Abstract
Upon sensing misfolded outer-membrane porins (OMPs) in the periplasm, the E. coli DegS protease cleaves RseA, a transmembrane regulator, transmitting a signal to activate cytoplasmic gene expression. Misfolding is detected by binding of normally inaccessible OMP sequences to the DegS-PDZ domain, which relieves allosteric inhibition and activates proteolysis. Here, we show that DegS stimulation can be regulated by OMP-peptide affinity for the active and for the inactive protease conformations, as well as by preferential substrate binding to active DegS. Based on the effects of mutations in the peptide-binding pocket of the PDZ domain and elsewhere, we suggest an allosteric pathway that links peptide binding to DegS activation. These results explain fast responses to envelope stress, demonstrate that the protein-unfolding response, even under catastrophic conditions, can be tailored by the peptide sequences that become accessible to DegS, and suggest strategies for control of related PDZ-proteases by allosteric effectors.
Introduction
PDZ proteases play important roles in protein quality control, stress responses, and virulence in bacteria, and mutations in human orthologs are frequently correlated with disease (Ehrmann and Clausen, 2004; Kim and Kim, 2005; Vande Walle et al., 2008). In general, these enzymes function as trimers or higher oligomers, and each subunit consists of a trypsin-like protease domain and one or two PDZ domains. The protease domain utilizes a conventional catalytic triad and oxyanion hole, and the PDZ domains, which typically bind C-terminal peptides, function in allosteric regulation of activity. Despite the widespread biological importance of PDZ proteases, the mechanisms that control the activities of these complex enzymes are only beginning to be understood.
DegS is a trimeric PDZ protease that plays a crucial role in initiation of the envelope-stress response in Escherichia coli and related bacteria (Alba and Gross, 2004). It is anchored, via an N-terminal segment, to the periplasmic side of the inner membrane. When protein folding in the periplasm is compromised, DegS cleaves the membrane-spanning protein RseA at a single site. This event initiates a cascade of additional cleavages by other proteases that destroy the cytoplasmic domain of RseA and liberate the σE transcription factor, which enhances expression of genes that encode periplasmic chaperones, proteases, and biosynthetic enzymes (Rhodius et al., 2006). Hence, a signal linked to protein misfolding in the periplasm is transduced across the inner membrane via a regulated proteolytic cascade, leading to adaptive changes in gene expression.
How is cleavage of RseA controlled? One mechanism involves peptide binding to the DegS PDZ domains. By itself, purified DegS cleaves RseA very slowly. However, peptides containing a C-terminal YXF sequence motif, which is present in many outer-membrane porins (OMPs), bind to the PDZ domains of DegS and enhance the RseA-cleavage rate substantially (Walsh et al., 2003; Sohn et al., 2007). OMPs are highly abundant envelope proteins, and simple overexpression of OMPs or fusion proteins containing their C-terminal sequences is sufficient to induce the envelope-stress response (Mecsas et al., 1993; Walsh et al., 2003). Moreover, the YxF motif is inaccessible in native assembled OMPs (Baslé et al., 2006). Thus, heat shock and other types of environmental stress appear to lead to accumulation of misfolded or unassembled OMPs, which then bind to and activate DegS (Walsh et al., 2003; Alba and Gross, 2004).
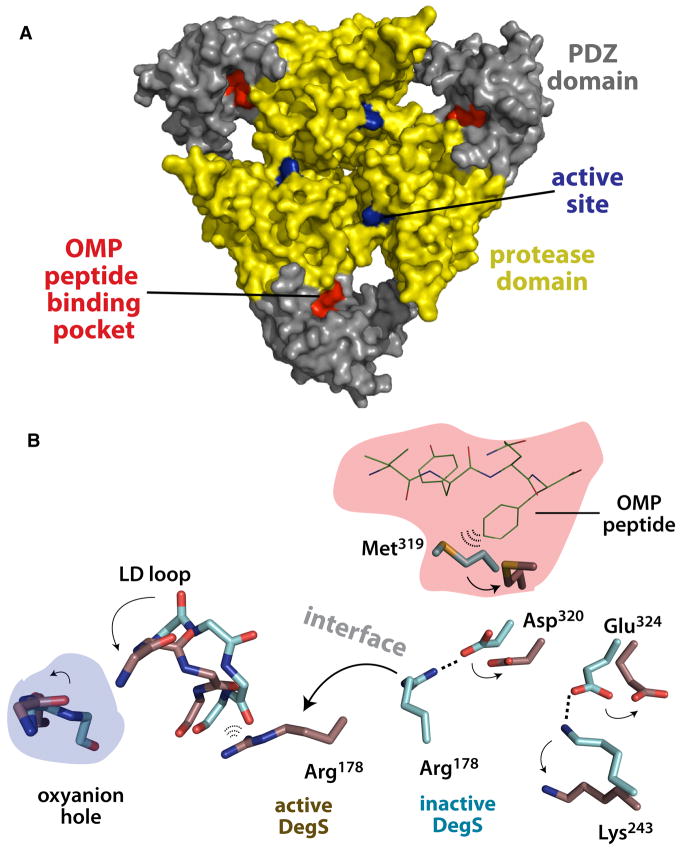
Crystal structures of unliganded DegS and DegS bound to an OMP peptide show two alternative conformations of the protease domain (Wilken et al., 2004; Zeth, 2004). In peptide-free DegS, the oxyanion hole of the active site is malformed, suggesting that this conformation is enzymatically inactive. In peptide-bound DegS, the structure of the oxyanion hole is that of an active protease. However, bound OMP peptides are more than 20 Å from the active sites in the DegS trimer (Fig. 1A), indicating that peptide must indirectly stabilize the catalytically active conformation. Indeed, a plausible communication path between the peptide-binding site and active site can be drawn (Fig. 1B). In this pathway, OMP-peptide binding changes the conformation of Met319, which helps break a salt-bridge between Asp320 in the PDZ domain and Arg178 in the protease domain, allowing the latter side chain to form a new set interactions that stabilize the functional oxyanion hole. Previously, we presented evidence for an allosteric model in which DegS equilibrates between inactive and active conformations, with OMP-peptide binding driving the equilibrium to favor the active state (Sohn et al., 2007). Evidence supporting this model included substantial increases in basal DegS activity and in DegS affinity for OMP peptides when salt bridges between the PDZ domains and the protease domain that stabilize the inactive conformation were eliminated by mutations. Moreover, deleting the entire PDZ domain resulted in an enzyme with a high level of peptide-independent RseA cleavage activity, and the crystal structure of DegS-ΔPDZ revealed a functional oxyanion hole. Thus, interactions between the unliganded PDZ domain and the protease domain must inhibit DegS activity, and OMP-peptide binding must counter this inhibition and stabilize active DegS. A model in which OMP peptides bind only to active DegS was consistent with the previously available experimental data. Moreover, prior experiments suggested that the positive cooperativity of substrate degradation arose from favorable substrate-substrate interactions and not from preferential binding of RseA to the active conformation of DegS.
Figure 1.
Conformational activation of DegS.
(A) Surface representation of inactive DegS (1TE0; Zeth (2004)) with the protease domains colored yellow and the PDZ domains gray. The active site and OMP peptide-binding pocket are colored blue and red, respectively. (B) Changes in the conformations of selected DegS residues between the inactive (cyan carbons; 1TE0) and active enzyme (brown carbons; 1SOZ; Wilken et al., 2004) are indicated by arrows. OMP peptide (VYQF) bound to active DegS is shown in line representation (green). In inactive DegS, the Met319 side chain appears to clash with bound OMP peptide. The strain introduced by this clash could help to break the Asp320•••Arg178 and Glu324•••Lys243 salt bridges that stabilize inactive DegS relative to active DegS. These disruptions, in turn, could allow repositioning of the Arg178 side chain, which is known to play a critical role in stabilizing the “active” conformation of the LD loop and the active oxyanion-hole conformation (Sohn et al., 2007).
