Abstract
We generated Prx1CreER-GFP transgenic mice that express tamoxifen-inducible Cre recombinase and GFP under the control of a 2.4 kb Prx1 promoter. The transgene is expressed in osteochondro progenitor cells in the developing limb buds and in a subpopulation of periosteal cells that is closely associated with the cortical bone. GFP-expressing cells isolated from the diaphyses of long bones by cell sorting express multiple markers of periosteal cells, including Prx-1, Fgf18, Tenascin-W, Periostin, and Thrombospondin 2. In addition, these cells undergo chondrogenic and osteogenic differentiation in culture upon induction. Cell fate analysis using the Rosa26 LacZ reporter indicated that transgene-expressing cells give rise to some of the chondrocytes and osteoblasts in the fracture callus. Collectively, these observations strongly suggest that the transgene-expressing cells are osteochondro progenitor cells in the periosteum. The established Prx1CreER-GFP mice would offer novel approaches for analyzing the functions of periosteal cells in vitro and in vivo.
Keywords: Periosteum, Perichondrium, Osteogenesis, Chondrogenesis, GFP, CreER, Tamoxifen, Osteochondro progenitor, Fracture healing
Introduction
Periosteum and perichondrium play central roles in skeletal development [1]. Periosteum and perichondrium cover most of the external surface of bone and cartilage and coordinate bone formation and cartilage development. In addition, periosteum contains osteochondro progenitor cells that participate in bone and cartilage formation during normal development and under pathologic conditions such as fracture healing [2,3]. Microscopically, periosteum is composed of at least 2 layers, outer fibrous layer and inner cambium layer [4]. The fibrous layer contains fibroblasts, while the cambium layer is believed to contain a number of cell types including mesenchymal progenitor cells, osteoprogenitor cells, osteoblasts, and fibroblasts. Although the importance of periosteal cells has long been recognized, periosteum still remains poorly characterized due to the lack of tools for molecular and cellular analyses. While mice provide a unique opportunity for utilizing numerous genetically engineered mouse models, their small size poses a considerable challenge in primary cell isolation. Recent analyses in chicks have indicated that there are several molecularly distinct domains within the periosteum and perichondrium [1]. The establishment of a method for isolating a pure population of mouse periosteal cells would greatly facilitate the molecular analysis of periosteum.
The in vivo analysis of periosteum is also hampered by the lack of genetic tools to express or recombine genes specifically in the periosteum. A 2.4 kb Prx1 promoter directs the transgene expression in undifferentiated mesenchyme in the developing limb buds as early as embryonic day 10.5 (E10.5) [5]. The transgene expression is extinguished in the condensing mesenchyme and chondrocytes, and the expression is confined to the periosteum and tendons of the limbs at E15.5. Therefore, the 2.4 kb Prx1 promoter would be useful for expressing genes in the periosteum after cartilage formation. Because Prx1-Cre transgenic mice that express Cre recombinase under the control of the 2.4 kb Prx1 promoter induce Cre-loxP mediated recombination in osteochondro progenitor cells in the developing limb bud [6,7], we hypothesized that periosteal cells, in which the 2.4 kb Prx1 promoter is active, represent osteochondro progenitor cells.
In the present study, we generated Prx1CreER-GFP transgenic mice that express CreER and GFP in the periosteum under the control of the 2.4 kb Prx1 promoter. CreER is a fusion molecule between Cre recombinase and the ligand-binding domain of estrogen receptor [8]. Upon induction with tamoxifen, CreER translocates into the nucleus and recombines genes at the loxP sites. While the expression of CreER allows timed recombination of the floxed allele, the expression of GFP allows isolation of transgene-expressing cells from the periosteum by cell sorting. We show here that the transgene is expressed in a subpopulation of periosteal cells, and transgene-expressing periosteal cells exhibit chondrogenic and osteogenic differentiation in culture. Furthermore, transgene-expressing cells give rise to some of the chondrocytes and osteoblasts in the fracture callus in vivo. These observations indicate that the Prx1CreER-GFP transgene is expressed in osteochondro progenitor cells in the periosteum. The established mouse line would be useful for studying the biology of periosteal cells.
