how paraspeckles are involved in the control of gene expression through retention of RNA in the nucleus (Prasanth et al., 2005). Ctn RNA is an alternative transcript generated from the mCAT2 gene (encoding the cationic amino acid transporter 2 protein). Ctn differs from the canonical mCAT2 mRNA in that it uses a different promoter and a distal poly(A+) site (producing a much longer 3′ untranslated region [UTR]) and is nuclear enriched (Fig. 2). However, similar to mCAT2, Ctn is spliced and contains the entire open reading frame of the mCAT2 protein. The fate of the two RNA species is quite different: although mCAT2 is exported and translated as normal, Ctn is retained in the nucleus and within paraspeckles in some cell types (Prasanth et al., 2005). The key to paraspeckle/nuclear retention lies in the long 3′ UTR of Ctn, which contains double-stranded RNA hairpins formed by inverted repetitive elements. These RNA hairpins were shown to be A to I hyperedited and associated with DBHS proteins in vivo, which is consistent with the previous study linking nuclear retention of inosine-containing RNA and DBHS proteins (Zhang and Carmichael, 2001). The fate of Ctn does not end in paraspeckles, as the long 3′ UTR is cleaved off (potentially via the paraspeckle-associated cleavage factor CFIm; Table I) as a response to a variety of stress signals. The cleavage event is associated with a concomitant rise in the shorter mCAT2 mRNA levels in the cytoplasm and a pulse of increased protein production. As the mCAT2 protein mediates uptake of precursors in the nitrous oxide response pathway, this retention–release mechanism allows the cell to rapidly mount a nitrous oxide response to the stress.
Figure 2.
The role of nuclear retention in gene regulation in differentiated cells. In the example of the mCat2 gene, two different promoters result in two alternative transcripts: the mCat2 mRNA, which is exported for translation of the mCAT2 cation transporter protein, and Ctn RNA, which is a longer transcript including the mCAT2 protein coding region and an extended 3′ UTR containing inverted repeats. These repeats undergo A to I hyperediting, resulting in Ctn binding to DBHS proteins and retention in paraspeckles. Stress signals mediated by IFN-γ–receptor (IFN-γ–IGR) and lipopolysaccharide–toll-like receptor 4 (LPS–TLR4) interaction result in a cleavage event that liberates a shorter Ctn, which is exported for translation. This up-regulation of the mCAT2 protein results in increased nitric oxide (NO) production as a response to the cellular stress. Although detail in this figure is specific to mCAT2 protein regulation, recent research hints at a more generic retention–release mechanism that exists for other transcripts containing hyperedited inverted repeats in their 3′ UTR (Chen et al., 2008; Chen and Carmichael, 2009; Osenberg et al., 2009).
Although Ctn is not present in humans, evidence supports widespread nuclear retention of RNA because up to half of human transcripts may have extended 3′ UTRs (Iseli et al., 2002). In addition, primates have the largest number of repetitive elements, many of which are inverted, and also have much more A to I hyperediting than other species, the bulk of which takes place at Alu sequences (repetitive elements that make up 10% of the human genome; Levanon et al., 2005; Chen et al., 2008). Moreover, nuclear retention of RNA also takes place when inverted repeats are taken out of their natural biological context and placed downstream of a reporter gene (Chen et al., 2008). P54NRB/NONO was also shown to associate in vivo with these nuclear-retained reporter RNA transcripts (Chen et al., 2008). The nuclear retention mechanism may also be mediated by inverted repeats in 5′ UTRs and retained introns, as many transcripts contain these features. Moreover, because there is at least one other example of a nuclear-retained RNA that does not have inverted repeats (Kay et al., 2005), it is also likely that additional RNA elements are mediating nuclear retention. Until very recently, Ctn was the only example of an RNA that underwent cleavage to be released from nuclear retention. However, a bioinformatic study has now shown evidence that many hundreds of human transcripts containing inverted repeats also exist in a shorter form in which the inverted repeats have been excised (Osenberg et al., 2009). This suggests that the excision of inverted repeats may be generally used as a mechanism for release of transcripts from nuclear retention.
Another very recent study has confirmed the importance of paraspeckles to the nuclear retention mechanism. In cells without paraspeckles such as human embryonic stem cells, mRNAs containing A to I hyperedited inverted repeats were able to efficiently overcome nuclear retention and were present in the cytoplasm at high levels (Chen and Carmichael, 2009). However, with cellular differentiation and the induction of paraspeckles, the ratio of nuclear to cytoplasmic A to I hyperedited mRNAs increased. Interestingly, DBHS protein expression did not vary between the two cellular contexts, instead, paraspeckle induction and increased RNA nuclear retention correlated with the expression of the paraspeckle-specific structural ncRNA species NEAT1.
