Abstract
Research embracing systems biology approaches and careful analysis of the critical host response has greatly expanded our understanding of infectious diseases. First-generation studies based on genomics and proteomics have made significant progress in establishing the foundation for network-based investigations on virus-host interactions. More recently, data from complementary high-throughput technologies, such as siRNA and microRNA screens and next-generation sequencing, are augmenting systems level analyses and are providing a more detailed and insightful multidimensional view of virus-host networks. Together with advances in data integration, systems biology approaches now have the potential to provide profound impacts on translational research, leading to the more rapid development of new therapeutics and vaccines for infectious diseases. In this review, we highlight new high-throughput technologies, a new philosophy for studying virus-host interactions, and discuss the potential of systems biology to facilitate bench-to-bedside research and create novel strategies to combat disease. Can we save the world using these approaches? Read on.
Introduction
For the past 50 years the discipline of virology has been overly focused on the pathogen itself. We now know that “the host response” is equally or more important in defining the eventual pathological outcome of infection. Based on recent vaccine failures and inadequate production of novel antiviral therapeutics it is clear that the virology/immunology community must take more daring and innovative approaches to help combat viral infection on multiple fronts. We believe that systems biology approaches, combined with sophisticated computational strategies, are required to combat the AIDS virus, other chronic and acute infections, and most recently a potential “swine flu” pandemic. Why is that? Systems biology analyzes all of the components, interactions, and dynamics of a biological system in a comprehensive, quantitative, and integrative fashion. To better understand human diseases, systems biology involves iterative cycles in which model organisms with different levels of complexity are perturbed and then measured using combinations of high-throughput technologies. After mining the multidimensional data, predictive computational models are developed, evaluated, and then refined based on the model predictions with new iterations of manipulation of the systems.
Systems biology, or more specifically components of a systems biology approach, have already greatly expanded our understanding of human diseases, including host responses to viral infection [1-3]. Only a few years ago, first-generation studies began to characterize patterns of gene expression on a genomic scale, distinguishing diseased states from normal states at the molecular level. With the development of new high-throughput technologies, and their successful application to infectious disease research, new-generation studies have demonstrated great potential for providing a more insightful multidimensional network view of virus-host interactions. Unfortunately, the full potential of systems wide approaches to study human diseases has not been fully realized.
In particular, as systems biology evolves, it is anticipated to have profound impacts on translational research. Not only does systems biology deliver scientific discoveries and analytical tools for translational research, it also challenges several shared barriers which also impact translational research. These include establishing efficient inter-disciplinary collaborations (indeed the cultures of any interdisciplinary team presents a multitude of cross culture communications and challenges), building integrated computational infrastructures for data management and data sharing, and developing sophisticated computational tools to translate large volumes of multidimensional data into better understanding of basic biology and disease. This review highlights new developments in high-throughput technologies for the systematic study of virus-host interactions and the value of systems biology in facilitating translational research (Figure 1).
Figure 1. Systems biology approaches for virus-host interactions and the implications on translational research.

Representation of the connection between systems biology approaches and the potential impact on translational research. As shown, the path leads from basic research to applying high-throughput technologies for better understanding the complex interactions occurring in a given model system. Through computational modeling and data integration, these technologies can have direct impact on translational research.
New high-throughput approaches
The interplay between the development of new high-throughput technologies and specific needs of biological and medical researchers is an essential component of systems biology. Microarray-based functional genomics, which provides a global view of transcriptional changes occurring in host cells during viral infection, has been the most common high-throughput method used to study virus-host interactions (reviews in [2,3]). In this section, we highlight several additional high-throughput technologies that have been recently applied to the study of virus-host interactions.