Materials and methods
The institutional animal care and use committee of Case Western Reserve University approved all animal procedures.
DNA construction, screening, and transgenic mice
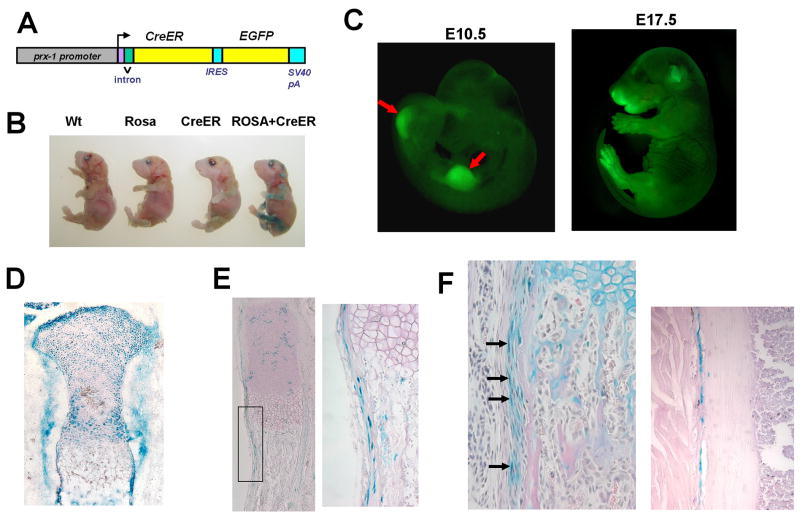
To express CreER and EGFP in periosteal cells, we cloned cDNAs for CreER and EGFP downstream of a 2.4 kb Prx1 promoter (Fig. 1A). CreER is a fusion molecule of Cre recombinase and ligand binding domain of a mutated estrogen receptor [8]. The internal ribosome entry site (IRES), EGFP cDNA, and SV40 polyadenylation signal were excised from pIRES2-EGFP (Clontech) and subcloned downstream of CreER cDNA. The construct was injected into the fertilized C57BL6 × SJL F2 hybrid eggs at the Case Transgenic and Targeting Facility. Transgenic founders were identified by Polymerase Chain Reaction (PCR) using specific primers for Cre recombinase (Supplementary Table 1). Transgene expression was examined by illuminating the limbs with a fluorescence-inducing flashlight.
Fig. 1.
(A) Schematic representation of the Prx1CreER-GFP transgene. cDNAs for CreER and EGFP were cloned downstream of a 2.4 kb Prx1 promoter. (B) X-gal staining of E18.5 embryos showing clear correlation with the genotype. Tamoxifen was injected at E15.5 and E16.5. Only the embryo harboring the Prx1CreER-GFP transgene and Rosa26 LacZ reporter shows positive staining in the limbs. Other embryos show minimal background. (C) GFP fluorescence of Prx1CreER-GFP embryos at E10.5 (left) and E17.5 (right). E10.5 embryo shows fluorescence in the forelimb and hindlimb buds (arrows). E17.5 embryo shows fluorescence in the limbs and some craniofacial regions. (D) X-gal staining of the proximal tibia of E17.0 Rosa26 LacZ; Prx1CreER-GFP embryo. Tamoxifen was injected at E9.0. (E) X-gal staining of the ulna of E18.5 Rosa26 LacZ; Prx1CreER-GFP embryo. Tamoxifen was injected at E15.5 and E16.5. The right panel shows higher magnification of the boxed area in the left panel. (F) X-gal staining of the radius (left) and tibia (right) of a Rosa26 LacZ; Prx1CreER-GFP mouse at P26. Tamoxifen was injected daily between P19–P23. Sections were counterstained with orcein (right) or hematoxylin, eosin, and alcian blue (left). In the radius, cells in the inner layer of the periosteum stain positive for X-gal (arrows). In the tibia, X-gal-stained periosteal cells are closely associated with the cortical bone.