New experiments presented here allow us to refine the DegS allosteric model in several important ways. We find that OMP peptides and RseA substrate bind both to inactive and active DegS, with the degree of activation depending on the relative affinities of these molecules for each state. As a consequence, different OMP peptides can maximally activate DegS to dramatically different extents. Moreover, we show that positive substrate cooperativity arises from preferential substrate binding to active versus inactive DegS and that changes in RseA cleavage caused by different OMP peptides or by a variety of DegS mutations can be quantitatively modeled using the concerted MWC formulation of allostery (Monod et al., 1965). Finally, we suggest a pathway of allosteric communication that allows OMP-peptide binding to stabilize active DegS. Our model helps explain how the biological response to envelope stress can occur so rapidly and suggests molecular strategies for regulating DegS and related PDZ proteases.
Results
If a concerted allosteric model is appropriate, then DegS protease activity should depend on the fraction of enzymes that assume the active (relaxed) conformation rather than the inactive (tense) conformation. This ratio will depend on the intrinsic equilibrium between these states and on the concentrations of activating peptide and substrate (Monod et al., 1965). Another important factor is whether peptides bind exclusively to the active enzyme or to both enzyme conformations. Below, we test these possibilities.
Different OMP peptides activate DegS to very different extents
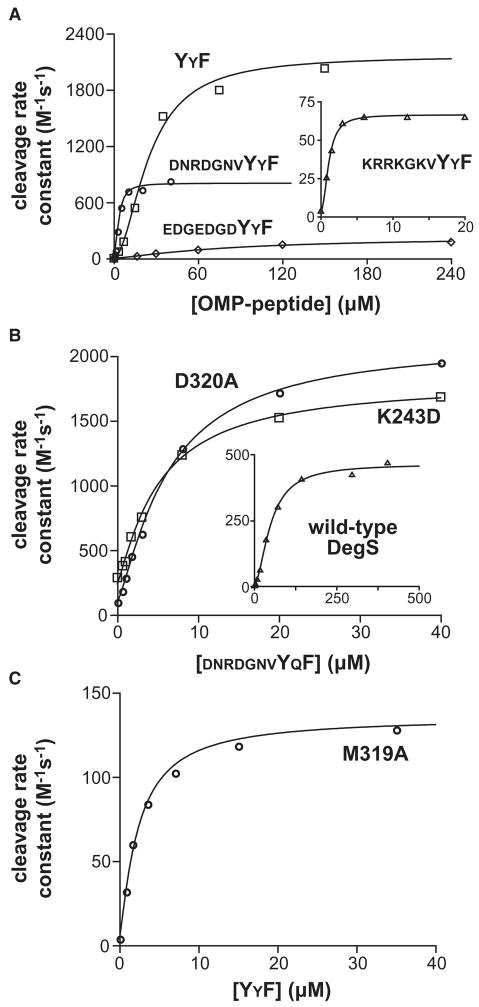
Although saturating concentrations of OMP peptides with different central residues in the C-terminal YXF sequence activated DegS cleavage of RseA similarly (Walsh et al., 2003; Sohn et al., 2007), we found that sequence changes prior to the YxF motif resulted in dramatically different maximal rates of substrate cleavage by DegS. For example, maximal activation by two tripeptides (YYF; YQF) and four decapeptides (DNRDGNVYYF; DNRDGNVYQF; EDGEDGDYYF; KRRKGKVYYF) differed over a 35-fold range (Fig. 2A; Table 1A). These results rule out a model in which peptides bind exclusively to relaxed DegS, as mass action of saturating concentrations of any OMP peptide would then drive all enzymes into the active conformation and result in the same maximal activity.
Figure 2.
OMP-peptide activation of DegS and mutants.
(A) Peptide activation of RseA cleavage by wild-type DegS (≤0.5 μM trimer). The DNRDGNVYYF data are from Sohn et al. (2007). (B) Activation of wild-type DegS and the D320A and K243D mutants (0.1 μM trimer) by DNRDGNVYQF. (C) YYF activation of the M319A DegS mutant. In all panels, the concentration of the RseA periplasmic domain was 200 μM, and the lines are ts to the equation: velocity = basal + max/(1+(Kact/[peptide])n), where n is the Hill constant.
Table 1.
Parameters for DegS Cleavage and/or OMP Peptide Binding Activity
| Activation Parameters |
||||
|---|---|---|---|---|
| Enzyme | OMP Peptide | Maximum Activity (M−1s−1) | Kact (μM) | Hill Constant |
| Wild-type | none | 2.9 ± 0.5 | n.a. | n.a. |
| Wild-type | DNRDGNVYYF | 730 ± 80 | 3.9 ± 0.8 | 1.8 ± 0.2 |
| Wild-type | DNRDGNVYQF | 590 ± 70 | 50 ± 5 | 1.6 ± 0.1 |
| Wild-type | EDGEDGDYYF | 290 ± 60 | 73 ± 10 | 1.3 ± 0.1 |
| Wild-type | KRRKGKVYYF | 70 ± 7 | ≤%1 μMa | ~1.2a |
| Wild-type | YYF | 2500 ± 550 | 29 ± 3 | 1.7 ± 0.1 |
| Wild-type | YQF | 2100 ± 200 | 260 ± 10 | 1.6 ± 0.1 |
| K243D | None | 210 ± 30 | n.a. | n.a. |
| K243D | DNRDGNVYYF | 1700 ± 150 | 0.3 ± 0.1 | b |
| K243D | DNRDGNVYQF | 1800 ± 200 | 5.3 ± 1.2 | 1.3 ± 0.1 |
| K243D | YYF | 3400 ± 550 | 6.4 ± 1.6 | 1.4 ± 0.2 |
| D320A | None | 100 ± 20 | n.a. | n.a. |
| D320A | DNRDGNVYYF | 2100 ± 200 | 0.2 ± 0.1 | b |
| D320A | DNRDGNVYQF | 2300 ± 200 | 6.3 ± 1.3 | 1.2 ± 0.1 |
| D320A | EDGEDGDYYF | 1020 ± 150 | 14 ± 2 | 1.3 ± 0.1 |
| D320A | KRRKGKVYYF | 810 ± 80c | n.d. | n.d. |
| D320A | YYF | 3600 ± 450 | 4.8 ± 1.2 | 1.4 ± 0.1 |
| M319A | None | 4.5 ± 1.0 | n.a. | n.a. |
| M319A | DNRDGNVYYF | 20 ± 2c | n.d. | n.d. |
| M319A | DNRDGNVYQF | 18 ± 2c | n.d. | n.d. |
| M319A | KRRKGKVYYF | 6 ± 1c | n.d. | n.d. |
| M319A | YYF | 120 ± 20 | 3.2 ± 0.4 | 1.1 ± 0.1 |
| K243D/M319A | None | 490 ± 50 | n.a. | n.a. |
| K243D/M319A | DNRDGNVYYF | 1100 ± 130c | n.d | n.d |
| K243D/M319A | YYF | 2700 ± 400 | 0.7 ± 0.2 | 1.2 ± 0.1 |