NEAT1: an architectural long ncRNA in paraspeckles.
We now know that the majority of our genome is transcribed to generate both protein- and nonprotein-coding RNA (Carninci et al., 2005). The numerous ncRNAs may be derived from introns of protein-coding genes or may be antisense or found between protein-coding genes (Mercer et al., 2009). Although our understanding of the roles and identity of different classes of small ncRNAs is substantial, we are only beginning to understand the varying roles that long ncRNAs may be playing in the cell (Prasanth and Spector, 2007). A long-held view in the field has been that there is a role for nuclear RNA as a structural component of nuclear organization.
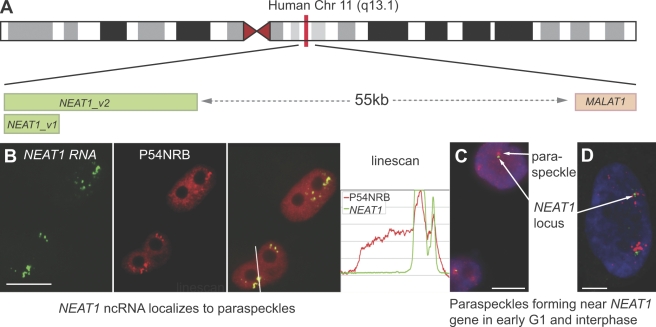
In 2007, a study to identify nuclear-enriched RNA transcripts found three major long ncRNA species: the XIST RNA (well-known for its role in X chromosome inactivation), and two nuclear-enriched autosomal transcripts termed NEAT1 (also reported in the literature as MEN-ϵ/β or VINC-1) and MALAT1 (also known as NEAT2; Hutchinson et al., 2007). Genes encoding NEAT1 and MALAT1 are typically found close together in mammalian genomes, some distance away from the nearest protein-coding gene. Besides both being nuclear-enriched ncRNA, another feature of NEAT1 and MALAT1 is that the long RNAs transcribed from each gene are both cleaved at their 3′ ends to produce an unusual small tRNA-like molecule that may be a hallmark of some nuclear ncRNAs (Wilusz et al., 2008; Sunwoo et al., 2009). Like XIST, both of these ncRNAs were shown to have defined subnuclear localization, MALAT1 within nuclear speckles and NEAT1 in subnuclear foci found abutting nuclear speckles (Hutchinson et al., 2007). Recently several groups have shown that these NEAT1 foci colocalize with paraspeckles and, moreover, that NEAT1 RNA is essential for paraspeckle integrity (Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009).
Two isoforms of NEAT1 are transcribed, NEAT1_v1 and NEAT1_v2 (also known as MEN-ϵ and MEN-β; Fig. 3), overlapping in ∼3–4 kb of sequence at the 5′ end, and both transcripts, either endogenous or overexpressed, localize in paraspeckles in mouse and human cells (Fig. 3 B). The confinement of NEAT1 to paraspeckles is greater than that seen with DBHS proteins such as P54NRB/NONO, which is also abundant in the nucleoplasm (Fig. 3 B). NEAT1 is essential for the formation and maintenance of paraspeckles: they do not reform in the absence of NEAT1 after temporary transcription inhibition (Sasaki et al., 2009; Sunwoo et al., 2009), and knocking down NEAT1 results in loss of paraspeckles (Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). Moreover, stable cell lines overexpressing NEAT1_v1 in NIH3T3 cells have more paraspeckles than control cells, suggesting that NEAT1 RNA is the limiting factor in paraspeckle formation (Clemson et al., 2009). NEAT1 is also likely to be the paraspeckle nucleating factor, as paraspeckles are observed forming in early G1 near to the NEAT1 gene locus and are often found clustered near the NEAT1 gene in interphase (Fig. 3, C and D; Clemson et al., 2009). Interestingly, in contrast to the essential role for NEAT1 in paraspeckles, MALAT1 is not required for nuclear speckle maintenance, as knockdown of MALAT1 has no effect on these or other subnuclear structures (Clemson et al., 2009).
Figure 3.
