Summary
Steroid hormone-activated receptor proteins are among the best understood class of factors for altering gene transcription in cells. Steroid receptors are of major importance in maintaining normal human physiology by responding to circulating concentrations of steroid in the nM range. Nonetheless, most studies of steroid receptor action have been conducted using the supra-physiological conditions of saturating concentrations (≥100 nM) of potent synthetic steroid agonists. Here we summarize the recent developments arising from experiments using two clinically relevant conditions: subsaturating concentrations of agonist (to mimic the circulating concentrations in mammals) and saturating concentrations of antagonists (which are employed in endocrine therapies to block the actions of endogenous steroids). These studies have revealed new facets of steroid hormone action that could not be uncovered by conventional experiments with saturating concentrations of agonist steroids, such as a plethora of factors/conditions for the differential control of gene expression by physiological levels of steroid, a rational approach for examining the gene-specific variations in partial agonist activity of antisteroids, and a dissociation of steroid potency and efficacy that implies the existence of separate, and possibly novel, mechanistic steps and cofactors.
Introduction
Steroid hormones are of major importance for the development, differentiation, and homeostasis of humans and vertebrates in general. Both agonist and antagonist steroids are involved, where antagonists or antisteroids are compounds that prevent the action of agonist steroids and are used in endocrine therapies to counter the actions of endogenous steroid. Five classes of steroid hormones are produced in humans (androgens, estrogens, glucocorticoids, mineralocorticoids, and progestins) and each preferentially binds to one of the 48 members of the steroid/nuclear receptor superfamily of intracellular proteins that are expressed in humans (1). Non-steroidal compounds, like 3,5,3′-triiodothyronine and vitamin D3, are the common ligands for the nuclear receptors but most nuclear receptors are “orphan receptors” with no known ligands. The conventional mode of action of steroid hormones begins by its binding to the ligand binding domain (LBD) of the cognate steroid receptor (Fig. 1). The resulting receptor-steroid complex binds to specific, biologically active DNA sequences (called hormone response elements, or HREs) of the nuclear chromatin, where a variety of cofactors are recruited both to modify the chromosomal organization to varying extents and to interact with the transcription complex, thus altering the rates of transcription of relatively nearby DNA sequences (2–4). Antagonists act by preventing agonist binding to receptors and by forming a comparatively transcriptionally inactive receptor-steroid complex. The time scale of these transcriptional effects on mRNA synthesis is on the order of 15–30 min. More recently, it has become clear that steroid hormones can initiate even more rapid responses via membrane-bound receptors and “non-genomic” pathways, so called because the receptor does not interact with genomic DNA. The properties of these “non-genomic” pathways will not be considered here.
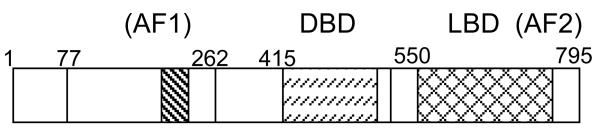
Fig. 1.
Schematic of domains of rat GR with numbers above the drawing indicating the amino acids of the LBD with AF2 (cross hatched), AF1 (core is striped), and DBD (dashes) domains. These domains are almost identical in GRs of other species and are similarly organized in the other steroid receptors (115).
Most studies of steroid-regulated gene transcription have been conducted at saturating, or pharmacological, concentrations of agonist steroids. Such studies assure maximal response and require a minimum of experimental points (i.e., vehicle and a single high concentration of steroid). However, physiology occurs at subsaturating concentrations of ligand for the steroid/nuclear receptors. It has been tacitly assumed that what happens at saturating concentrations of steroid also occurs at subsaturating concentrations. However, recent studies in rats indicate a much more rapidly cycling nuclear localization of GR with physiological vs. higher concentrations of the natural glucocorticoid, corticosterone (5). Furthermore, it is obviously impossible for studies with a single high agonist steroid concentration to yield answers about physiologically important steps that depend upon steroid concentration. For example, steroid potency or the positioning of the dose-response curve, which gives the amount of response for steroid concentrations causing 0–100% occupancy of the cognate receptor, cannot be determined from experiments with a saturating concentration of steroid that bind all of the receptors. Likewise, the causes for the frequent observation that gene induction by GRs requires a 10-fold higher steroid concentration than gene repression, even though the same receptor-steroid complex is involved (6–9), will not be elucidated by studies at saturating steroid concentrations. Also, an antisteroid, by definition, blocks the action of an agonist steroid but usually retains some residual activity of its own. What dictates the amount of residual agonist activity of an antisteroid cannot be elucidated from studies with only an agonist steroid. The current model of steroid hormone action suggests that the dose-response curve for steroid binding to a receptor determines the dose-response curve for steroid-activated, receptor-mediated gene induction (or repression) (4, 10, 11). This predicts that all responses regulated by a given receptor-steroid complex in an organism will display both the same potency for an agonist and equal amounts of partial agonist activity for an antisteroid, when expressed as percent of maximal agonist activity under the same conditions (= percent partial agonist activity). Abundant evidence has established that this view is incorrect. The immediate question then is how can the same receptor-steroid complex produce different dose-response curves for agonists, or unequal amounts of percent partial agonist activities for antagonists?
An equally important question is how much difference in the positioning of the dose-response curve (or the amount of partial agonist activity) is required to elicit physiologically relevant changes? The answer appears to be “not much”. A central tenet of developmental biology is that relatively small differences in the concentrations of appropriate factors induce different developmental fates in cells [Wolpert, L, J. Theor. Biology, 25, 1–47 (1969) from Nature, July 26, 420–421 (2007) = news and views]. It has been calculated that a 10% difference in the concentration of Bicoid is sufficient for morphogenic effects in the Drosophila embryo (12), although arguments have been made for the necessity of slightly larger changes (13). Small differences in Myc levels are adequate for numerous cell-specific effects (14) with a 50% reduction in c-Myc causing teleomer-independent senescence in human fibroblasts (15). The developmental stages of Drosophila larvae are exquisitely sensitive to changes in the level of the hormone ecdysone (16). A three-fold variation in the concentrations of Decapentaplegic or activin is adequate for unequal developmental responses in Drosophila and Xenopus respectively (reviewed in 17) while a 2- to 5-fold alteration in fibroblast growth factor is sufficient to trigger the differentiation of presomitic mesoderm into somites in mouse embryos (18). Thus, there is ample precedent that small differences in protein expression, achieved with subsaturating concentrations of steroid hormone, can have significant physiological consequences.
The objectives of this article are to review some of the transcription properties that transpire behind the “closed doors” of high agonist steroid concentrations and are missed when looking only at these saturating steroid concentrations. We summarize the evidence that these parameters of gene induction (and repression) are not static but can be modulated. Advances in elucidating the mechanisms by which factors modulate these parameters are presented, with an emphasis on the last two years. Finally, the physiological relevance of this modulation is discussed. Most of the data involves the classical steroid hormone receptors but some with nuclear receptors are available.