In vitro screens using siRNA
Small interfering RNAs (siRNAs) are a class of double-stranded short RNA molecules that are endogenously expressed by host cells in response to the nucleic acids of foreign organisms, including viruses. A siRNA specifically induces the cleavage of foreign RNAs through RNA interference (RNAi). By introducing a pool of synthetic siRNAs into cells, genome-wide siRNA screening provides a powerful (but potentially flawed) forward-genetic approach for identifying cellular genes with certain functions [4]. For example, recent reports have demonstrated the efficacy of large-scale siRNA screens in identifying cellular factors important for infection by and replication of a variety of viruses, including influenza virus [5], hepatitis C virus (HCV) [6], West Nile virus [7] and human immunodeficiency virus-1 (HIV-1) [8-10] (see Table 1 for a summary). Although some of the identified host factors may be system-specific [8-10], by identifying hundreds of infection-related host factors simultaneously, this approach has great utility in systematically locating key components of protein interaction networks. However, one of the key limiting factors is the need to define and enforce data-reporting standards to facilitate data dissemination and re-analysis by the scientific community (Minimum Information About an RNAi Experiment, URL: http://miare.sourceforge.net/HomePage), and to make the structured data available in a public repository, analogous to what has been advocated for by the microarray field [11,12]. Even with such a centralized database, it will require years (or even decades) of intensive biological confirmatory assays to prove the real worth of these RNAi screens.
Table 1. Summary of recent studies using high-throughput in vitro screen to identify viral infection related host factors.
| Virus | Host system | Description | Year | Reference | ||
|---|---|---|---|---|---|---|
| siRNA | Number of host genes targeted by siRNA library | Number of host genes identified | ||||
| Influenzaa | Drosophila D-Mel2 cells | ∼13,000 Drosophila genes | ∼100b | 2008 | 5 | |
| HCV replicons | Huh7-derived EN5-3 cells | ∼4,000 human genes | 9 | 2007 | 6 | |
| WNV | HeLa cells | ∼21,000 human genes | 305 | 2008 | 7 | |
| HIV | Hela-derived TZM-bl cells | ∼21,000 human gene | ∼250 | 2008 | 10 | |
| HIV | Human 293T cell | ∼20,000 human genes | 295 | 2008 | 9 | |
| HIV | HeLa P4/R5 cells | ∼20,000 human genes | ∼310 | 2008 | 8 | |
| Virus | Host system | Description | Year | Reference | ||
| Yeast two-hybrid | Number of viral proteins | Number of viral proteins | ||||
| EBV | human spleen AD-cDNA library | 85 EBV proteins | 112 human and 40 EBV proteins, 173 EBV-human protein interactions | 2007 | 13 | |
| HCV | Human fetal brain and spleen AD-cDNA libraries | 11 HCV proteins | 278 human proteins, 314 HCV–human protein interactions | 2008 | 14 | |
Modified to infect Drosophila cells
Three selected human homologues were confirmed in intro.
HCV: Hepatitis C virus. WNV: West Nile virus. HIV: Human immunodeficiency virus EBV: Epstein–Barr virus
In vitro screens with the yeast two-hybrid assay
Physical interactions between host and pathogen proteins are critical to establishing an infection and influencing pathogenesis. Until recently, large-scale interaction studies have primarily focused on intra-species interactions. Interactions between host and virus proteins have been identified from small directed experiments, focused on particular pathogen proteins or pathways of interest. Two recent studies have used a high-throughput yeast two-hybrid approach to identify physical interactions between human and viral proteins from Epstein-Barr virus (EBV) [13] and HCV [14], respectively (Table 1). After integrating their data with previously cataloged human protein interactions, both studies noted that human proteins interacting with viral proteins tended to be hubs (proteins with many interacting partners) and bottlenecks (proteins that are central to many paths in the network) in human protein interaction networks, hinting at a conserved mechanism among viral systems for efficiently controlling and manipulating host cellular processes [15]. The yeast two-hybrid datasets, when integrated with other ‘omics’ data, can potentially provide mechanistic insights into the changes observed at the transcriptional and proteome levels. However, the same challenges associated with tedious biological validation studies will nonetheless delay any true progress associated with these approaches.
microRNA profiling
Similar to siRNAs described above, microRNAs (miRNAs) are a group of endogenously expressed single-stranded small non-coding regulatory RNA molecules. Unlike a siRNA, however, a miRNA represses its target gene expression by inducing not only RNAi but also translational inhibition. During some virus infections, the virus and cell mutually regulate each other's gene expression through self-encoded miRNAs [16]. To date, over 5,000 microRNAs have been identified, including approximately 800 human microRNAs. Various hybridization- and PCR-based strategies for profiling miRNA expression have been developed [17] and applied to the study of virus-host interactions.