Paraspeckles contain NEAT1 ncRNA and form near the NEAT1 gene. (A) NEAT1 and MALAT1 gene loci on human chromosome (Chr) 11 q13.1. Two transcripts are produced from the NEAT1 gene, 3.7-kb NEAT1_v1 and 23-kb NEAT1_v2 in humans. (B) RNA FISH against NEAT1 ncRNA (green) and immunofluorescence against P54NRB/NONO (red) shows that they colocalize in paraspeckles. The line scan is taken over a line as indicated in the merged image. (C) HeLa cells in early G1 stained with PSPC1 (red) to mark the first forming paraspeckles and DNA FISH (green) against 11q13. (D) Interphase HeLa cell with NEAT1 RNA FISH to mark paraspeckles (red) and DNA FISH to mark chromosome 11 q13.1 (green). Panels B–D are reproduced from Clemson et al. (2009) with permission from Elsevier. Bars: (B and C) 10 µm; (D) 5 µm.
Like Ctn, NEAT1 associates with DBHS proteins in vivo, as immunoprecipitation of PSF/SFPQ, P54NRB/NONO, and PSPC1 all copurify NEAT1 RNA to varying levels (Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). In contrast to Ctn, NEAT1 shows no evidence of A to I hyperediting, suggesting that the DBHS proteins use more than one mode of RNA binding within paraspeckles. The presence of only very sporadic short regions of conservation between mammalian NEAT1 sequences raises the possibility that DBHS proteins bind an RNA structure rather than a sequence. However, a previous study of DBHS proteins binding to U5SnRNA supports both sequence- and structure-based aspects to binding (Peng et al., 2002). There is some discrepancy as to the relative roles of the short and long NEAT1 isoforms in paraspeckle formation and maintenance: NEAT1_v1 overexpression alone can increase paraspeckle number in NIH3T3 cells (Clemson et al., 2009) and NEAT1_v1 is present in paraspeckles when NEAT1_v2 is knocked down (Sunwoo et al., 2009) and is bound by recombinant DBHS proteins in vitro (Clemson et al., 2009). However, other data suggests that the final 10 kb of NEAT1_v2 may be required for exogenous rescue of the NEAT1 knockdown (Sasaki et al., 2009). Detailed studies of the molecular interactions between the DBHS proteins and NEAT1 should address these issues.
Paraspeckle formation
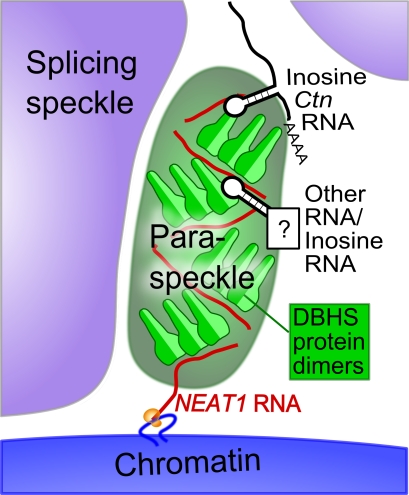
Now that we have a grasp of the main protein and RNA components of paraspeckles, it is possible to put this information together to form a picture of paraspeckle formation, beginning with the production of NEAT1 in daughter nuclei after cell division (Fig. 4). Before newly made NEAT1 has a chance to diffuse away from its gene locus, it is rapidly targeted by DBHS protein dimers, and together, the RNA–protein complex builds up the paraspeckle particle. The finished paraspeckle likely consists of multiple copies of NEAT1 RNA–DBHS protein complexes, which form a structural scaffold that is nevertheless dynamic, in that individual DBHS protein molecules can exchange with the nucleoplasm. It is possible that the oligomerization propensity of the DBHS proteins (Kiesler et al., 2003; Myojin et al., 2004; Fox et al., 2005) contributes to the paraspeckle structural framework. Interestingly, cytoplasmic P bodies have many parallels with nuclear paraspeckles, including being linked to the control of gene expression through mRNA storage. A recent structural study of Edc3, a protein involved in P-body formation, shows that the dimerization properties of this protein increase the RNA-binding potential of the complex and may also contribute to P-body assembly by linking messenger RNPs together (Ling et al., 2008).
Figure 4.
Model of paraspeckle formation. The schematic shows a paraspeckle forming in the interchromatin space in between two nuclear speckles. The paraspeckle forms near the NEAT1 gene locus through the interactions between newly transcribed NEAT1 RNA and DBHS protein dimers. RNA containing A to I hyperedited inverted repeats, as well as potentially other RNA, is regulated within paraspeckles via interaction with DBHS proteins.