| Michaelis-Menten Parameters |
||||
|---|---|---|---|---|
| Enzyme | OMP Peptide | Vmax (s−1enz−1) | KM (μM) | Hill Constant |
| Wild-type | DNRDGNVYYF | 1.1 ± 0.2 | 750 ± 120 | 1.6 ± 0.2 |
| Wild-type | YYF | 2.6 ± 0.2 | 370 ± 40 | 1.4 ± 0.2 |
| K243D | DNRDGNVYYF | 1.8 ± 0.2 | 580 ± 30 | 1.3 ± 0.1 |
| K243D | YYF | 2.3 ± 0.1 | 440 ± 30 | 1.3 ± 0.1 |
| D320A | DNRDGNVYYF | 2.5 ± 0.3 | 520 ± 90 | 1.1 ± 0.2 |
| D320A | YYF | 3.1 ± 0.3 | 430 ± 50 | 1.2 ± 0.1 |
| K243D/D320A | DNRDGNVYYF | 2.0 ± 0.3 | 630 ± 100 | 1.1 ± 0.1 |
| K243D/M319A | DNRDGNVYYF | 1.4 ± 0.1 | 840 ± 90 | 1.2 ± 0.1 |
| K243D/M319A | YYF | 2.1 ± 0.2 | 490 ± 40 | 1.2 ± 0.1 |
| OMP Peptide Binding |
|
|---|---|
| Enzyme | KD (μM) |
| Wild-type | 4.6 ± 0.3 |
| M319A | 0.14 ± 0.03 |
| K243D/M319A | 0.18 ± 0.02 |
| Fitted MWC Allosteric Parameters |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Enzyme | OMP Peptide | kr (s−1) | L0 | KRS (μM) | KTS (μM) | KRP(μM) | KTP (μM) | KTP/KRP | L3 Peptide |
| Wild-type | DNRDGNVYYF | 3.3 | 15000 | 150 | 505 | 0.20 | 1.36 | 6.8 | 50 |
| Wild-type | KRRKGKVYYF | 3.3 | 15000 | 150 | 505 | 0.23 | 0.76 | 3.3 | 420 |
| Wild-type | DNRDGNVYQF | 3.3 | 15000 | 150 | 505 | 2.1 | 12.8 | 6.1 | 61 |
| Wild-type | YYF | 3.3 | 15000 | 150 | 505 | 1.51 | 18.6 | 12.3 | 8 |
| K243D | DNRDGNVYYF | 3.3 | 180 | 150 | 505 | 0.07 | 0.14 | 2.0 | 21 |
| K243D | DNRDGNVYQF | 3.3 | 180 | 150 | 505 | 1.64 | 3.19 | 1.9 | 25 |
| K243D | YYF | 3.3 | 180 | 150 | 505 | 1.24 | 3.76 | 3.0 | 6.5 |
| D320A | DNRDGNVYYF | 3.3 | 350 | 150 | 505 | 0.06 | 0.2 | 3.3 | 11 |
| D320A | EDGEDGDYYF | 3.3 | 350 | 150 | 505 | 4.02 | 8.5 | 2.1 | 37 |
| D320A | DNRDGNVYQF | 3.3 | 350 | 150 | 505 | 1.09 | 2.98 | 2.8 | 16.4 |
| D320A | YYF | 3.3 | 350 | 150 | 505 | 0.9 | 3.7 | 4.1 | 5 |
| M319A | YYF | 3.3 | 5500 | 150 | 505 | 0.34 | 0.91 | 2.7 | 290 |
| K243D/M319A | YYF | 3.3 | 90 | 150 | 505 | 0.33 | 0.68 | 2.1 | 10.3 |
The fitted Kact was too close to the enzyme concentration (0.5 μM monomer) to determine reliable “free” peptide, and thus a Hill constant.
Binding was too tight to determine a reliable Hill constant.
Complete titration curves were not determined, but near saturation was confirmed by testing at least two peptide concentrations that differed by a 2-fold minimum.
Activation parameters were determined by experiments like those shown in Figures 2A and 2B. Values in italics are from Sohn et al. (2007); n.d., not determined; n.a., not applicable. In “OMP Peptide Binding,” the binding affinities are for the peptide fluoresceine-β-alanine-KKDNRDGNYYF. Experimental values are an average of two or more independent determinations. Errors were calculated as where n is the number of independent trials. In “Fitted MWC Allosteric Parameters,” L3 is the equilibrium constant relating the peptide-saturated tense and relaxed states (see Figure 5).
Maximal activation did not correlate with the peptide concentration required for 50% activation or with peptide length. The YYF and YQF tripeptides activated DegS to similar maximal extents but with activation curves displaced almost 10-fold in concentration. In addition, the peptide with the highest apparent affinity (KRRKGKVYYF) resulted in the lowest maximal activation. Although the tripeptides activated DegS better than the decapeptides, large differences were also observed among the decapeptides. Moreover, peptides of 10 and 20 residues that activated DegS as well as the best tripeptides were identified in other studies (R. Chaba, B.M. Alba, B.C. Nmezi, J.S., R.T.S., and C.A. Gross, in preparation). These results show that peptide sequences N-terminal to the YXF portion of OMP peptides can strongly influence the extent of DegS activation.
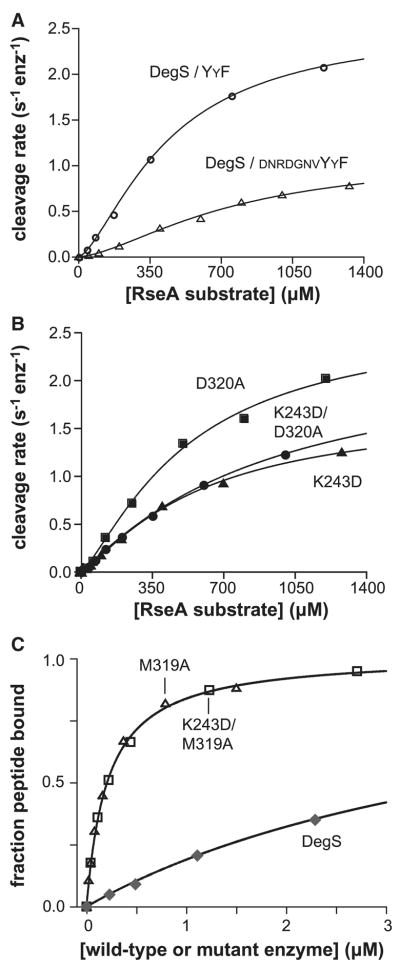
Why is YYF•DegS a more efficient enzyme than DNRDGNVYYF•DegS? When we determined steady-state kinetic parameters for RseA cleavage in the presence of saturating YYF tripeptide, KM was 370 ± 40 μM and the turnover number (Vmax/DegStotal) was 2.6 ± 0.2 s−1 (Fig. 3A). For saturating DNRDGNVYYF peptide, by contrast, KM was about two-fold higher and the turnover number was about two-fold lower (Table 1; Sohn et al., 2007). Thus, any model for peptide activation needs to explain how the chemical identity of the bound OMP peptide can change KM and the turnover number.
Figure 3.
Substrate binding and control of activity.