Theoretical background
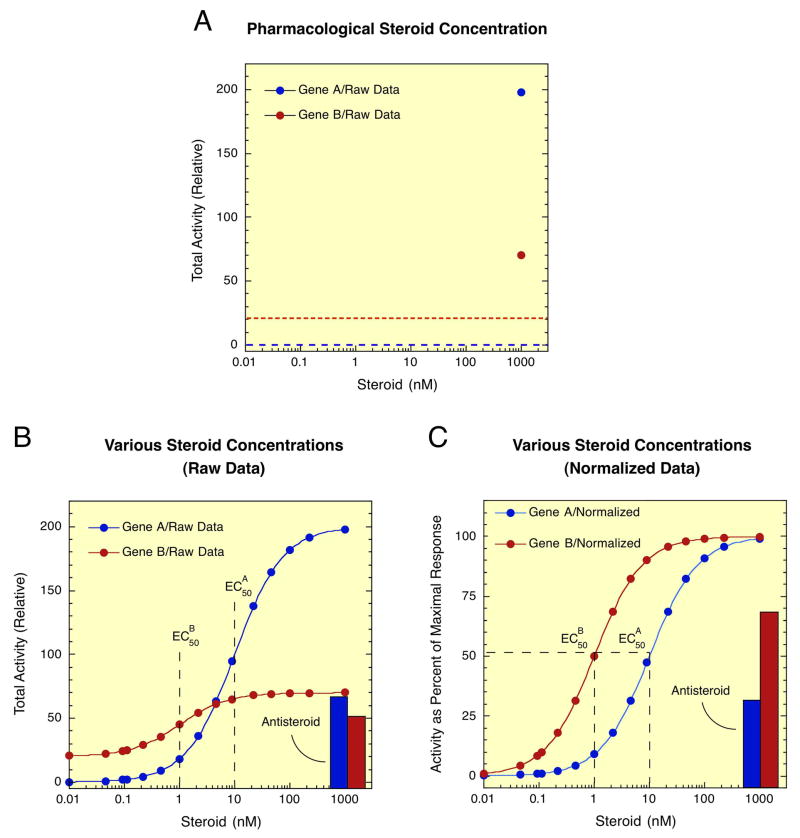
The experimental protocols used over the last 40+ years to unravel the mysteries of steroid hormone action almost invariably employed a single saturating, or pharmacological, concentration of agonist steroid. This approach yields only the maximum amount of steroid-regulated gene expression, as shown in Fig. 2A for the induction of two genes A and B. For the purposes of this review, we call the maximum activity the Amax. This value is also called the efficacy of the steroid but due to some confusion as to its precise meaning we will use Amax here. More information is obtained if lower concentrations of agonist steroid are included to examine steroid potency (Fig. 2B). Under these circumstances, the concentration of steroid required for half-maximal induction, or EC50, can be determined. As was found many years ago, the concentration of circulating cortisol in humans (80–600 nM) is about the same as the EC50 for some cortisol-induced effects (19). Importantly, changing the EC50 for steroid-regulated gene expression will obviously alter the response of that gene. Thus, Gene B, with a lower EC50, will be maximally induced at a lower steroid concentration than gene A, which is important for the differential induction of gene B vs. A during development, differentiation, and homeostasis. This also means that the activity with 100 nM agonist should not be assumed to be maximal. A right shift in the dose-response curve can easily move the EC50 to values ≥100 nM (20, 21). It should also be noted that there is no observed, or theoretical, correlation between Amax (Fig. 2A) and EC50 (Fig. 2B) so that studies organized as in Fig. 2A are unable to give any information about the EC50 of the gene of interest.
Fig. 2.
Data plots for experiments on steroid hormone action. (A) Induction of gene expression of transiently transfected luciferase reporter genes A and B with a saturating concentration of agonist steroid. Raw luciferase data are plotted. Background levels with no added steroid are indicated by the dashed lines. (B) Dose-response curves for induction of transiently transfected luciferase reporter genes A and B by varying concentrations of agonist steroid, and bar graphs of residual agonist activity of an antisteroid with each gene. Raw luciferase data are plotted. (C) Normalized data for gene induction data of panel B. For each gene, the data are expressed as percent of maximal induction.
Additional information is obtained if pharmacological concentrations of an antagonist, or antisteroid, are included (Fig. 2B). Antisteroids are used at these concentrations during endocrine therapies to block the actions of agonist steroids. However, the amount of residual agonist activity of each antisteroid is currently impossible to predict and must be experimentally determined. A useful parameter, when discussing the residual agonist activity of antisteroids, is the percent partial agonist activity.
While the precise values for the EC50, or potency, and percent partial agonist activity can be calculated from plots of the raw data, as in Fig. 2B, it is much easier if the data are first replotted as percent of maximal induction for each gene, as in Fig. 2C. In this case, the EC50 and percent partial agonist activity can be readily read off of the graph. Such plots also clearly demonstrate that the EC50 is independent of the Amax of gene induction. The importance of comparing the activities of antagonist and agonist under the same conditions cannot be overemphasized. The residual activity of the antisteroid is about the same for genes A and B (Fig. 2B) but that the percent partial agonist activity is much greater with gene B than with gene A (Fig. 2C). Replotting the data as in Fig. 2C will also immediately clarify whether or not reports of increased sensitivity (i.e, more activity at low concentrations of agonist), or more response with high levels of antisteroid, actually reflect a change in EC50, or percent partial agonist activity respectively.
The current model of steroid hormone action says little about the determinants of the EC50. Early observations of a close correlation between the Kd for steroid binding to receptors and the EC50 for steroid-regulated gene induction were viewed as evidence that the steroid was acting via binding to its cognate receptor. In fact, it has been known for many years that such a correlation is not general (22 and reviewed in 23, 24). It should also be noted that high steroid binding affinity does not assure high efficacy of biological response (Amax). RU486 has a high affinity for GR (25) but a negligible Amax for gene induction under most conditions (26, 27). Under conditions where the affinity label Dexamethasone 21-mesylate (Dex-Mes) covalently binds to >90% of the receptors, it displays ≤7% of maximal activity seen with Dex (28). Thus, detailed studies of the EC50 with sub-saturating concentrations of agonists, and the percent partial agonist activity with saturating concentrations of antagonists, are required to gain an understanding of the determinants of these relevant parameters. Finally, as is described below, such experiments have revealed that the EC50, and percent partial agonist activity, are not constant even for a single gene but can be modulated by a variety of factors.
Modulatory factors and their mechanism of action
Glucocorticoid modulatory element (GME)
The first example of a modulator of the EC50, and percent partial agonist activity, of GR-regulated gene induction was a cis-acting 21 bp element, called the GME, at −3.6 kb of the rat TAT gene (29). Interestingly, the GME modulates the EC50, and percent partial agonist activity, for GR induction of an exogenous reporter (GREtkLUC) in transiently transfected mouse mammary adenocarcinoma cells (1470.2 cells) but has no effect on these parameters for progesterone receptor (PR)-mediated induction of the same gene in the same cells (30). Thus there is receptor-selectivity of GME actions in 1470.2 cells.
GME-binding proteins 1 and 2 (GMEB-1 and GMEB-2)
An oligomeric complex of two proteins, GMEB-1 and -2, was found to participate in the actions of the GME (31). Increasing amounts of each GMEB, either alone or in combination in CV-1 and 1470.2 cells, cause a progressively larger right-shift to higher steroid concentrations for the GR dose-response curve of a GMEGREtkLUC reporter, and decrease in percent partial agonist activity, presumably due to squelching (30, 32). In contrast, low concentrations of GMEB-2 with PRs produce a left-shift in the dose-response curve to lower steroid concentrations and an increase in the percent partial agonist activity of antiprogestin. At higher GMEB-2 concentrations, PR and GR respond similarly (30). This suggests that PR is less sensitive to GMEB-2 than GR and needs higher GMEB-2 concentrations to see similar effects. This would also explain why the GME, with endogenous levels of GMEB-2, affects GRs but not PRs.
Changing concentrations of steroid receptor
The Kd for steroid (S) binding to receptor (R) is defined as [S][R]/[RS complex]. Just as the concentration of steroid receptor does not affect the binding affinity of steroid, different receptor concentrations would not be expected to alter the EC50, and percent partial agonist activity, for gene induction. This prediction, however, is wrong. Since the initial report that GR concentration influences not only the Amax but also the EC50, and percent partial agonist activity, of GR-induced transcription (33), the same phenomenon has been observed for each of the classical steroid receptors (reviewed in 23, 24). Gene repression by steroid receptors usually proceeds via a slightly different mechanism involving receptor-steroid complexes that are tethered to DNA-bound proteins as opposed to binding directly to DNA, as seen in induction (34, 35). Nonetheless, it has been shown that increasing GR protein also causes the same decrease in EC50 to lower steroid concentrations, and increase in percent partial agonist activity of antiglucocorticoids, in gene repression (36, 37) as observed for gene induction.
Posttranslationally modified receptors
The possibility that posttranslational modification of receptors and factors could influence the dose-response curve (or potency or EC50) and/or percent partial agonist activity is a very attractive hypothesis that would dramatically expand the number and variety of control mechanisms. We are aware of only one possible example in which the hepatitis B virus nonstructural protein, X protein (HBx), causes an increase in both EC50 and Amax. HBx did not physically interact with nuclear androgen receptor (AR)-ligand complexes. Instead, the authors speculate that increased AR activity occurs by elevating AR phosphorylation through HBx-mediated activation of the c-Src kinase signaling pathway, although a direct response with increased AR phosphorylation remains to be established (38).