Microarray-based miRNA profiling revealed that specific miRNA signatures may correlate with CD4+ T-cell counts and viral loads in HIV-infected individuals [18]. Similarly, we have found multiple cellular miRNAs in transplanted human liver biopsies that may regulate of host responses at the mRNA level during HCV infection (Peng et al, submitted).
With continued refinement in miRNA profiling technologies [19], the greater challenge is to study the biological functions of a miRNA, mainly because functional targets of a specific miRNA are difficult to identify. While computational target predictions may at best achieve a 50% success rate, target identification through mRNA expression profiling alone will overlook targets regulated mainly at protein level. To experimentally enrich functional miRNA targets, regardless of the regulatory mechanisms used, Hannon et al. used an assay coupling RNA-induced silencing complex (RISC) immuno-precipitation with microarray analysis of bound mRNAs [20]. In this approach, even miRNA targets exclusively regulated at the translational level would be detected by microarray analysis downstream, as miRNAs and their corresponding target mRNAs would be incorporated into the RISC before immuno-precipitation.
Next-generation sequencing
Next-generation sequencing is dramatically accelerating biomedical research by enabling comprehensive analyses of genomic variation, mRNA and small RNA expression and discovery, and DNA-protein interactions (see recent reviews on the technology and its applications [21,22]). This technology may allow investigators to conduct studies on virus-host interactions which were previously not feasible or affordable, such as the identification of important alternate splice isoforms, miRNA discovery and profiling, and expression profiling in organisms for which a complete genomic sequence has not been determined. RNA-seq [23], a new method for whole transcriptome analysis based on next-generation sequencing technology, offers a much greater dynamic range than microarrays, and therefore a better platform to quantify low-abundance transcripts. Compared to microarrays, prior sequence information requirements are less exacting. This is particularly important for studies on those organisms with very limited or low-coverage genome sequences, such as nonhuman primates, for which the current standard for expression profiling is to use rhesus or human microarrays. Although these microarrays provide useful information for a large number of genes, there are complications in analyzing the data due to potential hybridization differences between species. However, sequence information from RNA-seq does not depend upon hybridization to a specific, predetermined probe.
Information obtained from RNA-seq can also be used to improve current microarrays through alternative splice form detection, gene and exon boundary mapping, and novel transcript discovery. Although the technology is still in the early stages of use, a variety of software tools are being developed to deal with the vast amount of data generated by next-generation sequencing (see reference [22] for a short summary). There are also ongoing efforts to establish the guidelines for reporting and archiving next-generation sequencing data. Public repositories such as the NCBI Short Read Archive (SRA, URL: http://0-www.ncbi.nlm.nih.gov.millennium.unicatt.it/Traces/sra) have been set up to store both raw sequencing data from different platforms and associated metadata regarding experimental annotations and instrument runs. As the technology matures, the major challenge will be to use new bioinformatics approaches and computational methods to extract biologically relevant information from the large volume of data.
Advances in data integration and multi-disciplinary collaboration
Data integration and systems analysis
Until recently, systems level analyses have mostly been performed on single-cell organisms, or in the case of virus-host interactions, using cell-line infection models. Although a great deal may be learned from this approach, knowledge of how a specific cell type responds in isolation to virus infection provides an incomplete picture what happens in the complex milieu of an infected animal. Therefore, in the context of virus-host interactions, systems biology must leverage not only multiple high-throughput data types but also multiple model systems and perturbations to gain a systems level view of pathogenesis and disease. As an example, Figure 2 illustrates a three-dimensional matrix of comparisons that might be performed when studying influenza virus infection using multiple infection models. Assessing phenotypic differences, from the gross to the molecular level and across model systems and viral strains, will allow researchers to define commonalities and differences in the host response and to identify both common and unique targets for therapeutic intervention.
Figure 2. Matrix of potential comparisons performed to systematically study influenza virus infection.