Tamoxifen injection, X-gal staining, and histological analysis
Tamoxifen-inducible Cre activity was evaluated using the Rosa26 LacZ reporter mice (Jackson Laboratories). 1 mg/100μl/day tamoxifen (Sigma) was injected daily for indicated time periods into the peritoneal cavity of the pregnant mother or offspring mice. Mice were sacrificed 2–8 days after the last tamoxifen injection. X-gal staining was performed as previously described [9]. X-gal-stained tissues were embedded in paraffin and sectioned in 7μm. Some of the embryos were directly embedded in OCT compound (Sakura) and cryosectioned in 7μm. Frozen sections were fixed in 2% formalin, 0.2% glutaraldehyde in PBS for 5 min and reacted with X-gal solution. Sections were counterstained with orcein or hematoxylin, eosin, and alcian blue.
Isolation of transgene-expressing periosteal cells
For isolation of periosteal cells, diaphyses of tibiae, radii, and ulnae were dissected out from 5–19-day old Prx1CreER-GFP mice. Surrounding soft tissues, such as skin, muscle, and epiphyseal cartilage, which also contained some transgene-expressing cells, were removed from each skeletal element prior to isolation. Collected diaphyses were placed in PBS and centrifuged for 1 min to remove cells in the bone marrow. Then, the diaphyses were digested with 3 mg/ml collagenase B (Roche) at 37C for 1.5 h. Next, the diaphyses were centrifuged, and the pelleted tissues were further digested in 0.25% Trypsin/EDTA (Invitrogen) at 37C for additional 1.5 h. The released cells were collected by centrifugation, resuspended in DMEM, 10% FCS, and plated in a culture dish. On the following day, cells were trypsinized, resuspended in PBS, 5% FCS, and filtered through a 70μm cell strainer. Then, the cells were sorted by fluorescence-activated cell sorting at Case Western Reserve University Comprehensive Cancer Center Facility. Only cells with a strong GFP signal were isolated as GFP positive, and cells without a GFP signal were collected as GFP negative. Cells with an intermediate signal were not collected for experiments.
Chondrogenic and osteogenic differentiation
For chondrogenic differentiation, an equal number of sorted GFP positive and negative cells (16,000–71,000) were resuspended in a 10 μl droplet and plated in micromass at the center of a 48-well plate. Cells were incubated for 1.5 h with 5% CO2 at 37C. Then, 200 μl of DMEM, 10% FCS were added onto the micromass droplet. Chondrogenic differentiation was induced by 250–500 ng/ml BMP2 (R&D). The medium was replaced every 2–3 days. After 5–11 days of chondrogenic induction, cells were processed for RNA extraction or stained overnight with 1% alcian blue in 3% acetic acid.
For osteoblast differentiation, sorted GFP-positive and negative cells were plated in a 6-well plate at 6,000–8,000 cells/cm2 and cultured in αMEM, 10% FCS with 5% CO2 at 37C. After reaching confluence, osteogenic differentiation was induced by supplementing αMEM, 10% FCS with 5 mM β-glycerophosphate, 100μg/ml ascorbic acid, with/without 100 nM dexamethasone. Medium was changed 3 times per week. After 4 weeks of induction, osteogenic differentiation was examined by alkaline phosphatase staining, alizarin red staining, and semi-quantitative RT-PCR. For alkaline phosphatase staining, cells were fixed in 100% methanol for 1 min and incubated with the substrate (ALP substrate kit, Vector). For alizarin red staining, cells were fixed with 70% ethanol for 15 min and stained in 1% alizarin red solution.
Semi-quantitative and quantitative PCR
RNA was extracted from freshly sorted cells or cells in culture using RNeasy micro kit (Qiagen). RNA was also extracted from MC3T3E1, 3T31L1, and RAW264.7 cells. RNA was reverse-transcribed to cDNA with High Capacity cDNA Reverse Transcription Kits (Applied Biosystems). For semi-quantitative analyses, PCR was performed on the Applied Biosystems GeneAmp PCR system 9700. cDNA was amplified 27–35 cycles, and the band intensity was compared while the band intensity and cycle numbers were linear. The PCR primers are listed in Supplementary Table 1. All PCR products corresponded to the expected molecular size. For newly designed primers, the identity of the PCR product was further confirmed by DNA sequencing. Quantitative PCR was performed on the Applied Biosystems 7500 Real-time PCR detection system. TaqMan probe sets were designed and synthesized by Applied Biosystems (Col2a1; Mm01309562_g1, Col10a1; Mm00487041_m1, Aggrecan; Mm00545794_m1, Gapdh; 4352932E). To compare gene expression levels, the comparative cycle threshold (Ct) method was used. Gapdh was used as an endogenous control.