Mutant receptors
Receptor mutations that affect steroid binding affinity would be expected to alter the EC50 of gene expression in a parallel fashion. However, the K303R mutation at the junction of the hinge and LBD of the human estrogen receptor alpha (ERα) produces a left-shift in dose-response curve for cell growth and increased affinity for the coactivators (TIF2 and AIB1) without any change in steroid binding affinity (39). Several mutations in the GR LBD have recently been described that increase the EC50 of induction of exogenous and endogenous genes by ≥100-fold while increasing the Kd of steroid binding by <6-fold (40). This dissociation of EC50 and Kd is not unique to the classical steroid receptors. The L422R mutation in the LBD of thyroid receptor beta 1 (TRβ1) alters the EC50 for gene induction independent of steroid binding affinity but dependent on the DNA sequence of the thyroid response element (41). TRβ2 has a longer N-terminal domain than TRβ1 but is otherwise identical. Both isoforms bind thyroid hormone with the same affinity. Nevertheless, the dose-response curve for TRβ2 induction of a transfected reporter is ~3 fold left-shifted from that of TRβ1. This appears to be due, at least in part, to TRβ2 having an ~3 fold higher affinity than TRβ1 for the coactivator TIF2 (42). Thus not only variations in the concentration of receptors, but also changes in receptor sequence, can differentially alter the properties of transactivation.
Coactivators and corepressors with classical steroid receptors
The p160 coactivators (SRC1, TIF2/GRIP1, and AIB1) are defined as factors with intrinsic transactivation activity that increase the Amax of gene induction by receptor-agonist complexes (43). The corepressors NCoR and SMRT were discovered from their ability to repress the transactivation of enhancer-bound, ligand-free nuclear receptors and were found to have high affinity for antagonist-bound receptors. Subsequently it was determined that coactivators and corepressors do not selectively bind to receptor-agonist and receptor-antagonist complexes respectively. Instead, coactivators and corepressors can each associate with both agonist -and antagonist-bound receptors. Consequently, the ratio of coactivators to corepressors is thought to be a critical determinant of final Amax activity (44–46).
Coactivators and corepressors also modulate the EC50 of agonist-induced transcription and the percent partial agonist activity of antagonists. Exogenous GRIP1/TIF2 and SRC-1 each causes a left-shift in the dose-response curve of AR (47, 48), GR (26, 49), mineralocorticoid receptor (MR) (50), and PRs (51, 52). The behavior with ERα is more complex. Overexpression of SRC-1 (but not TIF2 or AIB1) made tamoxifen (but not raloxifen) an agonist in MCF-7 cells while SRC-1 small-interfering RNA (siRNA) eliminated tamoxifen but not estradiol agonist activity in Ishikawa cells (53). More recently, SRC-1 increased the total activity of tamoxifen in Cos-7 cells but not as much as for estradiol so that the percent partial agonist activity actually decreased (54). An attractive mechanistic explanation for the seemingly general behavior of coactivators (23, 24) may be the ability of coactivator to increase receptor-steroid complex binding to HREs. The coactivator PGC-1β has been found to increase both the EC50 and Amax of reporter gene induction by ERα in Cos-7 cells (Anastasia Kralli, personal communication). It has recently been shown that TIF2 stabilizes the cooperative binding of agonist-bound PR-A complexes to a tandem repeat of PREs to cause a 10-fold left-shift in the binding isotherm (55) while no TIF2 binding is observed to ligand-free GRs bound to GREs (21). The M770I rat GR mutation displays increased affinity for GRIP1 and produces a left-shift in the dose-response curve (56). The corepressors NCoR and SMRT interact with both agonist- and antagonist-bound ERαs, GRs, and PRs (44, 45, 57–60) to shift the position of the dose-response curve of agonists and amount of percent partial agonist activity of antisteroids (26, 44, 58, 60–62). Interestingly, the direction of the changes produced by the corepressors can be receptor-and cell-dependent (26, 62). Inhibition of the class III histone deacetylase, Sirt1, by either nicotinamide or Sirt1 siRNA was found to increase the Amax and decrease the EC50 of AR gene induction of an exogenous reporter in LNCaP cells (63).
There are several differences between gene induction and repression, the most obvious of which being that repression decreases, while induction increases, gene expression. This suggests at least some divergent mechanistic pathways for the two processes. Nevertheless, five modulators (coactivators TIF2 [GRIP1, SRC-2] and SRC-1 ± the comodulator STAMP, corepressor SMRT, and comodulator Ubc9) altered the Amax, EC50, and percent partial agonist activity of endogenous and exogenous repressed genes similarly to that for GR-regulated gene induction. These five factors interact with different domains of GR and thus are sensitive topological probes of GR action. This suggests that GR-mediated induction and repression share many of the same molecular interactions and that the causes for different levels of gene transcription arise from more distal downstream steps (37). The actions of SRC-1 by itself appear to be promoter and/or cell dependent as exogenous SRC-1 is reported to cause a decrease (64) or increase in EC50 (37) in two different systems.
The observations of equilibrium interactions of coactivators and corepressors with GRs have led to the hypothesis that the ratio, as opposed to the absolute amount, of coactivators and corepressors is a major determinant of the final EC50 and percent partial agonist activity values (26, 58, 59). These competitive interactions do not appear to be unique for GRs (23, 24). The coactivator TIF2 inhibits PR association with the corepressor NCoR in two-hybrid assays (59). Endogenous SMRT co-immunoprecipitates with both agonist and antiandrogen-bound AR while TIF2 and SMRT competitively inhibit the actions of each other with agonist- but not antagonist-bound ARs (65). The coactivators SRC-1 and CBP functionally compete with NCoR in transactivation, 3-hybrid, and nuclear colocalization assays with ARs in a manner that depends upon the ratio of coactivators to corepressors (46). Similar competitive binding to agonist-bound ARs has been inferred from the increased binding of the coactivator SRC-1 to the promoter region of three endogenous AR-regulated genes in ChIP assays with siRNAs to NCoR and SMRT (66). Interestingly, corepressors are recruited equally to the enhancers of these genes (66), and of a transfected reporter gene (60), by both agonist- and antagonist-bound ARs and PRs respectively. Coactivators and corepressors were initially found to associate with a common region in the LBD of thyroid receptors (67–69). Recent studies have confirmed competitive binding of coactivators and corepressors to the GR LBD (70, 71) and extended this competitive binding to include a second region in the N-terminal domain of both GRs and PRs (60).
IL-1β causes the loss in ChIP assays of NCoR and all NCoR-associated factors with antiandrogen-bound ARs, apparently by MEKK1 recruitment to TAB2. IL-1β also increases the percent partial agonist activity of ERα and PR antagonist complexes (72).
Coactivators and corepressors with nuclear receptors
Reports of modulatory activity with coactivators and corepressors was initially restricted to the classical steroid receptors but are becoming more common with the nuclear receptors. Equilibrium competitive binding of coactivators and corepressors to thyroid receptors (TRs) and retinoic acid receptors (RARs) in vivo has been described along with SMRT binding to a RAR response element in ChIP assays ± retinoic acid in a manner that is competed by added SRC-1 (73). These observations can explain the ability of SMRT to decrease the amount of thyroid hormone (T3) -induced transactivation by TRs. Furthermore, column-purified steroid-free TR/retinoid X receptor (RXR) complexes contain corepressors that are not eliminated by the addition of the agonist T3 (74).
Unexpectedly, SMRT but not NCoR appears to be required for full activity of agonists in a receptor- and cell-specific manner. With exogenous reporter genes, SMRT siRNA decreases ERα transactivation in MCF-7 and HeLa cells (but not HepG2 cells) but increases transactivation by vitamin D receptors, (VDRs), TRs, and ARs in HeLa cells. With MCF-7 cells, SMRT siRNA reduces expression of many but not all of the estrogen-regulated endogenous genes examined and reduces cell growth (75).
RIP140 usually functions as a corepressor. In an example of equilibrium binding, the ratio of RIP140 to the coactivator PCAF was found to determine amount of each factor binding to complexes of RAR/RXR/coactivator GRIP1 ± retinoids (76). Collectively, the above reports illustrate that the concept of competitive cofactor binding, determined by the ratio of competing cofactors, is more general than initially proposed for GRs (26) and also occurs with a variety of the classical steroid receptors and the nuclear receptors. Whether it is a property of, and will affect the EC50 and partial antagonist activity of, all steroid/nuclear receptors remains to be established.
When a coactivator is not a coactivator
Coactivators, as described above, were initially defined by their ability to increase gene transactivation by a receptor-agonist complex. When discussing receptor-mediated repression, where an agonist steroid causes a decrease in the total amount of gene expression, the expected behavior of a coactivator may not be immediately obvious. For the purposes of this review, we define a coactivator as a transcriptionally active cofactor that augments the activity of a receptor-agonist complex, whether it be to cause a further decrease (as in repression) or increase (as in induction) in gene expression. Thus, by this definition, the report of TIF2 accentuating the repression by GR-agonist complexes of an AP-1 induced gene in U2OS.rGR cells is an example of TIF2 acting as a coactivator in gene repression (77).