Without the production of NEAT1 RNA, paraspeckles fail to form, explaining why paraspeckles are not observed when Pol II transcription is inhibited nor in cell types that do not express NEAT1 or in organisms that do not contain the NEAT1 gene. Conversely, without abundant DBHS proteins, paraspeckles are also not observed. In light of this model, it is reasonable that, in the future, a molecule may only be labeled as paraspeckle associated once it is shown to localize with both DBHS proteins and NEAT1. The first paraspeckles form very close to the NEAT1 gene locus, and paraspeckles also remain closely associated with the NEAT1 gene in interphase; however, just how the association is maintained is not known. Interestingly, although there is a tendency for paraspeckles to cluster near the NEAT1 gene during interphase, it is not an absolute correlation; many nuclei also have paraspeckles at other nuclear locations in addition to a NEAT1 cluster (Fig. 3 D; Clemson et al., 2009). It is possible that these locations are the site of production for RNA species that are being regulated in paraspeckles such as mRNAs containing A to I hyperedited inverted repeats (for example, Ctn).
Cellular function of paraspeckles
A recent study brings together the contributions of the three main types of protein and RNA molecules within paraspeckles in an important area of cell biology: pluripotency and differentiation (Chen and Carmichael, 2009). In this study, the authors draw the conclusion that paraspeckle formation is associated with a loss of pluripotency in embryonic stem cells. Furthermore, they suggest that a lack of NEAT1 and paraspeckles can be used as a marker for pluripotency. Paraspeckles are most likely contributing to differentiation by changing the gene expression profile through nuclear retention of A to I hyperedited mRNA. Interestingly, another study linking NEAT1 to paraspeckle formation also showed a trend of NEAT1/paraspeckle induction with differentiation: in this case, the differentiation of myoblasts into myotubes is associated with a threefold up-regulation of NEAT1 and an increase in paraspeckle size and number (Sunwoo et al., 2009). It will be interesting in the future to determine how widespread changes in NEAT1 levels and paraspeckle abundance correlate with different models of differentiation and the identity of those transcripts being controlled by nuclear retention in these systems.
Another potential role for paraspeckles in cell biology is in the response to certain viruses. An earlier study reported that VINC-1 (since shown to correspond to NEAT1) is up-regulated in the central nervous system of mice upon infection with Japanese encephalitis or rabies viruses (Saha et al., 2006). Given that many of the aforementioned studies have proven a link between NEAT1 up-regulation and paraspeckle formation, it is possible that these viruses may trigger an increase in paraspeckle size and number, either as a cellular defense mechanism or for viral processing. The link between a viral response and paraspeckles is consistent with the speculation that HIV RNA may hijack the RNA processing pathways mediated by DBHS proteins, as PSF/SFPQ and P54NRB/NONO bind elements within HIV mRNAs and regulate their processing and nuclear export (Zolotukhin et al., 2003).
It is likely that the most telling analyses of the biological role for paraspeckles will arise from the generation and analysis of a NEAT1-null mouse, which should be devoid of paraspeckles. However, it should also be considered that there may be primate-specific functions for paraspeckles, given the abundance of A to I hyperedited inverted repeats specifically transcribed from primate genomes.
Paraspeckles as a paradigm for a class of subnuclear bodies
Paraspeckles are not the only subnuclear foci formed via the specific interactions between nuclear proteins and nuclear-retained ncRNA. A clear example occurs in myotonic muscular dystrophy, in which RNA transcribed from mutated genes with expanded CTG repeats is bound by the muscleblind-like family of proteins and retained in the nucleus within subnuclear foci (O'Rourke and Swanson, 2009). These myotonic muscular dystrophy foci do not colocalize with paraspeckles (Clemson et al., 2009). However, the similarities between these foci and paraspeckles suggest that the cell has common themes in nuclear retention of RNA that are apparent in both normal cell function and disease.
Paraspeckle markers localize to the IGAZ in TEM sections. Given the heterogeneity in IGAZ composition (Puvion-Dutilleul et al., 1995), it is possible that this compartment seen under EM is actually composed of numerous different subnuclear bodies akin to paraspeckles, i.e., containing distinct structural ncRNA and specific RNA-binding proteins and having different species of RNA regulated/retained within them. Indeed, there already exist two examples of other long ncRNA localizing to unique subnuclear foci, although in these cases, their appearance is very cell type specific, and no corresponding protein partners are known (Royo et al., 2007; Sone et al., 2007). The lack of identified marker ncRNAs and proteins for other subnuclear bodies may have so far prevented their detection and characterization. Further studies on subcellular localization of the many thousands of new long ncRNAs being currently discovered from high throughput genomics analyses will, in the future, provide a fuller picture of these structures and their roles in the cell.