Creation of fractures and histological analysis
Unilateral fracture of the ulna or femur was created in 4—7-week-old Rosa26 LacZ;Prx1CreER-GFP mice under ketamine, xylazine, and acepromazine anesthesia. A mid-diaphyseal fracture of the ulna was created using a nail clipper through an open incision, leaving the radius intact. No internal or external fixation was applied postoperatively. The femoral fracture was created using the fracture device as described previously [10,11]. Tamoxifen was injected into the peritoneal cavity before and/or after fracture. Fracture callus was harvested 5–10 days after fracture for histological analysis. Tissues were processed for cryosectioning and X-gal staining as described previously [3].
Results and Discussion
Screening of transgenic mice
Among 39 founder mice, we identified 11 transgenic founders by PCR. Three out of 11 mouse lines expressed the transgene as judged by the fluorescence in the limbs. These transgenic lines were further crossed with the Rosa26 LacZ reporter mice, and tamoxifen-inducible Cre activity was evaluated in offspring embryos/mice by X-gal staining. Two of the 3 lines showed intense X-gal staining upon induction with tamoxifen. Out of 2 lines that showed similar Cre activity, 1 line was chosen for further analysis. X-gal staining of offspring embryos showed clear correlation between X-gal staining and the genotype (Fig. 1B). The selected line was further mated to homozygosity. Both heterozygous and homozygous mice developed and bred normally, indicating that the transgene did not disrupt genes that are essential for normal development.
Transgene is expressed in the osteochondro progenitor cells in developing limb buds, and its expression persists in the perichondrium/periosteum after cartilage development
Expression domains of the transgene were further analyzed by the fluorescence of GFP and tamoxifen-inducible Cre recombinase activity. At E10.5, GFP fluorescence was observed in the developing limb buds (Fig. 1C). At E17.5, GFP fluorescence was also observed in some of the craniofacial regions in addition to the limbs.
Tamoxifen injection into the pregnant mother at E9.0 resulted in widespread recombination of the Rosa26 LacZ reporter allele at E17.0 (Fig. 1D). X-gal staining of the limbs indicated recombination in chondrocytes, osteoblasts, and periosteal and perichondrial cells. These observations indicate that the transgene is expressed in the osteochondro progenitor cells in the developing limb bud. When tamoxifen was injected into the pregnant mother at E15.5 and E16.5, most of the chondrocytes stained negative for X-gal at E18.5, while recombination in the perichondrial and periosteal cells persisted (Fig. 1E). The tamoxifen-induced recombination in the epiphyseal cartilage was further reduced postnatally. Daily tamoxifen injection between postnatal day 19 (P19) and P23 induced recombination in the periosteum and perichondrium, while most of the epiphyseal chondrocytes stained negative for X-gal at P26 (Fig. 1F). Interestingly, where the periosteum showed morphologically distinct inner and outer layers, recombined cells were predominantly found in the inner cambium layer (Fig. 1F, left). In other regions where the two layers were not distinct, recombined cells were closely associated with the outer surface of the cortical bone (Fig. 1F, right). In addition to the periosteum and perichondrium, some of the articular chondrocytes and tendon cells also showed recombination (data not shown). In contrast, very few cells showed recombination in the endosteum and in the bone marrow (Fig. 1F). Tamoxifen-induced recombination in the periosteum was observed at least up to P35 (data not shown).