Coactivators can bind to receptors but not exert the usual responses of increased Amax, decreased EC50, or increased percent partial agonist activity. The coactivator SRC-1 appears to associate with ARs bound by the antiandrogens cyproterone acetate and bicalutamide. However, SRC-1 increases the percent partial agonist activity of only cyproterone acetate-bound ARs (78). Point mutations in the GR LBD have been described to eliminate the activity of added TIF2 in the presence of the agonist dexamethasone (Dex) while maintaining the ability of TIF2/GRIP1 to interact with GR-Dex complexes (40). In both cases, the data support a mechanism where the steroid or point mutation causes minor changes in receptor-coactivator interactions that have little effect on binding but are magnified by the time they are propagated to the outer surface of the coactivator and thus have major consequences on its ability to modify transcriptional processes. This scenario is similar to the report that phosphorylation of the human ERα at Ser305, which is just before the LBD, alters the orientation (and presumably activity) of bound coactivator SRC-1 without affecting the affinity of SRC-1 binding (79).
Other factors that modulate EC50 and percent partial agonist activity of receptors
Ubc9 was initially found to modulate the properties of GR (23, 24) and was recently shown to affect PRs (30). β-catenin increases the percent partial agonist activity of antiandrogens with ARs (80). Squirrel monkey FKBP51 causes an ≈10-fold right-shift in the dose-response curve of both human and squirrel monkey GRs (81). Vav3 is localized to membranes and does not bind to ARs or alter AR protein levels. However, Vav3 causes a left-shift in AR dose-response curve in a manner that requires the AR AF1 domain along with the PH domain of Vav3 (82). STAMP is a new co-modulator that binds SRC-1 and TIF2 and enhances their ability to alter the EC50 and percent partial agonist activity of AR, GR, and PR complexes in gene induction (30, 77). The activity of STAMP with GRs requires TIF2 and STAMP binding to GR via their respective receptor interaction domains (RIDs). STAMP (in the presence of TIF2) and Ubc9 both retain the same modulatory activity in GR-mediated induction and repression (37). The dose-response curve for androgen induction of three transfected reporters is left-shifted to a lower EC50 in C4-2B cells (an androgen independent subline of LNCaP cells) compared to LNCaP cells even though the AR levels are the same (83). The factors responsible for this subline-dependent behavior are not yet known.
Sur2 is another modulator of GR transcriptional properties but no new data has appeared since the original report (84). Kinases have long been suspected of altering receptor transcriptional properties and several substantial effects have been documented (85). CDK5 and p25 together repress GR transactivation but with no obvious change in the EC50 of either exogenous (MMTV) or endogenous (SGK) genes (86).
Selectivity of Modulatory Factor Actions
Receptor-selective effects
The same DNA sequence is capable of mediating gene induction by most of the classical steroid receptors (i.e., AR, GR, MR, and PR). Thus, much of the specificity gained by steroids complexing with cognate receptors appears to be lost after binding DNA. The first indication that modulatory factors could restore some of the lost specificity was seen with the opposite responses of GRs and PRs to the corepressors NCoR and SMRT. Thus, in the same cell with the same exogenous reporter gene, NCoR induces a decrease in the EC50, and an increase in percent partial agonist activity, with PRs but an increase in EC50, and a decrease in percent partial agonist activity with GRs (62). Similar diversities in response of PRs vs. GRs are seen with Ubc9 and GMEB-2 and with the GME DNA element (30). The behavior of PR and GR with STAMP ± TIF2 are the same but separate regions of STAMP are involved, suggesting that activity differences may be revealed under other conditions (30). hZimp7 increases AR transactivation, with an apparent left-shift in dose-response curve but not with GRs or PRs (87). COBRA1 causes a right-shift in the dose-response curve of AR gene induction. COBRA1 binds ARs and GRs in a co-IP assay much more avidly than PRs (88). Thus, COBRA1 is predicted not to significantly alter the EC50 for PR-regulated induction in the same cells. The factor ELL reduces the EC50 for gene induction by MR by a factor of ≈10 and increases both the Amax for aldosterone and the percent partial agonist activity of the antagonist spironolactone while exhibiting no effect on ARs or PRs and decreasing the Amax for GRs (89).
Tissue selectivity of modulatory actions
The GME is active with GRs in Fu5-5 rat hepatoma (29), CV-1 (90) and 1470.2 cells but inactive with PRs in 1470.2 cells (30). GMEB-2 modifies the properties of GRs in CV-1 (32) and of GRs and PRs in 1470.2 cells. The initial activity of Ubc9 with GRs in CV-1 cells (21, 91) is reduced for GRs in 1470.2 cells but is very similar for PRs in both cell lines (30). STAMP modulatory activity with GRs has been reported in CV-1 and U2OS human osteosarcoma cells (77) and with PRs in CV-1 cells (30). Thus, the effects of these factors are not limited to a single steroid receptor or a single cell line.
Dissociation of Amax, EC50, and percent partial agonist activity
The current model of steroid hormone action gives no guidance as to whether changes in Amax, EC50, and/or percent partial agonist activity are linked. Recent reports suggest that it is possible to selectively modulate each parameter.
Separation of Amax (efficacy) and EC50 (potency)
The S530A mutation of PR causes about 10-fold right-shift in dose-response curve with no change in maximal induction or progesterone binding affinity both in cell-free extracts and in whole cells at 37 °C (92). Amino acids 41–64 of human ERα influence the EC50 but not the Amax with estradiol (93). The GR polymorphisms N363S and R23K cause increased or decreased transcription respectively of the endogenous GILZ gene (but not the repressed IL-2 gene) with no change in EC50 of the induced gene (94). Ubc9 increases the Amax of GR-regulated induction of an exogenous reporter in the presence of both low and high concentrations of GR but apparently affects the EC50 of gene induction only with high GR levels (21, 91). Removal of N- and C-terminal domains of TIF2 reduces the coactivator-mediated increase in Amax more than the shift of the dose-response curve to lower steroid concentrations (95). The deacetylase inhibitor TSA increases the Amax of transfected gene induction by GRs in the presence of the corepressor SMRT without altering the EC50 (58, see also references in 30). These results are consistent with the Amax (efficacy) and EC50 (potency) of gene induction being controlled by different pathways, at least under some conditions.
Separation of Amax and percent partial agonist activity
The S118A mutation in human ERα often blocks receptor phosphorylation by ERK1/2. With several endogenous genes for which ERα-mediated induction is decreased by the S118A mutation, the absolute amount of partial agonist activity is unchanged, which means that the percent partial agonist activity increases (96). Similar consequences were seen for the activity of the antagonist RU486 with the double mutant M770A/L771A in rat GR (27). Amino acids 41–64 of human ERα are necessary for percent partial agonist activity of antisteroids but not transactivation by estradiol (93). SMRT siRNA reduces the activity of ERα-agonist complexes but increases the activity of receptors bound by the antiestrogen 4-hydroxytamoxifen (75). Together these results suggest that the mechanisms governing the percent agonist activity of antisteroids can be different from those controlling the Amax of agonists.
Separation of EC50 and percent partial agonist activity
The percent partial agonist activity goes up when the EC50 goes down, and vice versa, under a wide variety of conditions (Fig. 2C; reviewed in 23). As there is no theoretical reason for this correlation, it is not surprising that exceptions are being found. The M578A mutation in rat GR increases the EC50 for Dex (and reduces the Amax) while converting the weak antiglucocorticoid cyproterone acetate (97) to a full agonist with lower EC50 than Dex (98). The deacetylase inhibitors VPA and TSA in the presence of high, but not low, GR concentrations cause little change in EC50 but increase the percent partial agonist activity (99).
Separation of EC50s in repression vs. induction
Receptor-regulated repression and induction are generally thought to proceed via two different mechanisms with repression employing receptors “tethered” to DNA by other DNA-bound proteins (34, 35). However, gene repression is not a property of tethered receptors, as seen by the numerous examples of gene activation by chimeras of receptors fused to a different DNA-binding protein. For GRs, the EC50 for gene induction is usually 6–10 fold higher than that for repression by the same receptor-steroid complex in the same cells (6–9). Elucidating the reasons for these differences will provide valuable insight into the mechanism of receptor-mediated gene expression in general and gene repression in particular.