Conclusion
Paraspeckles lie at the nexus of two expanding areas of interest in the control of gene expression in mammals: the roles of functional long ncRNAs and the effects of RNA editing of transcripts. An exciting prospect is finding new subcellular complexes by studying the localization of newly identified ncRNAs. Potentially, paraspeckles can be used as a model system for studying protein–ncRNA interactions and dynamics in such complexes. Within paraspeckles, the mechanism of RNA nuclear retention may be critical for controlling gene expression in a variety of cellular contexts. The discovery of the identity and function of the many key molecules that may be regulated in paraspeckles through this mechanism will be of great interest in the years to come.
Acknowledgments
We would like to thank Sam Swift and Silvana van Koningsbruggen (University of Dundee, Dundee, Scotland, UK) for help generating fluorescence micrographs and Sylvie Souquere and Gerard Pierron for providing TEM images. We would also like to thank members of our laboratories for their helpful comments during the preparation of this manuscript.
A.H. Fox and C.S. Bond are funded by Project Grant 513880 from the National Health and Medical Research Council, Australia.
Footnotes
Abbreviations used in this paper:
- DBHS
- Drosophila melanogaster behavior, human splicing
- IGAZ
- interchromatin granule–associated zone
- ncRNA
- noncoding RNA
- Pol II
- polymerase II
- TEM
- transmission EM
- UTR
- untranslated region
References
- Amelio A.L., Miraglia L.J., Conkright J.J., Mercer B.A., Batalov S., Cavett V., Orth A.P., Busby J., Hogenesch J.B., Conkright M.D. 2007. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway.Proc. Natl. Acad. Sci. USA. 104:20314–20319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J.S., Lyon C.E., Fox A.H., Leung A.K.L., Lam Y.W., Steen H., Mann M., Lamond A.I. 2002. Directed proteomic analysis of the human nucleolus.Curr. Biol. 12:1–11 [DOI] [PubMed] [Google Scholar]
- Brown S.A., Ripperger J., Kadener S., Fleury-Olela F., Vilbois F., Rosbash M., Schibler U. 2005. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator.Science. 308:693–696 [DOI] [PubMed] [Google Scholar]
- Cardinale S., Cisterna B., Bonetti P., Aringhieri C., Biggiogera M., Barabino S.M. 2007. Subnuclear localization and dynamics of the Pre-mRNA 3′ end processing factor mammalian cleavage factor I 68-kDa subunit.Mol. Biol. Cell. 18:1282–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. 2005. The transcriptional landscape of the mammalian genome.Science. 309:1559–1563 [DOI] [PubMed] [Google Scholar]
- Chen L.L., Carmichael G.G. 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA.Mol. Cell. doi:10.1016/j.molcel.2009.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.L., DeCerbo J.N., Carmichael G.G. 2008. Alu element-mediated gene silencing.EMBO J. 27:1694–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A., Lawrence J.B. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles.Mol. Cell. 33:717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettwiler S., Aringhieri C., Cardinale S., Keller W., Barabino S.M. 2004. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization.J. Biol. Chem. 279:35788–35797 [DOI] [PubMed] [Google Scholar]
- Dong B., Horowitz D.S., Kobayashi R., Krainer A.R. 1993. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6.Nucleic Acids Res. 21:4085–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Shylnova O., Challis J.R., Lye S.J. 2005. Identification and characterization of the protein-associated splicing factor as a negative co-regulator of the progesterone receptor.J. Biol. Chem. 280:13329–13340 [DOI] [PubMed] [Google Scholar]
- Dutton J.R., Lahiri D., Ward A. 2006. Different isoforms of the Wilms' tumour protein WT1 have distinct patterns of distribution and trafficking within the nucleus.Cell Prolif. 39:519–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A.H., Lam Y.W., Leung A.K., Lyon C.E., Andersen J., Mann M., Lamond A.I. 2002. Paraspeckles: a novel nuclear domain.Curr. Biol. 12:13–25 [DOI] [PubMed] [Google Scholar]