Transgene-expressing cells isolated by cell sorting are periosteal cells
To further characterize the transgene-expressing periosteal cells, GFP-expressing cells were isolated by cell sorting from the diaphyses of Prx1CreER-GFP mice. We obtained approximately 9,000 cells/mouse from heterozygous mice and 25,000 cells/mouse from homozygous mice on the average. GFP-positive cells constituted 0.3–9.2% of cells released from the diaphyses. Because very few cells express the transgene in the endosteum and bone marrow, sorted GFP-positive cells would be predominantly periosteal cells. To further support this notion, we examined the expression of genes that are preferentially expressed in the periosteum. RT-PCR analyses indicated that the sorted GFP-positive cells indeed express multiple markers of periosteal cells (Fig. 2A). These markers include Col1a1, Col1a2, Prx-1, Fgf18, Tenascin-W (TN-W), Alcam, Periostin, and Thrombospondin 2 (Tsp2). The expression profile is unique in that none of the control cell lines MC3T3E1, 3T3L1, RAW 264.7 expressed all the periosteal markers examined. Collectively, these observations strongly suggest that the sorted GFP-positive cells are periosteal cells.
Fig. 2.

Semi-quantitative PCR analyses of sorted Prx1CreER-GFP-positive cells, MC3T3E1 (osteoblastic cell line), 3T31L1 (preadipocyte cell line), and RAW264.7 cells (macrophage cell line). (A) RT-PCR for molecular markers of the periosteum. (B) RT-PCR for hormone and growth factor receptors and transcription factors for chondrocyte and osteoblast differentiation. (TN-W) Tenascin-W; (Tsp2) Thrombospondin 2; (Alcam) Activated leukocyte cell adhesion molecule; (Smo) Smoothened; (Ptc) Patched; (Osx) Osterix.
Transgene-expressing periosteal cells express master transcription factors for chondrocyte and osteoblast differentiation and receptors for FGF, Ihh, and PTH
Because periosteum consists of heterogeneous cell populations, the roles of transgene-expressing periosteal cells are of utmost interest. Since the periosteum is known to contain osteochondro progenitor cells, we examined the expression of transcription factors that are essential for chondrocyte and osteoblast differentiation. The sorted GFP-positive cells express Sox9, a master transcription factor for chondrocyte differentiation, and Runx2 and Osterix, two master transcriptional regulators for osteoblast differentiation, suggesting chondrogenic and osteogenic potential (Fig 2B). We also examined receptors for known regulators of bone formation, FGF, Indian hedgehog (Ihh), parathyroid hormone (PTH), and PTH-related protein (PTHrP). RT-PCR analyses indicated the expression of Fgfr1, Fgfr2, Fgfr3, Smoothened (Smo), Patched (Ptc), and type I PTH receptor (Pthr1) in the sorted GFP-positive cells (Fig. 2B). These observations suggest that the transgene-expressing periosteal cells are a target of FGF, Ihh, PTH, and PTHrP.
Chondrogenic and osteogenic differentiation of transgene-expressing periosteal cells
To examine the chondrogenic potential of GFP-positive periosteal cells, we plated GFP-positive and negative cells in micromass and treated the cells with BMP2 or its vehicle. Chondrogenic differentiation was analyzed by alcian blue staining and quantitative PCR. After 8 days of BMP2 treatment, GFP-positive cells showed the most intense staining with alcian blue, while BMP2-treated GFP-negative cells showed weak staining (Fig. 3A). Consistent with this observation, Col2a1 and Aggrecan expression was dramatically increased in BMP2-treated GFP-positive cells, while the increase was fairly modest in GFP-negative cells (Fig. 3B,C). These observations indicate that GFP-positive cells can differentiate into chondrocytic cells. We further examined the time course of Col2a1 and Col10a1 expression in GFP-positive cells in the presence or absence of BMP2 (Fig. 3D,E). While Col2a1 expression gradually decreased in BMP2-treated GFP-positive cells between Day 5 and Day 11, Col10a1 expression showed a gradual increase during the same period. It is possible that continuous BMP2 treatment induced hypertrophic chondrocyte differentiation in GFP-positive cells.
Fig. 3.