Parallel changes in Amax and EC50
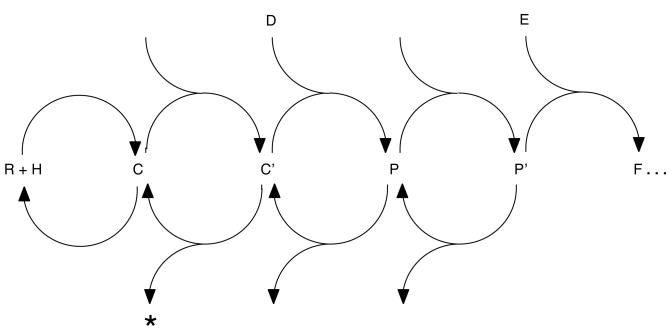
A recently proposed mathematical model of gene induction by steroid hormones involving a series of linked, cyclical steps (Fig. 3) can explain most changes in both Amax and EC50 as resulting from alterations in a single step downstream of steroid binding affinity for receptors (99) (Ong et al., in preparation). Only when the Amax and EC50 both change in the same direction (e.g., increase in Amax and right-shift in dose-response curve to higher steroid concentrations) does it appear that effects on two separate parameters or steps must be invoked. Such parallel changes in Amax and EC50 have been reported with GRs with CBP (33), with SMRT in a cell-specific manner (62), with GMEB-2 and PR and the GMEGREtkLUC reporter (30), and for the R629Y mutant (rat GR) in a steroid-specific manner (40). Parallel changes have also been reported for ARs with COBRA1 (88) and hepatitis B virus protein X protein (HBx) (38), and for PR with NCoR (62) and with GMEB-2 in a manner that depends upon PR concentration (30).
Fig. 3.
Cyclical model of steroid hormone regulated gene transcription. Each circle of two reactions (single semi-circular arrow) represents a proposed, but usually unidentified, reaction in the currently uncharacterized series of steps from steroid (H) binding to receptor (R) to give the initial receptor-steroid complex (C) to the production of mRNA (beyond F). Other species correspond to both known (e.g., C′ = activated complex; * = biologically inert “activated complex”) and unknown (e.g., P′, E, etc.) factors and intermediates. This cyclical model predicts sigmoidal dose-response curves under a variety of conditions (from ref. 99).
Mechanistic considerations of independence of parameters
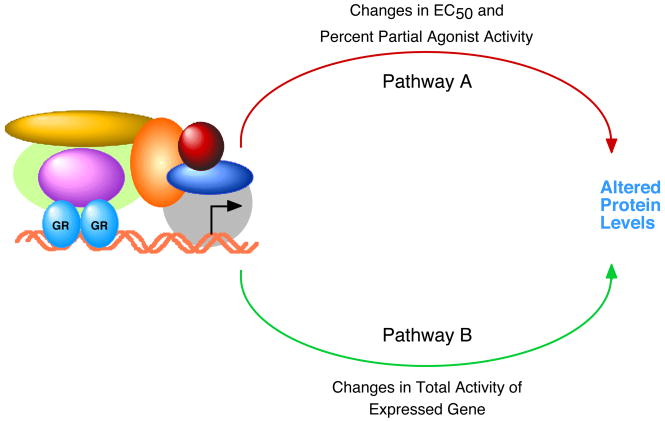
The separate regulation of EC50 and/or percent partial agonist activity from Amax might have been predicted because graphical displays of EC50 and percent partial agonist activity omit any mention of Amax (Fig. 2C). This plus the mathematical model for modifying Amax and EC50, suggests that different pathways or steps may be recruited under appropriate conditions to modulate these two parameters (Fig. 4). Thus, mechanisms and cooperating factors may emerge during investigations of the determinants of EC50 that would go undetected in studies of Amax. Some of the pathways that appear to be influenced by transcription factors, and may participate in the dissociation of Amax and EC50, are steps downstream of receptor binding to DNA, including transcription initiation (100), RNA pol II elongation (99, 101), relief of supercoiling (99), and mRNA splicing (100, 102–104). This hypothesis is consistent with the view that transcription is not a series of individual, isolated steps but rather reflects the net product of a series of interconnected reactions (105, 106).
Fig. 4.
Proposed model by which changes in total activity (Amax) and in EC50 and percent partial agonist activity can sometimes be expressed via different steps or pathways. The promoter-bound complex with GR is depicted as being a single multimeric species only for ease of drawing. The start of transcription is designated by the bent arrow. The mathematical model suggests that the various molecules, including GR, enter and leave the promoter regions sequentially so that only a few components are present at any one moment. The depicted arrows for pathways A and B may, depending upon the conditions, represent entirely different sequences in the reactions of Fig. 3 from receptor binding of steroid to production of protein product. Alternatively, A and B may represent other single cyclical steps in the overall pathway of Fig. 3, or anything in between. Similar different steps/processes may also dictate independent changes in percent partial agonist activity, although no mathematical model has yet been constructed for this.
Physiological relevance of modulating the EC50 and percent partial agonist activity
Examples of physiological consequences of modulating the concentrations of relevant factors, and thus the EC50 and percent partial agonist activity, are becoming more numerous in proportion to the frequency of studying these parameters. A two-fold increase in GR gene dosage in mice, achieved by using a yeast artificial chromosome, causes a 60% increase in receptor protein and GR mRNA but a >10-fold decrease in EC50 for induction of thymocyte apoptosis (107). The naturally occurring I747M mutation in human GR causes a two-fold decrease in affinity and Amax and a 20–30 fold increase in EC50. Thus the steroid concentration sufficient for 50% induction by the wild type GR affords 5-fold less activity with the mutant GR (49). Androgen resistance correlates with increased AR levels in prostate tumor xenograph models (108), as expected from tissue culture studies where increases in AR decrease the EC50 for gene induction and increased the percent partial agonist activity of antiandrogens. It should be remembered, though, that only 1/3 of androgen-insensitive prostate cancers are associated with elevated AR (109), so that other mechanisms are clearly involved (110). The estrogen-insensitivity of many breast tumors with increased levels of AIB1 (111) is consistent with the predicted ability of elevated AIB1 to decrease the EC50 for endogenous estrogens and increase the percent partial agonist of antiestrogens (50). However, it now appears that these effects of AIB1 involve being a coactivator of the cell cycle regulator E2F1 and are independent of ERαs (112). At least part of the syndrome of pituitary (or central) resistance to thyroid hormone (RTH) can be explained by TRβ2, found primarily in the pituitary/hypothalamus, displaying an enhanced response to T3, relative to the TRβ1 isoform, due to increased affinity for the coactivator TIF2. This causes a lower EC50, which makes TRβ2 respond more robustly to physiological levels of hormone (42). During development in Drosophila larvae, the temporal regulation of gene expression by the steroid hormone ecdysone is such that the first genes expressed have the lowest EC50 (16). A more comprehensive discussion of physiologically relevant changes in factor concentration can be found in a recent review (24).
Conclusions
Dramatic advances have been made over the last decade in unraveling the mechanism of steroid hormone action. Nevertheless, our understanding of the differential control of gene expression by the single circulating concentration of steroid hormone during development, differentiation, and homeostasis remains fragmentary at best. These difficulties derive in part from the relative scarcity of experiments run with physiological (i.e., sub-saturating) concentrations of agonist steroid. Those studies that have been performed have yielded the attractive hypothesis that increasing concentration differences among cells of factor(s) that interact with receptor-steroid complexes act like a rheostat to progressively change the position of the dose-response curve for agonists, and the percent partial agonist activity of antagonists. Abundant support for this hypothesis is now available from tissue culture studies and physiological evidence is beginning to mount. These factor gradients could easily be established if the extra-cellular concentration of possible inducers (or repressors) of the genes for each factor, or regulators of factor mRNA stability, are found to change. This will require detailed studies of the type recently performed for the receptors themselves, where it was found that the level of steroid receptors varies both by at least a factor of 20 between tissues (113) and in a circadian rhythm (114). New techniques, such as ChIP, ChIP-reChIP, ChIP-on-chip, chromatin conformation capture, siRNA, and transgenic mice with knock-out and knock-in mutations provide powerful new tools to examine the mechanisms behind the changes in EC50 and percent partial agonist activity. It is now possible to look at the specific factors that are recruited to specific regions of exogenous and endogenous regulated genes, to ask if two (or more) factors are present at a given region of the gene at the same time or sequentially, to determine whether the factors are covalently modified and/or move to new DNA locations under the different conditions, to investigate the possible communication between proteins bound at separated DNA sites, to define the precise nuances that are becoming commonplace between endogenous genes, and to examine the functional consequences of varying the amounts of specific factors (or their mutated forms) within the physiologically encountered levels. The net results from such studies should provide a quantum leap forward in our understanding of steroid hormone action at physiologically important levels of both steroid and protein factors.
Table 1.