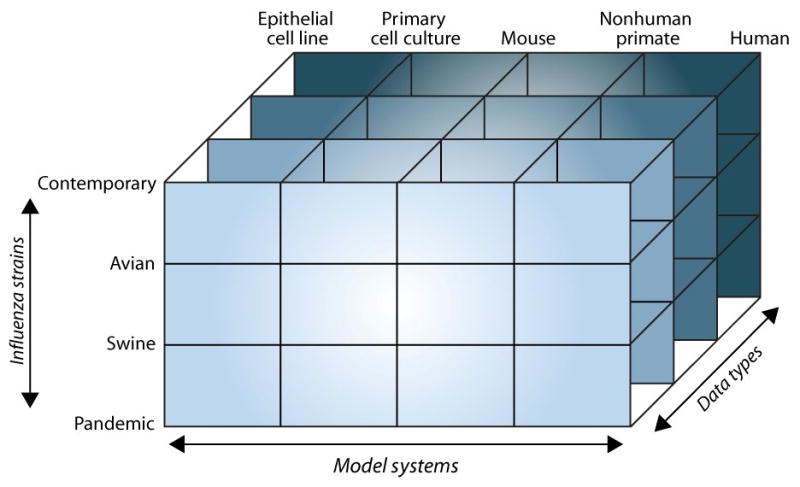
Illustration highlighting the multiple comparisons possible when taking a systems approach to studying Influenza virus infection. High-throughput technologies have enabled many of these comparisons to be possible and now it is up to the research community to design studies in a multi-disciplinary fashion and implement new data integration techniques to best leverage this wealth of data.
The real strength, but potentially the biggest challenge of this approach, will come from integrating vast amounts of data into a single systems-level view. This challenge is already being met in other fields where new computational tools have been developed to take advantage of the data generated by high-throughput technologies. For example, Gunsalus et al. [24] have integrated transcriptomics, protein interaction, and RNAi data to provide a multidimensional view of how molecular machines work together in the early embryogenesis in Caenorhabditis elegans. In the same light, Chuang et al. [25] have developed a network-based classifier by integrating gene expression profiles of metastatic and non-metastatic cancer patients with the human protein interaction network. Similarly, Pujana et al. [26] combined gene expression profiling with functional genomic and proteomic data from different species to identify genes associated with a higher risk of breast cancer.
One goal of virus-host systems biology is to make predictions on host responses and dynamic interactions between viruses and hosts by computational modeling. As structures and interactions of virus-host networks unearthed by high-throughput studies are often static, predictive computational modeling attempts to identify key network components and interactions by predicting the dynamics of the whole network during viral infections, in turn revealing potential translational targets for assessments. Depending on the scope of the study and the availability of experimental data, modeling may be done at different levels of abstraction: genes, proteins, cells, and organisms.
Several recent studies have attempted to model the dynamic host immune responses to different pathogens such as influenza A virus [27], vesicular stomatitis virus [28], and Bordetellae bacteria [29]. In these cell-based models, molecular-level descriptions of host response were not available; yet, in order to generate more testable hypotheses, relevant molecular details such as key signaling pathways and regulatory networks should be included, especially the identification of host targets for antiviral therapies. As an interesting example, Zhang et al. [30] proposed a multi-scale model to simulate cancer heterogeneity, integrating a simplified epidermal growth factor receptor gene-protein network coupled with a cell cycle module. While computational modeling of host responses to viral infection is still in its infancy, continued development of predictive models will have significant impacts on identifying host cell factors for antiviral therapies.
Infrastructure for multi-disciplinary collaboration
Effective multi-disciplinary collaboration is key to the success of both systems biology and translational research (Box 1) as is attested to by funding for translational research infrastructure through the Clinical and Translational Science Awards (CSTAs; URL: http://www.ctsaweb.org/) from the National Institutes of Health. To promote the use of systems biology approaches for delivering translational discoveries with clinical impacts, investments in organizational infrastructure in the public sector and public-private partnerships will be necessary. Computational infrastructure presents a major challenge for utilizing systems biology approaches in translational research, as both basic researchers and clinical scientists need ready access to clinical data and biomolecular data, a significant challenge from both technological and regulatory standpoints. In this regard, increasing awareness in both the research community and funding agencies has transformed the conduct of biomedical research in multi-disciplinary collaborations. For example, researchers from University of Washington and their collaborators are building strong inter-disciplinary research teams through several large research programs (NIDA Center for Functional Genomics; URL: http://nida.viromics.washington.edu/; Systems Virology; URL: https://www.systemsvirology.org).