Chondrogenic and osteogenic differentiation of sorted GFP-positive and negative cells. (A–E) Chondrogenic differentiation. (A) Alcian blue staining of GFP-positive and negative cells. GFP-positive cells showed the most intense staining after 8 days of BMP2 treatment at 500 ng/ml. (B–E) Quantitative PCR for chondrogenic markers after BMP2 treatment. BMP2-treated GFP-positive cells showed highest Col2a1 (B) and Aggrecan (C) expression after 8 days of BMP2 treatment at 500 ng/ml. (D,E) Time course of Col2a1 (D) and Col10a1 (E) expression in GFP-positive cells cultured in the presence or absence of 250 ng/ml BMP2. RNA was extracted at days 5, 8 and 11. Col2a1 expression decreased, while Col10a1 expression increased between days 5–11 in BMP2-treated GFP-positive cells. (F–H) Osteogenic differentiation. (F) Alkaline phosphatase staining showing strong alkaline phosphatase activity in GFP-positive cells after 4 weeks of osteogenic treatment. (G) Alizarin red staining showing massive calcium deposition in cells with 4 weeks of osteogenic treatment. (H) Semi-quantitative PCR for osteoblastic markers in sorted GFP-positive cells with or without osteogenic induction. (Osx) Osterix; (Bsp) Bone sialoprotein; (Alp) Alkaline phosphatase.
We also examined the osteogenic potential of GFP-positive and negative cells by inducing osteogenic differentiation with β-glycerophosphate and ascorbic acid. After 4 weeks of induction, GFP-positive cells showed more intense staining for alkaline phosphatase activity compared with GFP-negative cells (Fig. 3F). GFP-positive cells also showed more intense staining with alizarin red, indicating increased matrix mineralization (Fig. 3G). Semi-quantitative PCR analyses of GFP-positive cells indicated that osteogenic induction caused increased expression of osteoblast markers Runx2, Osterix (Osx), Bone sialoprotein (Bsp), and Alkaline phosphatase (Alp) (Fig. 3H). These observations indicate that GFP-positive cells can differentiate into osteoblastic cells in vitro.
Altogether, these observations indicate that GFP-positive cells show higher efficiency for chondrogenic and osteogenic differentiation compared with GFP-negative cells, suggesting that GFP-positive cells are enriched in osteochondro progenitor cells. The apparent low efficiency of GFP-negative cells may be explained by the reduced chondrogenic and osteogenic potential of periosteal cells that do not express the transgene. It is also possible that the presence of non-chondrogenic and non-osteogenic cells from surrounding tissues further decreased the overall efficiency of chondrogenic and osteogenic differentiation of GFP-negative cells.
Transgene-expressing cells in the periosteum are osteochondro progenitor cells in vivo
To examine the chondrogenic and osteogenic differentiation of transgene-expressing periosteal cells in vivo, tamoxifen was injected daily into Rosa26 LacZ; Prx1CreER-GFP mice for 10 consecutive days starting at P28. Diaphyseal fractures were created in the ulna at P33. Tamoxifen was last injected at P37 and discontinued before overt cartilage formation at the fracture site. Fracture callus was harvested 7 days after creating the fracture, and recombination of the Rosa26 LacZ reporter allele was examined by X-gal staining (Fig. 4A–D). Histological analysis indicated that cells in the periosteum overlying the fracture callus (Fig. 4C,D arrowheads), undifferentiated cells at the fracture site (Fig. 4B), some of the chondrocytes in the cartilaginous callus (Fig. 4C), and osteoblasts lining the bone trabeculae in the newly formed subperiosteal bone (Fig. 4D) stained positive for X-gal. Similar results were also obtained in a model of femoral fracture, in which fracture callus was harvested 10 days after creating the fracture (Fig. 4E,F). These observations strongly suggest that transgene-expressing periosteal cells differentiated into chondrocytes and osteoblasts in the fracture callus and further support the notion that transgene-expressing cells are osteochondro progenitor cells. Furthermore, these experiments also indicated that numerous cells in the fracture callus remain negative for X-gal despite repeated tamoxifen injection, suggesting the presence of osteochondro progenitor cells that do not express the Prx1CreER-GFP transgene. Future generation of transgenic mice that target GFP-negative cell population will provide further information about the cellular origin of cells in the fracture callus.
Fig. 4.