Entities that modulate the EC50 and/or percent partial agonist activity of receptors
| Factor | Receptor | Reference |
|---|---|---|
| β-catenin | AR | (80) |
| Cell sub-line | AR | (83) |
| Coactivators | AR, ERα, GR, MR, PR, TR | (23, 24, 27, 40, 42, 47, 48, 53, 54, 56, 62, 77, 78, 93) |
| COBRA1 | AR | (88) |
| Corepressors | AR, ERα, GR, MR, PR | (23, 24, 44, 45, 57, 60, 61, 75) |
| Deacetylase inhibitors | GR | (99) |
| ELL | MR | (89) |
| FKBP51 | GR | (81) |
| GME | GR, PR | (29, 30) |
| GMEB-2 | GR, PR | (30, 32) |
| Hbx | AR | (38) |
| IL-1β | ERα, PR | (72) |
| PGC-1β | ERα | (A. Kralli, personal communication) |
| Receptor mutations | ERα, GR, TR | (39–42, 49, 56, 61, 96, 98) |
| STAMP | AR, GR, PR | (30, 77) |
| Steroid receptor | AR, ERα, GR, MR, PR | (23, 24, 36) |
| Sur2 | GR | (84) |
| Transcription mode* | GR | (6–9) |
| Ubc9 | GR, PR | (23, 24, 30) |
| Vav3 | AR | (82) |
| hZimp7 | AR | (87) |
Induction vs. Repression
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, NIDDK. We thank Greti Aguilera, Michael Beaven, Carson Chow, Sam John, Karen Ong, and members of the Steroid Hormones Section for critical review of this work.
Abbreviations
- Amax
maximal activity
- AR
androgen receptor
- Dex
dexamethasone
- Dex-Mes
Dex-21-mesylate
- DBD
DNA binding domain
- EC50
effective concentration for 50% of maximal response
- ER
estrogen receptor
- GME
glucocorticoid modulatory element
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- HBx
hepatitis B virus nonstructural protein, X protein
- HRE
hormone response element
- LBD
ligand binding domain
- MR
mineralocorticoid receptor
- MMTV
mouse mammary tumor virus
- PR
progesterone receptor
- RAR
retinoic acid receptor
- RXR
retinoid X receptor
- siRNA
small-interfering RNA
- TR
thyroid receptor
- TRβ1
thyroid receptor beta 1
References
- 1.Zhang Z, Burch PE, Cooney AJ, Lanz RB, Pereira FA, Wu J, Gibbs RA, Weinstock G, Wheeler DA. Genomic analysis of the nuclear receptor family: new insights into structure, regulation, and evolution from the rat genome. Genome Res. 2004;14:580–590. doi: 10.1101/gr.2160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Kinyamu HK, Archer TK. Changes in attitude, changes in latitude: nuclear receptors remodeling chromatin to regulate transcription. Mol Endocrinol. 2006;20:1–13. doi: 10.1210/me.2005-0192. [DOI] [PubMed] [Google Scholar]
- 4.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 5.Conway-Campbell BL, McKenna MA, Wiles CC, Atkinson HC, de Kloet ER, Lightman SL. Proteasome-dependent down-regulation of activated nuclear hippocampal glucocorticoid receptors determines dynamic responses to corticosterone. Endocrinology. 2007;148:5470–5477. doi: 10.1210/en.2007-0585. [DOI] [PubMed] [Google Scholar]
- 6.Wei P, Ahn YI, Housley PR, Alam J, Vedeckis WV. Modulation of hormone-dependent glucocorticoid receptor function using a teracycline-regulated expression system. J Steroid Biochem Mol Biol. 1998;64:1–12. doi: 10.1016/s0022-1759(97)89907-3. [DOI] [PubMed] [Google Scholar]
- 7.Nye AC, Rajendran RR, Stenoien DL, Mancini MA, Katzenellenbogen BS, Belmont AS. Alteration of large-scale chromatin structure by estrogen receptor. Mol Cell Biol. 2002;22:3437–3449. doi: 10.1128/MCB.22.10.3437-3449.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koubovec D, Ronacher K, Stubsrud E, Louw A, Hapgood JP. Synthetic progestins used in HRT have different glucocorticoid agonist properties. Mol Cell Endocrinol. 2005;242:23–32. doi: 10.1016/j.mce.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Smit P, Russcher H, de Jong FH, Brinkmann AO, Lamberts SW, Koper JW. Differential regulation of synthetic glucocorticoids on gene expression levels of glucocorticoid-induced leucine zipper and interleukin-2. J Clin Endocrinol Metab. 2005;90:2994–3000. doi: 10.1210/jc.2004-2298. [DOI] [PubMed] [Google Scholar]
- 10.Robyr D, Wolffe AP, Wahli W. Nuclear hormone receptor coregulators in action: diversity for shared tasks. Mol Endocrinol. 2000;14:329–347. doi: 10.1210/mend.14.3.0411. [DOI] [PubMed] [Google Scholar]
- 11.McKenna NJ, O’Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 12.Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinitz J. Developmental biology: a ten per cent solution. Nature. 2007;448:420–421. doi: 10.1038/448420a. [DOI] [PubMed] [Google Scholar]
- 14.Levens DL. Reconstructing MYC. Genes Dev. 2003;17:1071–1077. doi: 10.1101/gad.1095203. [DOI] [PubMed] [Google Scholar]
- 15.Guney I, Wu S, Sedivy JM. Reduced c-Myc signaling triggers telomere-independent senescence by regulating Bmi-1 and p16(INK4a) Proc Natl Acad Sci U S A. 2006;103:3645–3650. doi: 10.1073/pnas.0600069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karim FD, Thummel CS. Temporal coordination of regulatory gene expression by the steroid hormone ecdysone. EMBO J. 1992;11:4083–4093. doi: 10.1002/j.1460-2075.1992.tb05501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurdon JB, Bourillot PY. Morphogen gradient interpretation. Nature. 2001;413:797–803. doi: 10.1038/35101500. [DOI] [PubMed] [Google Scholar]
- 18.Dubrulle J, Pourquie O. fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature. 2004;427:419–422. doi: 10.1038/nature02216. [DOI] [PubMed] [Google Scholar]
- 19.Hackney JF, Gross SR, Aronow L, Pratt WB. Specific glucocorticoid-binding macromolecules from mouse fibroblasts growing in vitro. A possible steroid receptor for growth inhibition. Mol Pharmacol. 1970;6:500–512. [PubMed] [Google Scholar]
- 20.Kaul S, Murphy PJM, Chen J, Brown L, Pratt WB, Simons SS., Jr Mutations at positions 547-553 of rat glucocorticoid receptors reveal that hsp90 binding requires the presence, but not defined composition, of a seven amino acid sequence at the amino terminus of the ligand binding domain. J Biol Chem. 2002;277:36223–36232. doi: 10.1074/jbc.M206748200. [DOI] [PubMed] [Google Scholar]
- 21.Cho S, Kagan BL, Blackford JA, Jr, Szapary D, Simons SS., Jr Glucocorticoid receptor ligand binding domain is sufficient for the modulation of both the dose-response curve of receptor-agonist complexes and the partial agonist activity of receptor-antisteroid complexes by glucocorticoid receptors, coactivator TIF2, and Ubc9. Mol Endo. 2005;19:290–311. doi: 10.1210/me.2004-0134. [DOI] [PubMed] [Google Scholar]
- 22.Mercier L, Thompson EB, Simons SS., Jr Dissociation of steroid binding to receptors and steroid induction of biological activity in a glucocorticoid responsive cell. Endocrinology. 1983;112:601–609. doi: 10.1210/endo-112-2-601. [DOI] [PubMed] [Google Scholar]
- 23.Simons SS., Jr The importance of being varied in steroid receptor transactivation. TIPS. 2003;24:253–259. doi: 10.1016/S0165-6147(03)00101-9. [DOI] [PubMed] [Google Scholar]
- 24.Simons SS., Jr How much is enough? Modulation of dose-response curve for steroid receptor-regulated gene expression by changing concentrations of transcription factor. Current Topics in Medicinal Chemistry. 2006;6:271–285. doi: 10.2174/156802606776173465. [DOI] [PubMed] [Google Scholar]
- 25.Cadepond F, Ulmann A, Baulieu EE. RU486 (mifepristone): mechanisms of action and clinical uses. Annu Rev Med. 1997;48:129–156. doi: 10.1146/annurev.med.48.1.129. [DOI] [PubMed] [Google Scholar]
- 26.Szapary D, Huang Y, Simons SS., Jr Opposing effects of corepressor and coactivators in determining the dose-response curve of agonists, and residual agonist activity of antagonists, for glucocorticoid receptor regulated gene expression. Mol Endocrinol. 1999;13:2108–2121. doi: 10.1210/mend.13.12.0384. [DOI] [PubMed] [Google Scholar]