Systems biology and translational research
Clearly, systems biology is a rapidly evolving discipline. To fulfill its promise to provide us with a raft of new drug targets, systems biology needs to address several major challenges: 1) Computation. To enable targeted search for antiviral therapeutics, it is necessary to develop predictive computational models that incorporate sufficient molecular details and make effective predictions of virus-host interactions for experimental testing. 2) Data. Relative to the complexity of biological systems being studied, the field is actually data-poor even though some may be overwhelmed by the sheer data volume. A body of self-consistent, comprehensive, and viral infection-specific experimental data is needed for building effective computational models. We are currently building a “virus-host genomic compendium” of both our own data and data of others; this effort will enable us to identify common and unique features of the host response to dozens of highly virulent viruses. 3) Money. Systems Biology is not cheap. It costs literally millions of dollars to establish the proper biological, clinical, and computational infrastructure. 4) Engagement and Dedication. Systems biology researchers need to be fully engaged in translational research, especially bridging bench and bedside scientists during the development of its own infrastructures and collaborations.
Box 1. Translational research
“It's easy for a biomedical researcher studying a disease to forget that his or her work someday actually needs to help treat an actual patient”.[31]
Translational research transforms scientific discoveries into clinical applications to prevent, detect and cure human disease. Typically such discoveries begin at “the bench” with basic researches at a molecular or cellular level, then progress to the patient's “bedside”, the clinical practice.
The ‘bench-to-bedside’ translational research is a two-way process. Researchers provide new analytic tools for use and new targets (biomarkers, drug targets, mechanisms of gene and pathway action) for assessment in patients. Clinicians make novel observations about disease progression and provide clinical samples and variables for laboratory investigation, which often requires detailed experimental studies using model systems. A key barrier to translational research is for both researchers and clinicians to have ready access to two types of data: 1) clinical data, including medical records, pathology reports, diagnostic results and clinical trial information; and 2) biomolecular data, including sequencing, genomics, proteomics and other high-throughput research data.
Acknowledgments
We thank Sean Proll for discussions and assistance with the preparation of the original figures. We thank Matt Dyer for insightful suggestions and assistance with the preparation of the manuscript. Research in the authors' laboratory is supported by Public Health Service grants R01AI022646, R01HL080621, R24RR016354, P30DA015625, P01AI058113, and P51RR00166 from the National Institutes of Health, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Xinxia Peng, Email: xinxiap@u.washington.edu.
Eric Y. Chan, Email: ericchan@u.washington.edu.
Yu Li, Email: liyu@u.washington.edu.
Deborah L. Diamond, Email: ddiamond@u.washington.edu.
Michael G. Katze, Email: honey@u.washington.edu.