(A) Substrate dependence of the steady-state rate of cleavage by wild-type DegS (0.5 μM trimer) with YYF peptide (300 μM). For comparison, rates in the presence of saturating DNRDGNVYYF peptide (Sohn et al., 2007) are also shown. (B) Substrate dependence of the cleavage by the K243D, D320A and K243D/D320A mutants of DegS (0.3 μM trimer) with DNRDGNVYYF peptide (30 μM). (C) Binding of DegS, the M319A mutant, and the K243D/M319A mutant to the OMP peptide fluoresceine-β–alanine-KKDNRDGNYYF (20 nM) were monitored by changes in fluorescence anisotropy. The lines are fits to a quadratic form of a hyperbolic binding isotherm. The data for wild-type DegS are from Sohn et al. (2007). In panels A & B, the lines are fits to the Hill form of the Michaelis-Menten equation: velocity = Vmax/(1+(KM/[substrate])n).
Testing a model for varied peptide activation
To rationalize these results in terms of an allosteric model, OMP peptides would need to bind to both relaxed and tense DegS. The degree of maximal activation would then depend on how tightly a given peptide bound to each conformation. One prediction if peptides bind both DegS states is that maximal peptide activation should depend on the intrinsic equilibrium distribution of the tense and relaxed enzyme states. Specifically, a peptide that activated wild-type DegS relatively poorly should result in better activation of a DegS mutant in which the free-energy difference between the active and inactive conformations was decreased.
The K243D and D320A mutations disrupt salt-bridges that stabilize the interface between the PDZ domain and protease domain in inactive but not active DegS (Fig. 1B; Wilken et al., 2004; Zeth, 2004). DegS variants with either mutation behave as if more unliganded enzymes adopt the active conformation, displaying increased activity in the absence of peptide and requiring less peptide for stimulation (Sohn et al., 2007). Here, we found that the K243D and D320A mutants cleaved a single sub-KM concentration of RseA about three-fold faster than did wild-type DegS in the presence of saturating DNRDGNVYQF peptide (Fig. 2B; Table 1A). Moreover, in Michaelis-Menten experiments performed with saturating DNRDGNVYYF, both mutants had higher Vmax values than wild-type DegS (Fig. 3A and 3B; Table 1B). Thus, both DNRDGNVYQF and DNRDGNVYYF, which activate wild-type DegS only moderately compared to some peptides (Table 1A), convert more of the K243D and D320A enzymes into the active state. Conversely, the YYF peptide, which was one of the best activators of wild-type DegS, activated these mutants only marginally better than wild-type DegS at a single substrate concentration (Table 1A) and resulted in Vmax values that were roughly within error for wild-type and the mutants (Table 1B). This result is expected if the degree of activation depends both on the binding preference of a given peptide for the two states of DegS and on the intrinsic equilibrium between these states. In other words, peptides that bind sufficiently tightly to active DegS compared to inactive DegS (e.g., YYF) can convert most enzymes into the active conformation, even when the conformational equilibrium is less favorable.
Compared to wild-type DegS, half-maximal activation (Kact) of the K243D and D320A mutant enzymes was observed at lower concentrations of a given OMP peptide and the activation curves had smaller Hill constants, indicating reduced positive cooperativity of activation (Fig. 2B; Table 1A). Tighter peptide binding and reduced positive cooperativity are expected if less of the peptide-binding energy is needed to drive the tense-to-relaxed conversion for the mutants compared to wild-type DegS. The KM’s and Hill constants for the RseA substrate were also lower for the K243D and D320A enzymes than for wild-type DegS (Table 1B). Indeed, with saturating DNRDGNVYYF, the substrate Hill constants for the D320A (1.1 ± 0.2) and K243D/D320A (1.1 ± 0.1) enzymes were within error of 1, whereas the value for wild-type DegS was 1.6 ± 0.2 (Table 1B). These results strongly suggest that the RseA substrate also binds preferentially to active versus inactive DegS and thus helps to stabilize the catalytically competent enzyme (see Discussion). By this model, positive cooperativity in substrate binding would only be expected under conditions where peptide binding alone was not sufficient to drive most DegS molecules into the active conformation.
A DegS mutation that reduces maximal activation
A model in which OMP peptides bind both to the tense and relaxed enzyme conformations also predicts that some DegS mutations should diminish activation by reducing peptide-binding preference for the active enzyme. We screened several DegS mutations that alter side chains that contact bound OMP peptide and found one mutant (M319A) that displayed markedly lower levels of peptide activation (Fig. 2C; Table 1A). For example, saturating YYF resulted in M319A activity that was 20-fold lower than the wild-type DegS value, and peptides that partially activated wild-type DegS resulted in even lower levels of M319A activation (Table 1A).
Because OMP peptides still activate M319A, they must bind somewhat more tightly to the relaxed than to the tense state of the mutant enzyme. Poor activation of M319A could be explained if peptide binding to the relaxed state of this mutant is only modestly better than to the tense state or if the mutation stabilizes the tense relative to the relaxed enzyme. Multiple results support the former explanation. First, the basal activity of the M319A mutant was slightly higher (4.5 ± 1.0 M−1s−1) than that of wild-type DegS (2.9 ± 0.5 M−1s−1). This result is inconsistent with tense-state stabilization, which predicts lower basal activity. Second, when we constructed and characterized the K243D/M319A double mutant, its basal activity (490 ± 50 M−1s−1) was about twice that of the K243D enzyme (210 ± 30 M−1s−1), indicating that the M319A mutation actually contributes to relaxed-state stabilization. Third, saturating concentrations of the YYF peptide activated the K243D/M319A double mutant to levels (2700 ± 400 M−1s−1) only modestly lower than those of the K243D enzyme (3400 ± 550 M−1s−1). This result shows that enzymes bearing the M319A mutation are capable of robust cleavage of RseA. Fourth, saturating concentrations of the DNRDGNVYYF peptide, which is a poorer activator than YYF, enhanced K243D/M319A activity to a level (1100 ± 130 M−1s−1) lower than that of the K243D enzyme (1700 ± 150 M−1s−1). Hence, the M319A mutation, alone or in combination with other mutations, reduces the ability of multiple peptides to activate DegS. Taken together, these results indicate that the M319A mutation has two effects, stabilizing the active relative to the inactive enzyme and decreasing the binding preference of OMP-peptides for active DegS. By itself, the first effect would enhance peptide activation. Because depressed activation was observed, however, the second effect must dominate.
The side chain of Met319 in wild-type DegS contacts the C-terminal phenylalanine of bound OMP peptide (Fig. 1B; Wilken et al., 2004), and thus M319A DegS might be expected to bind OMP peptides more weakly. Exactly the opposite result was observed. This mutant was half-maximally activated by the YYF tripeptide at a 10-fold lower concentration than wild-type DegS (Table 1A). Moreover, in direct binding assays, both the M319A and K243D/M319A mutants bound roughly 35-fold more tightly than wild-type DegS to a fluorescent OMP peptide (Fig. 3C). These increases in peptide-binding affinity are substantially greater than can be explained by modest stabilization of the relaxed enzyme by the M319A mutation. Indeed, modeling suggests that tense M319A DegS binds YYF peptide about 20-fold more tightly than does tense wild-type DegS (see below). In the Discussion, we propose a role for unfavorable contacts between OMP peptides and the Met319 side chain in tense DegS in biasing the allosteric equilibrium towards the active conformation.