- 27.Sarlis NJ, Bayly SF, Szapary D, Simons SS., Jr Quantity of partial agonist activity for antiglucocorticoids complexed with mutant glucocorticoid receptors is constant in two different transactivation assays but not predictable from steroid structure. J Steroid Biochem Molec Biol. 1999;68:89–102. doi: 10.1016/s0960-0760(99)00021-7. [DOI] [PubMed] [Google Scholar]
- 28.Sistare FD, Hager GL, Simons SS., Jr Mechanism of dexamethasone 21-mesylate antiglucocorticoid action: I. Receptor-antiglucocorticoid complexes do not competitively inhibit receptor-glucocorticoid complex activation of gene transcription in vivo. Mol Endocrinol. 1987;1:648–658. doi: 10.1210/mend-1-9-648. [DOI] [PubMed] [Google Scholar]
- 29.Oshima H, Simons SS., Jr Modulation of transcription factor activity by a distant steroid modulatory element. Mol Endocrinol. 1992;6:416–428. doi: 10.1210/mend.6.3.1584217. [DOI] [PubMed] [Google Scholar]
- 30.Szapary D, Song L-N, He Y, Simons SS., Jr Differential modulation of glucocorticoid and progesterone receptor transactivation. Mol Cell Endocrinol. 2008;283:114–126. doi: 10.1016/j.mce.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshima H, Szapary D, Simons SS., Jr The factor binding to the glucocorticoid modulatory element of the tyrosine aminotransferase gene is a novel and ubiquitous heteromeric complex. J Biol Chem. 1995;270:21893–21910. doi: 10.1074/jbc.270.37.21893. [DOI] [PubMed] [Google Scholar]
- 32.Kaul S, Blackford JA, Jr, Chen J, Ogryzko VV, Simons SS., Jr Properties of the glucocorticoid modulatory element binding proteins GMEB-1 and -2: potential new modifiers of glucocorticoid receptor transactivation and members of the family of KDWK proteins. Mol Endocrinol. 2000;14:1010–1027. doi: 10.1210/mend.14.7.0494. [DOI] [PubMed] [Google Scholar]
- 33.Szapary D, Xu M, Simons SS., Jr Induction properties of a transiently transfected glucocorticoid-responsive gene vary with glucocorticoid receptor concentration. J Biol Chem. 1996;271:30576–30582. doi: 10.1074/jbc.271.48.30576. [DOI] [PubMed] [Google Scholar]
- 34.Webster JC, Cidlowski JA. Mechanisms of glucocorticoid-receptor-mediated repression of gene expression. Trends Endocrinol Metab. 1999;10:396–402. doi: 10.1016/s1043-2760(99)00186-1. [DOI] [PubMed] [Google Scholar]
- 35.Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem. 2001;276:13615–13621. doi: 10.1074/jbc.M008384200. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Q, Pang J, Favata MF, Trzaskos JM. Receptor density dictates the behavior of a subset of steroid ligands in glucocorticoid receptor-mediated transrepression. Int Immunopharmacol. 2003;3:1803–1817. doi: 10.1016/j.intimp.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Sun Y, Tao Y, Kagan BL, He Y, Simons SS., Jr Modulation of transcription parameters in glucocorticoid receptor-mediated repression. Mol Cell Endocrinol. 2008 doi: 10.1016/j.mce.2008.05.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiu CM, Yeh SH, Chen PJ, Kuo TJ, Chang CJ, Chen PJ, Yang WJ, Chen DS. Hepatitis B virus X protein enhances androgen receptor-responsive gene expression depending on androgen level. Proc Natl Acad Sci U S A. 2007;104:2571–2578. doi: 10.1073/pnas.0609498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuqua SA, Wiltschke C, Zhang QX, Borg A, Castles CG, Friedrichs WE, Hopp T, Hilsenbeck S, Mohsin S, O’Connell P, Allred DC. A hypersensitive estrogen receptor-alpha mutation in premalignant breast lesions. Cancer Res. 2000;60:4026–4029. [PubMed] [Google Scholar]
- 40.Tao Y-G, Xu Y, Xu HE, Simons SSJ. Mutations of glucocorticoid receptor differentially affect AF2 domain activity in a steroid-selective manner to alter the potency and efficacy of gene induction and repression. Biochemistry. 2008 doi: 10.1021/bi800472w. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velasco LF, Togashi M, Walfish PG, Pessanha RP, Moura FN, Barra GB, Nguyen P, Rebong R, Yuan C, Simeoni LA, Ribeiro RC, Baxter JD, Webb P, Neves FA. Thyroid hormone response element organization dictates the composition of active receptor. J Biol Chem. 2007;282:12458–12466. doi: 10.1074/jbc.M610700200. [DOI] [PubMed] [Google Scholar]
- 42.Wan W, Farboud B, Privalsky ML. Pituitary resistance to thyroid hormone syndrome is associated with T3 receptor mutants that selectively impair beta2 isoform function. Mol Endocrinol. 2005;19:1529–1542. doi: 10.1210/me.2005-0014. [DOI] [PubMed] [Google Scholar]
- 43.Onate SA, Tsai SY, Tsai M-J, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 44.Jackson TA, Richer JK, Bain DL, Takimoto GS, Tung L, Horwitz KB. The partial agonist activity of antagonist-occupied steroid receptors is controlled by a novel hinge domain-binding coactivator L7/SPA and the corepressors N-CoR or SMRT. Mol Endocrinol. 1997;11:693–705. doi: 10.1210/mend.11.6.0004. [DOI] [PubMed] [Google Scholar]
- 45.Smith CL, Nawaz Z, O’Malley BW. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y, Kawate H, Ohnaka K, Nawata H, Takayanagi R. Nuclear compartmentalization of N-CoR and its interactions with steroid receptors. Mol Cell Biol. 2006;26:6633–6655. doi: 10.1128/MCB.01534-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregory CW, He B, Johnson RT, Ford OH, Mohler JL, French FS, Wilson EM. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res. 2001;61:4315–4319. [PubMed] [Google Scholar]
- 48.Agoulnik IU, Vaid A, Bingman WEr, Erdeme H, Frolov A, Smith CL, Ayala G, Ittmann MM, Weigel NL. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res. 2005;65:7959–7967. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- 49.Vottero A, Kino T, Combe H, Lecomte P, Chrousos GP. A novel, C-terminal dominant negative mutation of the GR causes familial glucocorticoid resistance through abnormal interactions with p160 steroid receptor coactivators. J Clin Endocrinol Metab. 2002;87:2658–2667. doi: 10.1210/jcem.87.6.8520. [DOI] [PubMed] [Google Scholar]
- 50.Wang Q, Richter WF, Anzick SL, Meltzer PS, Simons SS., Jr Modulation of transcriptional sensitivity of mineralocorticoid and estrogen receptors. J Steroid Biochem Molec Biol. 2004;91:197–210. doi: 10.1016/j.jsbmb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 51.Rowan BG, Weigel NL, O’Malley BW. Phosphorylation of steroid receptor coactivator-1. Identification of the phosphorylation sites and phosphorylation through the mitogen-activated protein kinase pathway. J Biol Chem. 2000;275:4475–4483. doi: 10.1074/jbc.275.6.4475. [DOI] [PubMed] [Google Scholar]
- 52.Giannoukos G, Szapary D, Smith CL, Meeker JEW, Simons SS., Jr New antiprogestins with partial agonist activity: potential selective progesterone receptor modulators (SPRMs) and probes for receptor- and coregulator-induced changes in progesterone receptor induction properties. Mol Endocrinol. 2001;15:255–270. doi: 10.1210/mend.15.2.0596. [DOI] [PubMed] [Google Scholar]
- 53.Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–2468. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- 54.Kressler D, Hock MB, Kralli A. Coactivators PGC-1beta and SRC-1 Interact Functionally to Promote the Agonist Activity of the Selective Estrogen Receptor Modulator Tamoxifen. J Biol Chem. 2007;282:26897–26907. doi: 10.1074/jbc.M705596200. [DOI] [PubMed] [Google Scholar]
- 55.Heneghan AF, Connaghan-Jones KD, Miura MT, Bain DL. Coactivator assembly at the promoter: efficient recruitment of SRC2 is coupled to cooperative DNA binding by the progesterone receptor. Biochemistry. 2007;46:11023–11032. doi: 10.1021/bi700850v. [DOI] [PubMed] [Google Scholar]