Reference List
- 1.Tan SL, Ganji G, Paeper B, Proll S, Katze MG. Systems biology and the host response to viral infection. Nat Biotechnol. 2007;25:1383–1389. doi: 10.1038/nbt1207-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katze MG, Fornek JL, Palermo RE, Walters KA, Korth MJ. Innate immune modulation by RNA viruses: emerging insights from functional genomics. Nat Rev Immunol. 2008;8:644–654. doi: 10.1038/nri2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andeweg AC, Haagmans BL, Osterhaus ADME. Virogenomics: the virus-host interaction revisited. Current Opinion in Microbiology. 2008;11:461–466. doi: 10.1016/j.mib.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nature Reviews Genetics. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- 5.Hao LH, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–U46. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng TI, Mo H, Pilot-Matias T, He Y, Koev G, Krishnan P, Mondal R, Pithawalla R, He W, Dekhtyar T, et al. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology. 2007;45:1413–1421. doi: 10.1002/hep.21608. [DOI] [PubMed] [Google Scholar]
- 7.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–U67. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou HL, Xu M, Huang Q, Gates AT, Zhang XHD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, et al. Genome-Scale RNAi Screen for Host Factors Required for HIV Replication. Cell Host & Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Konig R, Zhou YY, Elleder D, Diamond TL, Bonamy GMC, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 11.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME) toward standards for microarray data. Nature Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 12.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles - database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, Hill DE, et al. Epstein-Barr virus and virus human protein interaction maps. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2007;104:7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Chassey B, Navratil V, Tafforeau L, Hiet MS, ublin-Gex A, Agaugue S, Meiffren G, Pradezynski F, Faria BF, Chantier T, et al. Hepatitis C virus infection protein network. Molecular Systems Biology. 2008;4 doi: 10.1038/msb.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyer MD, Murali TM, Sobral BW. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottwein E, Cullen BR. Viral and cellular MicroRNAs as determinants of viral pathogenesis and immunity. Cell Host & Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong W, Zhao JJ, He L, Cheng JQ. Strategies for Profiling MicroRNA Expression. Journal of Cellular Physiology. 2009;218:22–25. doi: 10.1002/jcp.21577. [DOI] [PubMed] [Google Scholar]
- 18.Houzet L, Yeung ML, de Lame V, Desai D, Smith SM, Jeang KT. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology. 2008;5 doi: 10.1186/1742-4690-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson PT, Wang WX, Wilfred BR, Tang GL. Technical variables in high-throughput miRNA expression profiling: Much work remains to be done. Biochimica et Biophysica Acta-Gene Regulatory Mechanisms. 2008;1779:758–765. doi: 10.1016/j.bbagrm.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 23.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 24.Gunsalus KC, Ge H, Schetter AJ, Goldberg DS, Han JDJ, Hao T, Berriz GF, Bertin N, Huang J, Chuang LS, et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436:861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- 25.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Molecular Systems Biology. 2007;3 doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pujana MA, Han JDJ, Starita LM, Stevens KN, Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nature Genetics. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 27.Hancioglu B, Swigon D, Clermont G. A dynamical model of human immune response to influenza A virus infection. Journal of Theoretical Biology. 2007;246:70–86. doi: 10.1016/j.jtbi.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 28.Haseltine EL, Lam V, Yin J, Rawlings JB. Image-guided modeling of virus growth and spread. Bulletin of Mathematical Biology. 2008;70:1730–1748. doi: 10.1007/s11538-008-9316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thakar J, Pilione M, Kirimanjeswara G, Harvill ET, Albert R. Modeling systems-level regulation of host immune responses. PLoS Computational Biology. 2007;3:1022–1039. doi: 10.1371/journal.pcbi.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Strouthos CG, Wang Z, Deisboeck TS. Simulating brain tumor heterogeneity with a multiscale agent-based model: Linking molecular signatures, phenotypes and expansion rate. Mathematical and Computer Modelling. 2009;49:307–319. doi: 10.1016/j.mcm.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutson S. HHMI's Med Into Grad Initiative expands. Nat Med. 2009;15:123. doi: 10.1038/nm0209-123b. [DOI] [PubMed] [Google Scholar]
Reference Annotations
Special interest (*)
Outstanding interest (**)
- [10] Brass et al. (*) – The authors were the first to demonstrate the importance of siRNA genomic screens in identifying host cellular factors vital for HIV viral replication.
- [13] Calderwood et al. (*) – This is the first study to apply a high-throughput yeast two-hybrid approach to identify physical interaction between Epstein Barr virus and human proteins. The authors use these interactions to suggest mechanism of virulence.
- [23] Mortazavi et al (**) – This is the first study to apply next-generation sequencing technology to analyze mammalian transcriptome. This study reported wide linear range of transcript detection, and the detection of alternate splice isoforms and novel transcripts.
- [30] Zhang et al. (*) – The authors reported a multi-scale agent-based computational model to simulate cancer heterogeneity. The model integrates a simplified epidermal growth factor receptor gene-protein network coupled with a cell cycle module.