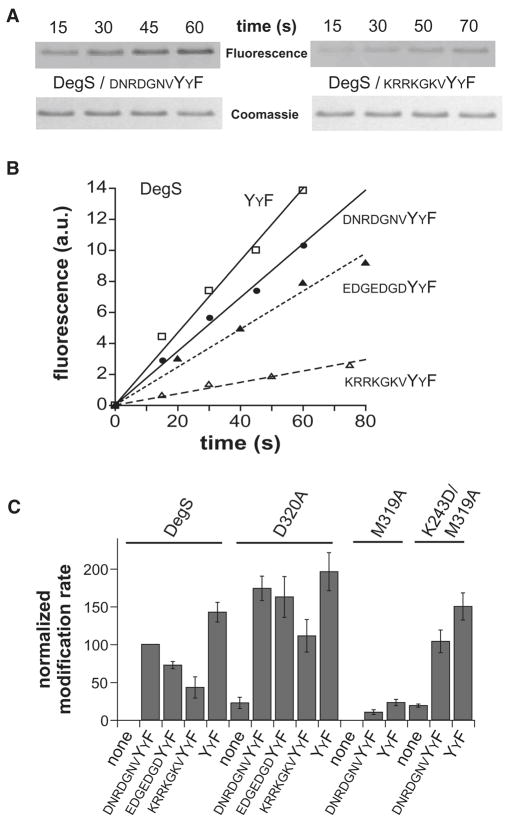
Peptide activation of active-site reactivity
As an independent test of the influence of peptide binding on the two competing DegS conformations, we used a fluorescent derivative of fluorophosphate called rhodamine-FP (Liu et al., 1999) and monitored enzyme modification of the active-site serine by fluorography after SDS-PAGE (Fig. 4A). Wild-type DegS did not detectably react with rhodamine-FP unless activating peptide was present, a result consistent with previous studies using diisopropylfluorophosphate (Sohn et al., 2007). Modification was fastest in the presence of saturating YYF peptide, slower with the DNRDGNVYYF peptide, slower still with the EDGEDGDYYF peptide, and slowest with the KRRKGKVYYF peptide (Fig. 4B). This order of reactivity was the same as observed for peptide activation of DegS cleavage of RseA. Hence, both assays show that the binding of different OMP peptides can result in substantially different degrees of partitioning between the active and inactive conformations of wild-type DegS.
Figure 4.
Peptide dependence of active-site modification.
(A) SDS-PAGE of wild-type DegS after reaction with rhodamine-FP in the presence of two OMP peptides. Fluorescence is shown in the upper strips, and Coomassie-blue staining in the lower strips. (B) Rhodamine-FP (20 μM) modification of wild-type DegS (0.9 μM trimer) in the presence of saturating concentrations of four OMP-like peptides. The lines are linear fits (R ≥ 0.975). (C) Rates of rhodamine-FP modification of wild-type DegS and the D320A, M319A, and K243D/M319A mutants without peptide or with saturating OMP peptides. Rates were normalized to an arbitrary value of 100 for wild-type DegS with saturating DNRDGNVYYF peptide. The error bars represent standard deviations based on three or more experiments.
In the absence of OMP peptide, the D320A mutant was modified, albeit slowly, by rhodamine-FP (Fig. 4C), whereas wild-type DegS was not modified. This result is expected if the D320A mutation increases the equilibrium population of the active enzyme. Moreover, different activating peptides enhanced rhodamine-FP modification of the D320A mutant in roughly the same order as observed for wild-type DegS but to higher levels of modification (Fig. 4C). For the best activating peptides, these differences in D320A activation were close to the error of the measurement. Because less peptide-binding energy is needed to convert D320A DegS into the active conformation, it is possible that all of these peptides activate D320A to roughly comparable extents.
In the presence of saturating OMP peptides, the M319A mutant showed much lower levels of rhodamine-FP modification than wild-type DegS or the D320A mutant but the K243D/M319A double mutant showed substantial modification (Fig. 4C). These results indicate that OMP peptides bind with a reduced preference to the relaxed state of the M319A variant, an effect that can be overcome by stabilization of the active enzyme form by an additional mutation. Overall, the results of rhodamine-FP modification support the idea that DegS activation depends both on the preference of individual peptides for binding to the relaxed and tense states and on the intrinsic equilibrium between these states.
Modeling
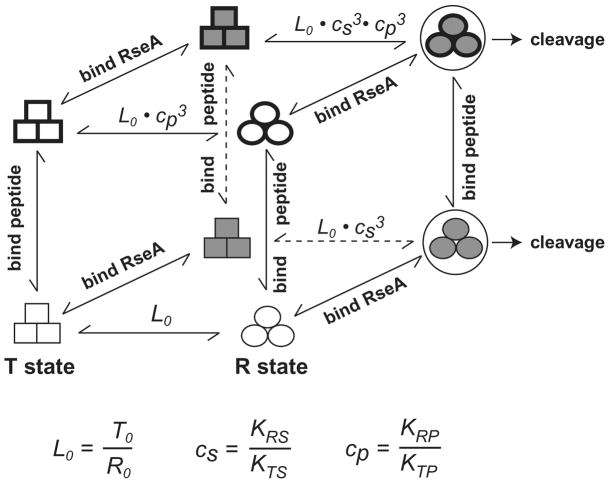
The results described above support an allosteric model in which DegS exists as an equilibrium mixture of active and inactive enzymes and OMP peptides alter RseA cleavage by binding with different affinities to these two enzyme conformations. To determine if the effects of peptide binding, substrate binding, and DegS mutations could be modeled, we performed global fitting to equations derived from the MWC model of allostery (Monod et al., 1965). Because DegS is a trimer, the MWC model predicts 16 relaxed and 16 tense equilibrium species, which differ in the number of bound OMP peptides and/or in the number of bound RseA substrates. The relative populations of these species at any given concentration of peptide and substrate can be calculated from the equilibrium constant (L0) for conversion of the unliganded relaxed (R) state to the tense (T) state, the microscopic equilibrium dissociation constants of OMP peptides for a T-state subunit (KTP) or an R-state subunit (KRP), and the microscopic equilibrium dissociation constants of substrate for a T-state subunit (KTS) or an R-state subunit (KRS). For example, Fig. 5 shows the equilibrium relationships between T-state and R-state trimers without ligands, with three peptides bound, with three substrates bound, and with three peptides and three substrates bound. Because a functional oxyanion hole is absent in peptide-free structures of DegS (Wilken et al., 2004; Zeth, 2004), we assumed that T-state species are enzymatically inactive. Thus, DegS cleavage activity is a function of the concentration of substrate-bound R-state subunits and the rate constant (kr) for cleavage of RseA by these subunits.
Figure 5.
Allosteric relationships.
Equilibria between relaxed DegS trimers (circles) and tense DegS trimers (squares) are perturbed by the binding of OMP peptides and RseA substrate. For simplicity, partially liganded states are not shown. Species that comprise the top face of the cube have three OMP peptides bound; species that comprise the back face of the cube have three RseA substrates bound. The front face of the cube consists of substrate-free trimers. The bottom face of the cube consists of peptide-free trimers. Horizontal arrows indicate conformational equilibria. Vertical arrows signify reactions in which three OMP-like peptides associate or dissociate from DegS. Arrows at an angle indicate reactions in which three molecules of the RseA substrate associate or dissociate from DegS. Cleavage of RseA only occurs from substrate-bound relaxed DegS.