- Fox A.H., Bond C.S., Lamond A.I. 2005. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner.Mol. Biol. Cell. 16:5304–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K., Nishimura R., Muramatsu S., Matsuda A., Matsubara T., Amano K., Ikeda F., Harley V.R., Yoneda T. 2008. Paraspeckle protein p54nrb links Sox9-mediated transcription with RNA processing during chondrogenesis in mice.J. Clin. Invest. 118:3098–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A. 2007. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains.BMC Genomics. 8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseli C., Stevenson B.J., de Souza S.J., Samaia H.B., Camargo A.A., Buetow K.H., Strausberg R.L., Simpson A.J., Bucher P., Jongeneel C.V. 2002. Long-range heterogeneity at the 3′ ends of human mRNAs.Genome Res. 12:1068–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Watanabe H., Yamamichi N., Kondo S., Tando T., Haraguchi T., Mizutani T., Sakurai K., Fujita S., Izumi T., et al. 2008. Brm transactivates the telomerase reverse transcriptase (TERT) gene and modulates the splicing patterns of its transcripts in concert with p54(nrb).Biochem. J. 411:201–209 [DOI] [PubMed] [Google Scholar]
- Kameoka S., Duque P., Konarska M.M. 2004. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes.EMBO J. 23:1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S., Rozenblatt-Rosen O., Meyerson M., Manley J.L. 2007. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination.Genes Dev. 21:1779–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay R.A., Ellis I.R., Jones S.J., Perrier S., Florence M.M., Schor A.M., Schor S.L. 2005. The expression of migration stimulating factor, a potent oncofetal cytokine, is uniquely controlled by 3′-untranslated region-dependent nuclear sequestration of its precursor messenger RNA.Cancer Res. 65:10742–10749 [DOI] [PubMed] [Google Scholar]
- Kiesler E., Miralles F., Ostlund Farrants A.K., Visa N. 2003. The Hrp65 self-interaction is mediated by an evolutionarily conserved domain and is required for nuclear import of Hrp65 isoforms that lack a nuclear localization signal.J. Cell Sci. 116:3949–3956 [DOI] [PubMed] [Google Scholar]
- Kuwahara S., Ikei A., Taguchi Y., Tabuchi Y., Fujimoto N., Obinata M., Uesugi S., Kurihara Y. 2006. PSPC1, NONO, and SFPQ are expressed in mouse Sertoli cells and may function as coregulators of androgen receptor-mediated transcription.Biol. Reprod. 75:352–359 [DOI] [PubMed] [Google Scholar]
- Levanon K., Eisenberg E., Rechavi G., Levanon E.Y. 2005. Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome.EMBO Rep. 6:831–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S.H., Decker C.J., Walsh M.A., She M., Parker R., Song H. 2008. Crystal structure of human Edc3 and its functional implications.Mol. Cell. Biol. 28:5965–5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Ippolito G.C., Wall J.K., Niu T., Probst L., Lee B.S., Pulford K., Banham A.H., Stockwin L., Shaffer A.L., et al. 2006. Functional studies of BCL11A: characterization of the conserved BCL11A-XL splice variant and its interaction with BCL6 in nuclear paraspeckles of germinal center B cells.Mol. Cancer. 5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur M., Tucker P.W., Samuels H.H. 2001. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors.Mol. Cell. Biol. 21:2298–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T.R., Dinger M.E., Mattick J.S. 2009. Long non-coding RNAs: insights into functions.Nat. Rev. Genet. 10:155–159 [DOI] [PubMed] [Google Scholar]
- Myojin R., Kuwahara S., Yasaki T., Matsunaga T., Sakurai T., Kimura M., Uesugi S., Kurihara Y. 2004. Expression and functional significance of mouse paraspeckle protein 1 on spermatogenesis.Biol. Reprod. 71:926–932 [DOI] [PubMed] [Google Scholar]
- O'Rourke J.R., Swanson M.S. 2009. Mechanisms of RNA-mediated disease.J. Biol. Chem. 284:7419–7423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenberg S., Dominissini D., Rechavi G., Eisenberg E. 2009. Widespread cleavage of A-to-I hyperediting substrates.RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton J.G., Porro E.B., Galceran J., Tempst P., Nadal-Ginard B. 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor.Genes Dev. 7:393–406 [DOI] [PubMed] [Google Scholar]
- Peng R., Dye B.T., Pérez I., Barnard D.C., Thompson A.B., Patton J.G. 2002. PSF and p54nrb bind a conserved stem in U5 snRNA.RNA. 8:1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani M., Lamond A.I. 2004. Nuclear organisation and subnuclear bodies.Prog. Mol. Subcell. Biol. 35:1–22 [DOI] [PubMed] [Google Scholar]