X-gal staining of fracture callus harvested from Rosa26 LacZ; Prx1CreER-GFP mice. (A) Fracture callus of the ulna at 7 days after fracture. 1 mg/100μl tamoxifen was injected daily into the peritoneal cavity between P28–37. Fracture was created in the diaphysis of ulna at P33. Fracture callus was harvested at P40. Sections were counterstained with hematoxylin, eosin and alcian blue. (B–D) Higher magnification of the boxed areas in (A). X-gal stained cells were observed in undifferentiated cells at the fracture site (B), periosteal cells overlying the fracture callus (arrowheads in C, D), chondrocytes in the cartilaginous callus (arrows in C), and osteoblasts lining the bone trabeculae (arrows in D). (E) Fracture callus of the femur at 10 days after fracture. Femoral fracture was created at P49, and fracture callus was harvested at P59. 1 mg/100μl tamoxifen was injected into the peritoneal cavity at P52 and P53. Sections were counterstained with hematoxylin, eosin and alcian blue. (F) Higher magnification of the boxed area in (E). X-gal-stained cells were observed in the cartilaginous callus.
In summary, we have established a novel transgenic mouse line expressing GFP and CreER in the periosteum. We have successfully isolated transgene-expressing periosteal cells by cell sorting. GFP-expressing periosteal cells showed chondrogenic and osteogenic potential in vitro and in vivo. Since no established methods are currently available for isolating a pure cell population from the periosteum, the established mouse line provides a unique resource for studying the phenotype of periosteal cells. In addition, the expression of tamoxifen-inducible Cre recombinase will allow genetic recombination in this particular cell population in the periosteum. The Prx1CreER-GFP mouse line offers a novel approach for studying the biology of periosteal cells.
Supplementary Material
Acknowledgments
We thank Drs. James Martin and Kazuhisa Nakashima for the Prx1 promoter and CreER cDNA. We also thank Mr. Amad Awadallah and Ms. Teresa Pizzuto for technical assistance, Dr. James Dennis for scientific discussion, and Ms. Valerie Schmedlen for editorial assistance. We thank the Case Transgenic and Targeting Facility for assistance with the generation of transgenic mice. This work was supported by March of Dimes Birth Defects Foundation, National Institutes of Health R21DE017406, and Musculoskeletal Transplant Foundation to S.M.. The Cytometry and Imaging Microscopy Core Facility of the Case Comprehensive Cancer Center was supported by P30CA43703 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bandyopadhyay A, Kubilus JK, Crochiere ML, Linsenmayer TF, Tabin CJ. Identification of unique molecular subdomains in the perichondrium and periosteum and their role in regulating gene expression in the underlying chondrocytes. Dev Biol. 2008;321:162–174. doi: 10.1016/j.ydbio.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakahara H, Bruder SP, Goldberg VM, Caplan AI. In vivo osteochondrogenic potential of cultured cells derived from the periosteum. Clin Orthop Relat Res. 1990;259:223–232. [PubMed] [Google Scholar]
- 3.Colnot C. Skeletal cell fate decisions within periosteum and bone marrow during bone regeneration. J Bone Miner Res. 2009;24:274–282. doi: 10.1359/jbmr.081003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanenbaum HC. An ultrastructural study of osteogenesis in chick periosteum in vitro. Bone. 1986;7:295–302. doi: 10.1016/8756-3282(86)90211-5. [DOI] [PubMed] [Google Scholar]
- 5.Martin JF, Olson EN. Identification of a prx1 limb enhancer. Genesis. 2000;26:225–229. [PubMed] [Google Scholar]
- 6.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre recombinase in the developing mouse limb bud driven by a Prx1 enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 9.Matsushita T, Wilcox WR, Chan Y, Kawanami A, Bükülmez H, Balmes G, Krejci P, Mekikian PB, Otani K, Yamaura I, Warman ML, Givol D, Murakami S. FGFR3 promotes synchondrosis closure and fusion of ossification centers through the MAPK pathway. Hum Mol Genet. 2009;18:227–240. doi: 10.1093/hmg/ddn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnarens F, Einhorn TA. Production of a standard closed fracture in laboratory animal bone. J Orthop Res. 1984;2:97–101. doi: 10.1002/jor.1100020115. [DOI] [PubMed] [Google Scholar]
- 11.Tsuji K, Bandyopadhyay A, Harfe BD, Cox K, Kakar S, Gerstenfeld L, Einhorn T, Tabin CJ, Rosen V. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet. 2006;38:1424–1429. doi: 10.1038/ng1916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.