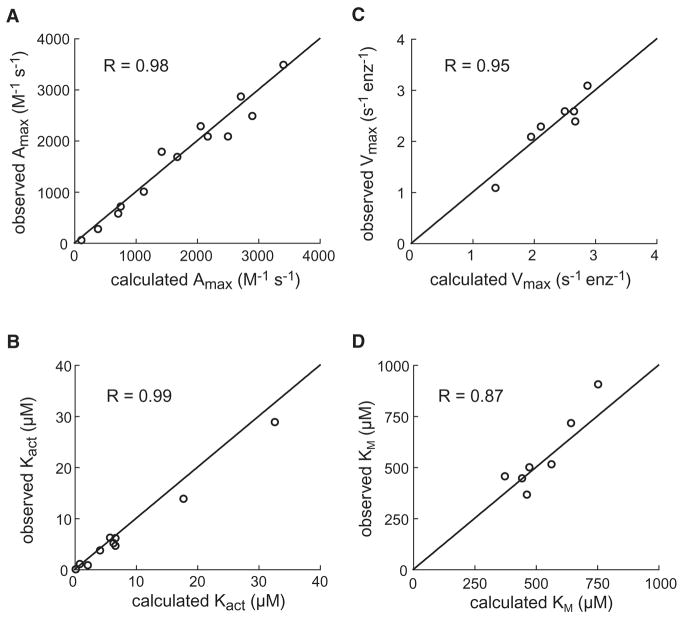
Good fits of the experimental data were obtained using the same values of KTS, KRS, and kr for wild-type DegS and all mutants; individual values of L0 for wild-type DegS, and the K243D, D320A, M319A, and K243D/M319A mutants; and KTP and KRP values specific for each peptide and enzyme (Table 1D). Using these parameters resulted in excellent correlations (R>0.98) between the predicted and experimental parameters that characterize the OMP-peptide dependence of RseA cleavage by DegS and its variants, including maximal activation and the concentration dependence of activation (Fig. 6A & B). The correlations between the predicted and experimental values of KM (R=0.87) and Vmax (R=0.95) for RseA cleavage by different DegS variants in the presence of saturating peptide were also good (Fig. 6C & D) given the relatively small range of these values. We conclude that the MWC model provides a good description of the allosteric regulation of DegS and mutant variants by the binding of OMP peptides and RseA substrate.
Figure 6.
Correlations between predicted and experimental parameters.
(A) Second-order rate constants for cleavage by DegS or mutants with saturating OMP-peptide and RseA substrate (200 μM). (B) OMP-peptide concentrations required for half-maximal activation of RseA (200 μM) cleavage by DegS or mutants. (C) Maximal rates of cleavage by DegS or mutants with saturated RseA substrate and OMP peptide. (D) Substrate concentration required for half-maximal cleavage by DegS or mutants with saturating OMP peptide. In each panel, predicted values were calculated from the MWC allosteric model using the parameters listed in Table 1D, and the correlation coefficients are for fits to the equation: y = x.
When both peptide and substrate are saturating, our model predict that the fractional activity of DegS equals 1/(1+L0/((KTP/KRP)3•(KTS/KRS)3)). However, the KTS/KRS value (~3.4) was constant for DegS and its mutants, indicating that RseA binds about 3-fold more tightly to relaxed than to tense DegS. The fractional-activity expression then simplifies to 1/(1+L0/((3.4•KTP/KRP)3)). Consequently, changes in the initial equilibrium constant (L0) and the ratio of the peptide-binding constants for tense and relaxed DegS ((KTP/KRP)3) determine how different OMP peptides, under saturating conditions, and mutations affect DegS activity. For wild-type DegS, the fitted L0 value was 15000 and the fitted KTP/KRP values for the YYF, DNRDGNVYYF, and KRRKGKVYYF peptides were 12.3, 6.8, and 3.3, respectively. Although these differences in binding preference seem modest, they are magnified in the peptide-saturated trimer because the equilibrium constant relating the tense and relaxed enzyme conformations (L3) is equal to L0/((KTP/KRP)3). Thus, in the presence of saturating substrate, saturating YYF drives 83% of DegS molecules into the active conformation, whereas DNRDGNVYYF and KRRKGKVYYF result in 45% and 9% active enzyme, respectively.
The MWC fitting results indicate that the K243D, M319A, and D320A mutations reduce L0 and also decrease KTP/KRP (Table 1D). Because these mutations affect residues near the PDZ-domain/protease-domain interface and the peptide-binding site (Fig. 1), it is not surprising that each mutation affects the allosteric equilibrium and the peptide-binding preference. However, a reduction in L0 enhances DegS activity whereas a reduction in KTP/KRP decreases peptide stimulation of activity. Thus, the phenotype caused by a given mutation depends on the degree to which each parameter is changed. Compared to wild-type DegS, L0 for the M319A mutant is reduced about three-fold but (KTP/KRP)3 for YYF is reduced almost 100-fold. The large decrease in the latter term outweighs the modest decrease in L0, and thus saturating YYF peptide and substrate drive only about 10% of the mutant M319A enzymes into the active conformation. For the K243D and D320A mutants, however, L0 is reduced ~40–80 fold and (KTP/KRP)3 for YYF binding is reduced ~30–70-fold. In these cases, the large decrease in L0 overwhelms the suppressing effects of the decreased peptide-binding preference, and thus saturating RseA and YYF peptide drive about 85% of the mutant K243D and D320A enzymes into the active conformation. Overall, these results show that the MWC allosteric model provides an explanation for the effects of mutations, peptide-binding preference, and peptide and substrate concentration in determining the proteolytic activity of DegS.
Discussion
Activation by OMP peptides
Our results demonstrate that saturating concentrations of different OMP peptides activate DegS to dramatically different degrees. For example, the rate at which DegS cleaved the periplasmic domain of RseA varied by as much as 35-fold, depending upon the activating peptide. Activation was also a function of the equilibrium between the active and inactive conformations of DegS. For a given peptide, higher levels of activation were observed for DegS mutants in which the conformational equilibrium was shifted towards the relaxed state. These results indicate that peptides bind to the active and inactive conformations of DegS, with the degree of activation depending upon the relative affinity of a given peptide for each conformation. This model has an interesting biological implication. Namely, that different activating OMP sequences could evolve to elicit graded responses in terms of DegS activation and subsequent induction of the envelope-stress response. Thus, it will be important to establish how different natural OMP sequences interact with DegS and the extent to which specific OMPs function in stress signaling. For example, differential sensitivity of individual OMPs to heat stress, acid shock, or oxidative stress might result in differential induction of the σE-stress response even under catastrophic conditions where the activating signals are saturating.
The C-terminal YXF motif of OMPs was initially identified as a sequence that bound to the PDZ domain of DegS and activated cleavage of RseA (Walsh et al., 2003). In the studies reported here, OMP peptides with identical C-terminal tripeptides resulted in highly varied levels of maximal activation. Thus, upstream residues before the YXF sequence can play important roles in activation. Presently, there are three structures of DegS or an ortholog bound to activating peptides, but none show contacts between the enzyme and peptide residues upstream of the four C-terminal residues (Wilken et al., 2004; Hasselblatt et al., 2007; Mohamedmohaideen et al., 2008). These upstream residues may interact with DegS electrostatically. Indeed we observed markedly worse activation when the upstream residues were highly basic as opposed to highly acidic. Alternatively, upstream residues might influence activation largely through contacts with inactive DegS, as all known peptide-bound structures represent the active enzyme conformation. Small differences in peptide binding can have significant functional effects. For example, our best activating peptides bind to a relaxed subunit of wild-type DegS about 12-fold better than to a tense subunit, and this preference is reduced to about 3-fold for the worst activating peptide. Differences of this magnitude correspond to a free-energy change of 0.7–1.5 kcal/mole per subunit and thus could depend on a small number of favorable or unfavorable contacts between the peptide and either conformational state of DegS. Because these small energy differences are magnified in the peptide-saturated DegS trimer, however, individual OMP peptides can differ greatly in activation potential.