- Prasanth K.V., Spector D.L. 2007. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum.Genes Dev. 21:11–42 [DOI] [PubMed] [Google Scholar]
- Prasanth K.V., Prasanth S.G., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L. 2005. Regulating gene expression through RNA nuclear retention.Cell. 123:249–263 [DOI] [PubMed] [Google Scholar]
- Proteau A., Blier S., Albert A.L., Lavoie S.B., Traish A.M., Vincent M. 2005. The multifunctional nuclear protein p54nrb is multiphosphorylated in mitosis and interacts with the mitotic regulator Pin1.J. Mol. Biol. 346:1163–1172 [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F., Besse S., Chan E.K., Tan E.M., Puvion E. 1995. p80-coilin: a component of coiled bodies and interchromatin granule-associated zones.J. Cell Sci. 108:1143–1153 [DOI] [PubMed] [Google Scholar]
- Rivera M.N., Kim W.J., Wells J., Stone A., Burger A., Coffman E.J., Zhang J., Haber D.A. 2009. The tumor suppressor WTX shuttles to the nucleus and modulates WT1 activity.Proc. Natl. Acad. Sci. USA. 106:8338–8343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo H., Basyuk E., Marty V., Marques M., Bertrand E., Cavaillé J. 2007. Bsr, a nuclear-retained RNA with monoallelic expression.Mol. Biol. Cell. 18:2817–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Murthy S., Rangarajan P.N. 2006. Identification and characterization of a virus-inducible non-coding RNA in mouse brain.J. Gen. Virol. 87:1991–1995 [DOI] [PubMed] [Google Scholar]
- Sasaki Y.T., Ideue T., Sano M., Mituyama T., Hirose T. 2009. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles.Proc. Natl. Acad. Sci. USA. 106:2525–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y., Zipori D. 2002. PSF and p54(nrb)/NonO—multi-functional nuclear proteins.FEBS Lett. 531:109–114 [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y., Blechman J., Darzacq X., Montagna C., Dye B.T., Patton J.G., Singer R.H., Zipori D. 2005. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition.Mol. Biol. Cell. 16:2395–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M., Hayashi T., Tarui H., Agata K., Takeichi M., Nakagawa S. 2007. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons.J. Cell Sci. 120:2498–2506 [DOI] [PubMed] [Google Scholar]
- Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., Spector D.L. 2009. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles.Genome Res. 19:347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N., Puvion-Dutilleul F., Bachellerie J.P., Puvion E. 1993. Intranuclear distribution of U1 and U2 snRNAs visualized by high resolution in situ hybridization: revelation of a novel compartment containing U1 but not U2 snRNA in HeLa cells.Eur. J. Cell Biol. 60:308–321 [PubMed] [Google Scholar]
- Weis K., Rambaud S., Lavau C., Jansen J., Carvalho T., Carmo-Fonseca M., Lamond A., Dejean A. 1994. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells.Cell. 76:345–356 [DOI] [PubMed] [Google Scholar]
- Wilusz J.E., Freier S.M., Spector D.L. 2008. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA.Cell. 135:919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S.Q., Martin S., Guillot P.V., Bentley D.L., Pombo A. 2006. Splicing speckles are not reservoirs of RNA polymerase II, but contain an inactive form, phosphorylated on serine2 residues of the C-terminal domain.Mol. Biol. Cell. 17:1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.S., Hanke J.H., Carayannopoulos L., Craft C.M., Capra J.D., Tucker P.W. 1993. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss.Mol. Cell. Biol. 13:5593–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.S., Yang M.C., Tucker P.W., Capra J.D. 1997. NonO enhances the association of many DNA-binding proteins to their targets.Nucleic Acids Res. 25:2284–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Carmichael G.G. 2001. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs.Cell. 106:465–475 [DOI] [PubMed] [Google Scholar]
- Zolotukhin A.S., Michalowski D., Bear J., Smulevitch S.V., Traish A.M., Peng R., Patton J., Shatsky I.N., Felber B.K. 2003. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression.Mol. Cell. Biol. 23:6618–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]