Other models for DegS activation by OMP peptide have been suggested. For example, it was proposed that the identity of the penultimate residue of the OMP peptide and the nature of its interactions with the L3 loop of DegS were critical determinants of activation (Wilken et al., 2004; Hasselblatt et al., 2007). Here and previously (Sohn et al., 2007), however, we found that saturating concentrations of OMP peptides with different penultimate residues activate DegS similarly. Indeed, maximal activation by peptides differing only at the penultimate residue varies from the average by less than 20%. It is likely that these small effects reflect minor differences in peptide affinity for active and inactive DegS, but we find no evidence to support a model in which specific contacts between the penultimate peptide side chain and DegS play crucial roles in activation. In this regard, we note that the penultimate side chain of the activating peptide in the structure of a DegS ortholog from M. tuberculosis makes no contacts with the protease domain (Mohamedmohaideen et al., 2008).
Activation of the envelope-stress response occurs in minutes in vivo, and OMP-peptide activation of DegS in vitro occurs even more rapidly (Ades et al., 1999; Sohn et al., 2007). OMP-peptide binding to both inactive and active DegS helps explain fast activation. Specifically, most OMP peptides would be expected to bind initially to inactive DegS molecules, with intramolecular conversion to the active enzyme occurring after binding of multiple peptides. By contrast, if OMP peptides bound exclusively to active DegS, then only a small fraction of total unliganded enzymes would serve as peptide receptors, and activation could easily be limited by slow initial binding. For example, if we assume that the rate constant for binding of OMP peptides to both conformation of DegS is the same, then binding to inactive DegS would occur at an initial rate about 15000-fold faster than to active DegS because of the difference in the equilibrium populations of both species.
Substrate-binding cooperativity revisited
Previously, we found that a DegS mutant lacking the PDZ domain (DegSΔPDZ) had essentially the same RseA-cleavage activity as wild-type DegS bound to DNRDGNVYXF peptides (Sohn et al., 2007). We interpreted this result as evidence that all DegSΔPDZ molecules assumed an active conformation, which suggested that the positive cooperativity observed in substrate cleavage by DegSΔPDZ and DegS arose from favorable substrate-substrate interactions rather than from preferential substrate binding to the relaxed enzyme. However, the results presented here show that wild-type DegS is not fully activated by DNRDGNVYXF peptides and thus cast doubt on the substrate-interaction model. One prediction of the substrate-substrate interaction model is retention of substantial positive cooperativity when almost all DegS enzymes are in the relaxed conformation. Our present results are inconsistent with this model. Specifically, when we stabilized R-state DegS by destabilizing the tense state with the K243D/D320A mutations, the Hill constant for substrate cleavage in the presence of saturating OMP peptide was 1.1 ± 0.1. Because a Hill constant of 1 indicates no cooperativity, we conclude that most positive cooperativity in substrate binding for the wild-type enzyme results from preferential binding of RseA to relaxed DegS. Indeed, fitting of our experimental data to the MWC model suggests that an RseA substrate binds 3.4-fold more tightly to an R-state DegS subunit than a T-state subunit. This difference would be magnified to approximately 40-fold in the substrate-saturated DegS trimer.
Allosteric control
In the absence of substrate and OMP peptides, our results indicate that the ratio of inactive:active DegS is about 15000:1. In the presence of saturating RseA substrate, this ratio changes to about 400:1, allowing a low rate of basal RseA cleavage. Our best OMP peptides shift the conformational equilibrium more dramatically. For example, saturating YYF tripeptide shifts the inactive:active ratio to 8:1 in the absence of substrate and to 1:5 with saturating RseA.
Some DegS cleavage of RseA is required for cell viability (Alba et al., 2001). Peptide-independent basal cleavage might satisfy this demand, or low levels of unassembled OMPs present under normal growth conditions might stimulate somewhat higher levels of cleavage. DegS and RseA are both bound to the inner membrane in E. coli. Hence, co-localization probably stabilizes substrate-enzyme binding, compared to the relatively weak binding observed in soluble assays in vitro. Importantly, however, our modeling indicates that higher effective concentrations of the substrate would not change the allosteric equilibrium significantly because both relaxed and tense DegS bind RseA. Thus, even if binding were saturated in the cell, the vast majority of RseA would bind inactive DegS, leaving the system poised for activation. Indeed, genetic experiments suggest that RseA binding to inactive DegS may prevent cleavage by downstream proteases in the σE-activation cascade until robust OMP-dependent activation occurs (Grigorova et al., 2004).
A pathway for peptide-stabilized changes in DegS conformation
In active DegS, the side chain of Met319 packs against the aromatic ring of the C-terminal phenylalanine in the OMP peptide (Fig. 1B; Wilken et al., 2004). In inactive DegS, this methionine assumes a different rotamer that would clash with the peptide in the “active” conformation. When we changed Met319 to Ala the affinity of OMP peptides for the inactive conformation of DegS increased more than 10-fold, indicating that the Met319 side chain normally hinders OMP-peptide binding to this conformation.
We propose that peptide-induced changes in the Met319 side-chain conformation are part of the mechanism by which peptide binding stabilizes active DegS. The adjacent residue, Asp320, makes a salt bridge with Arg178 in inactive DegS, stabilizing this conformation relative to active DegS (Fig. 1B; Wilken et al. 2004; Zeth, 2004; Sohn et al., 2007). OMP-peptide binding to inactive DegS should force movement of the Met319 side chain, and our mutant results indicate that the resulting conformation is strained. This initial binding strain, in turn, should favor conformational changes that break the Asp320•••Arg178 salt bridge, allowing Arg178 to move. In active DegS, Arg178 is known to make a new set of interactions with the protease domain that help to stabilize the active oxyanion-hole conformation (Fig. 1B; Wilken et al. 2004; Sohn et al., 2007). Hence, unfavorable contacts between the Met319 side chain and OMP peptide in inactive DegS seem to be an important part of the network of interactions that determine the relative stabilities of the peptide-bound tense and relaxed conformations of DegS. We note, however, that the M319A mutant is still activated, albeit poorly, by OMP-peptide binding. Thus, additional peptide-mediated interactions must also contribute to altering the energy balance between allosteric conformations.
Allosteric Models
The concerted MWC model of allostery provides a basis for understanding regulation of DegS protease activity by OMP peptide and substrate binding. Moreover, this model in combination with the equilibrium and kinetic parameters listed in Table 1D provides a good quantitative fit of a wide range of experimental data. We anticipate that this information will help in modeling of the envelope-stress response in the cell. Our results do not rule out more complicated allosteric models, including sequential models in which hybrid mixtures of T-state and R-state subunits are allowed in a single DegS trimer (Koshland et al., 1966) or models in which more than two allosteric conformations of the enzyme are allowed. Indeed, crystallographic studies reveal a structural intermediate between the peptide-bound and peptide-free conformations of DegS (Wilken et al., 2004). However, different structures that are nearly isoenergetic will approximate a single functional state of the system. Hence, we believe that the relative simplicity of the MWC model is well suited for describing allosteric regulation of DegS.
Regulatory implications for related PDZ proteases
DegS is part of an enzyme family with siblings in organisms ranging from bacteria to humans (Kim and Kim, 2005). These enzymes, which can serve as stress-sensing proteases and/or as molecular chaperones, affect myriad processes including microbial virulence and apoptotic regulation in mammalian cells. It seems likely that the activities of many of these proteases will be controlled in ways similar to DegS, specifically by peptide binding to both active and inactive enzyme conformations. Indeed, the results presented here suggest that it should be possible to select or design peptides or mimics that act as potent activators or inhibitors of these enzymes. It will also be interesting to determine if any of these systems are subject to negative biological regulation by peptide signals that bind preferentially to the inactive enzyme.
Experimental procedures
Proteins and peptides
E. coli DegS (residues 27–355) lacking the wild-type membrane anchor and containing an N-terminal His6 tag, a related His6-tagged DegS variant without the PDZ domain (residues 27–256), and a 35S-labeled variant of the periplasmic domain of E. coli RseA (residues 121–216) with a C-terminal His6 tag were expressed and purified as described (Walsh et al., 2003; Cezairliyan and Sauer, 2007; Sohn et al., 2007). Mutations were generated by the QuikChange method (Stratagene) and confirmed by DNA sequencing. Mutant proteins were expressed and purified by the same methods used for the wild-type counterparts. Wild-type and mutant DegS proteins eluted as trimers in the gel-filtration step of purification. Proteins were stored in 50 mM NaHPO4 (pH 8.0), 200 mM NaCl, 10% glycerol, and 2 mM EDTA. Peptides were synthesized by the MIT Biopolymer Laboratory, purified by HPLC, and the expected molecular mass was confirmed by MALDI-TOF mass-spectrometry. All peptides had free α-amino and α-carboxyl groups, except those modified by addition of an N-terminal fluoresceine during synthesis.
Enzymatic and biochemical assays
Unless noted, assays were performed at room temperature (23 ± 1 °C) in 150 mM NaHPO4 (pH 8.3), 380 mM NaCl, 10% glycerol, and 4 mM EDTA. DegS cleavage of 35S-labeled periplasmic domain of RseA was quantified by scintillation counting after separation of the N-terminal cleavage fragment from uncleaved substrate and the C-terminal fragment by differential acid solubility (Sohn et al. 2007). The binding of DegS or mutant variants to a fluorescent OMP peptide was assayed by changes in fluorescence anisotropy (excitation 480 nm; emission 520 nm), after correction for protein scattering. Binding curves, Michealis-Menten curves, and peptide-activation curves were fitted to appropriate equations using the non-linear-least-squares subroutine in KaleidaGraph (Synergy software).
Rhodamine-FP (Liu et al., 1999) was a gift from C. Salisbury, E. Weerapana, and B. Cravatt (Scripps Institute). Modification of DegS or variants (0.9 μM trimer) with Rhodamine-FP (20 μM) was performed in the presence or absence of OMP-like peptides. After quenching reactions by addition of an equal volume of 3X Laemmli sample buffer and boiling, samples were analyzed by SDS-PAGE and fluorescence was quantified using an Amersham Typhoon imager (excitation 532 nm; emission 588 nm). Initial rates were determined from the linear phase of the reaction (generally 0–2 min). Rhodamine-FP modification was not observed for a DegS variant in which the active-site serine was mutated (S201A).
Fitting and modeling
Steady-state kinetic parameters were obtained by fitting data to the Hill form of the Michaelis-Menten equation using the non-linear-least-squares subroutine in the program KaleidaGraph (Synergy software). Equations from the MWC model of allostery were also used to fit experimental data using subroutines in ORIGIN (global fit; OriginLab) and MATLAB (lsqnonlin; MathWorks), assuming that R-state but not T-state subunits are enzymatically active. The substrate dependence of DegS cleavage of RseA was fit to the equation:
| (eq. 1) |
where kr is the rate constant for RseA cleavage, αis [substrate]/KRS, cs is KRS/KTS, and L0 is T0/R0. To fit data obtained in the presence of saturating OMP-like peptides, equation 1 was modified by substituting LPapp (L0•cp3) for L0, where cp is KRP/KTP. The peptide dependence of DegS cleavage of RseA was fit to the equation:
| (eq. 2) |
where A0 is basal peptide-independent activity, Amax is kr[DegS3]α/(1−α)−A0, β is [peptide]/KRP, and LSapp is L0(1+csα)3/(1+α)3.
Acknowledgments
We thank C. Salisbury, E. Weerapana, and B. Cravatt for the gift of rhodamine-FP, S. Hahn for assistance with MATLAB, and B. Cezairliyan, R. Chaba, E. Gur, and C. Gross for helpful discussions and sharing unpublished data. Supported by NIH grant AI-16892. J. Sohn was supported by an NIH postdoctoral fellowship (AI-074245-01A1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ades SE, Connolly LE, Alba BM, Gross CA. The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- Baslé A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. Crystal structure of osmoporin OmpC from E. coli at 2.0 Å. J Mol Biol. 2006;362:933–942. doi: 10.1016/j.jmb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Cezairliyan BO, Sauer RT. Inhibition of regulated proteolysis by RseB. Proc Natl Acad Sci USA. 2007;104:3771–3776. doi: 10.1073/pnas.0611567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrmann M, Clausen T. Proteolysis as a regulatory mechanism. Annu Rev Genet. 2004;38:709–724. doi: 10.1146/annurev.genet.38.072902.093416. [DOI] [PubMed] [Google Scholar]
- Flynn JF, Levchenko I, Sauer RT, Baker TA. Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev. 2004;18:2292–2301. doi: 10.1101/gad.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorova IL, Phleger NJ, Mutalik VK, Gross CA. Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. Proc Natl Acad Sci USA. 2006;103:5332–5337. doi: 10.1073/pnas.0600828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorova IL, Chaba R, Zhong HJ, Alba BM, Rhodius V, Herman C, Gross CA. Fine-tuning of the Escherichia coli sigmaE envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev. 2004;18:2686–2697. doi: 10.1101/gad.1238604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselblatt H, Kurzbauer R, Wilken C, Krojer T, Sawa J, Kurt J, Kirk R, Hasenbein S, Ehrmann M, Clausen T. Regulation of the sigmaE stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress. Genes Dev. 2007;21:2659–2670. doi: 10.1101/gad.445307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Kim KK. Structure and function of HtrA family proteins, the key players in protein quality control. J Biochem Mol Biol. 2005;38:266–274. doi: 10.5483/bmbrep.2005.38.3.266. [DOI] [PubMed] [Google Scholar]
- Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci USA. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamedmohaideen NN, Palaninathan SK, Morin PM, Williams BJ, Braunstein M, Tichy SE, Locker J, Russell DH, Jacobs WR, Jr, Sacchettini JC. Structure and function of the virulence-associated high-temperature requirement A of Mycobacterium tuberculosis. Biochemistry. 2008;47:6092–6102. doi: 10.1021/bi701929m. [DOI] [PubMed] [Google Scholar]
- Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: A Plausible Model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J, Grant RA, Sauer RT. Allosteric activation of DegS, a stress sensor PDZ protease. Cell. 2007;131:572–583. doi: 10.1016/j.cell.2007.08.044. [DOI] [PubMed] [Google Scholar]
- Vande Walle L, Lamkanfi M, Vandenabeele P. The mitochondrial serine protease HtrA2/Omi: an overview. Cell Death Differ. 2008;15:453–460. doi: 10.1038/sj.cdd.4402291. [DOI] [PubMed] [Google Scholar]
- Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T. Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease. Cell. 2004;117:483–494. doi: 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
- Zeth K. Structural analysis of DegS, a stress sensor of the bacterial periplasm. FEBS Lett. 2004;569:351–358. doi: 10.1016/j.febslet.2004.06.012. [DOI] [PubMed] [Google Scholar]