- 56.Ricketson D, Hostick U, Fang L, Yamamoto KR, Darimont BD. A conformational switch in the ligand-binding domain regulates the dependence of the glucocorticoid receptor on Hsp90. J Mol Biol. 2007;368:729–741. doi: 10.1016/j.jmb.2007.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webb P, Nguyen P, Kushner PJ. Differential SERM effects on corepressor binding dictate ERalpha activity in vivo. J Biol Chem. 2003;278:6912–6920. doi: 10.1074/jbc.M208501200. [DOI] [PubMed] [Google Scholar]
- 58.Wang Q, Blackford JA, Jr, Song L-N, Huang Y, Simons SS., Jr Equilibrium interactions of corepressors and coactivators modulate the properties of agonist and antagonist complexes of glucocorticoid receptors. Mol Endocrinol. 2004;18:1376–1395. doi: 10.1210/me.2003-0421. [DOI] [PubMed] [Google Scholar]
- 59.Wang D, Simons SS., Jr Corepressor binding to progesterone and glucocorticoid receptors involves the AF-1 domain and is inhibited by molybdate. Mol Endo. 2005;19:1483–1500. doi: 10.1210/me.2005-0012. [DOI] [PubMed] [Google Scholar]
- 60.Wang D, Wang Q, Awasthi S, Simons SS., Jr Amino-terminal domain of TIF2 is involved in competing for corepressor binding to glucocorticoid and progesterone receptors. Biochemistry. 2007;48:8036–8049. doi: 10.1021/bi7004575. [DOI] [PubMed] [Google Scholar]
- 61.Yamamoto Y, Wada O, Suzawa M, Yogiashi Y, Yano T, Kato S, Yanagisawa J. The tamoxifen-responsive estrogen receptor alpha mutant D351Y shows reduced tamoxifen-dependent interaction with corepressor complexes. J Biol Chem. 2001;276:42684–42691. doi: 10.1074/jbc.M107844200. [DOI] [PubMed] [Google Scholar]
- 62.Song L-N, Huse B, Rusconi S, Simons SS., Jr Transactivation specificity of glucocorticoid vs. progesterone receptors: role of functionally different interactions of transcription factors with amino- and carboxyl-terminal receptor domains. J Biol Chem. 2001;276:24806–24816. doi: 10.1074/jbc.M102610200. [DOI] [PubMed] [Google Scholar]
- 63.Dai Y, Ngo D, Forman LW, Qin DC, Jacob J, Faller DV. Sirtuin 1 is required for antagonist-induced transcriptional repression of androgen-responsive genes by the androgen receptor. Mol Endocrinol. 2007;21:1807–1821. doi: 10.1210/me.2006-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van der Laan S, Lachize SB, Vreugdenhil E, de Kloet ER, Meijer OC. Nuclear receptor coregulators differentially modulate induction and glucocorticoid receptor-mediated repression of the corticotropin-releasing hormone gene. Endocrinology. 2008;149:725–732. doi: 10.1210/en.2007-1234. [DOI] [PubMed] [Google Scholar]
- 65.Liao G, Chen LY, Zhang A, Godavarthy A, Xia F, Ghosh JC, Li H, Chen JD. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J Biol Chem. 2003;278:5052–5061. doi: 10.1074/jbc.M206374200. [DOI] [PubMed] [Google Scholar]
- 66.Yoon HG, Wong J. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol Endocrinol. 2006;20:1048–1060. doi: 10.1210/me.2005-0324. [DOI] [PubMed] [Google Scholar]
- 67.Hu X, Lazar MA. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 68.Nagy L, Kao H-Y, Love JD, Li C, Banayo E, Gooch JT, Krishna V, Chatterjee K, Evans RM, Schwabe JWR. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes and Develop. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, Lambert MH, Milburn MV, Glass CK, Rosenfeld MG. Molecular determinants of nuclear receptor-corepressor interaction. Genes and Develop. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frego L, Davidson W. Conformational changes of the glucocorticoid receptor ligand binding domain induced by ligand and cofactor binding, and the location of cofactor binding sites determined by hydrogen/deuterium exchange mass spectrometry. Protein Sci. 2006;15:722–730. doi: 10.1110/ps.051781406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kroe RR, Baker MA, Brown MP, Farrow NA, Gautschi E, Hopkins JL, Lafrance RR, Kronkaitis A, Freeman D, Thomson D, Nabozny G, Grygon CA, Labadia ME. Agonist versus antagonist induce distinct thermodynamic modes of co-factor binding to the glucocorticoid receptor. Biophys Chem. 2007;128:156–164. doi: 10.1016/j.bpc.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 72.Zhu P, Baek SH, Bourk EM, Ohgi KA, Garcia-Bassets I, Sanjo H, Akira S, Kotol PF, Glass CK, Rosenfeld MG, Rose DW. Macrophage/cancer cell interactions mediate hormone resistance by a nuclear receptor derepression pathway. Cell. 2006;124:615–629. doi: 10.1016/j.cell.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 73.Sohn YC, Kim SW, Lee S, Kong YY, Na DS, Lee SK, Lee JW. Dynamic inhibition of nuclear receptor activation by corepressor binding. Mol Endocrinol. 2003;17:366–372. doi: 10.1210/me.2002-0150. [DOI] [PubMed] [Google Scholar]
- 74.Jeyakumar M, Liu XF, Erdjument-Bromage H, Tempst P, Bagchi MK. Phosphorylation of thyroid hormone receptor-associated nuclear receptor corepressor holocomplex by the DNA-dependent protein kinase enhances its histone deacetylase activity. J Biol Chem. 2007;282:9312–9322. doi: 10.1074/jbc.M609009200. [DOI] [PubMed] [Google Scholar]
- 75.Peterson TJ, Karmakar S, Pace MC, Gao T, Smith CL. The silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) corepressor is required for full estrogen receptor alpha transcriptional activity. Mol Cell Biol. 2007;27:5933–5948. doi: 10.1128/MCB.00237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gupta P, Park SW, Farooqui M, Wei LN. Orphan nuclear receptor TR2, a mediator of preadipocyte proliferation, is differentially regulated by RA through exchange of coactivator PCAF with corepressor RIP140 on a platform molecule GRIP1. Nucleic Acids Res. 2007;35:2269–2282. doi: 10.1093/nar/gkl1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He Y, Simons SS., Jr STAMP: a novel predicted factor assisting TIF2 actions in glucocorticoid receptor-mediated induction and repression. Mol Cell Biol. 2007;27:1467–1485. doi: 10.1128/MCB.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hodgson MC, Astapova I, Hollenberg AN, Balk SP. Activity of androgen receptor antagonist bicalutamide in prostate cancer cells is independent of NCoR and SMRT corepressors. Cancer Res. 2007;67:8388–8395. doi: 10.1158/0008-5472.CAN-07-0617. [DOI] [PubMed] [Google Scholar]
- 79.Zwart W, Griekspoor A, Berno V, Lakeman K, Jalink K, Mancini M, Neefjes J, Michalides R. PKA-induced resistance to tamoxifen is associated with an altered orientation of ERalpha towards co-activator SRC-1. EMBO J. 2007;26:3534–3544. doi: 10.1038/sj.emboj.7601791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Truica CI, Byers S, Gelmann EP. Beta-catenin affects androgen receptor transcriptional activity and ligand specificity. Cancer Res. 2000;60:4709–4713. [PubMed] [Google Scholar]
- 81.Westberry JM, Sadosky PW, Hubler TR, Gross KL, Scammell JG. Glucocorticoid resistance in squirrel monkeys results from a combination of a transcriptionally incompetent glucocorticoid receptor and overexpression of the glucocorticoid receptor co-chaperone FKBP51. J Steroid Biochem Mol Biol. 2006;100:34–41. doi: 10.1016/j.jsbmb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 82.Lyons LS, Burnstein KL. Vav3, a Rho GTPase guanine nucleotide exchange factor, increases during progression to androgen independence in prostate cancer cells and potentiates androgen receptor transcriptional activity. Mol Endocrinol. 2006;20:1061–1072. doi: 10.1210/me.2005-0346. [DOI] [PubMed] [Google Scholar]
- 83.Jia L, Shen HC, Wantroba M, Khalid O, Liang G, Wang Q, Gentzschein E, Pinski JK, Stanczyk FZ, Jones PA, Coetzee GA. Locus-wide chromatin remodeling and enhanced androgen receptor-mediated transcription in recurrent prostate tumor cells. Mol Cell Biol. 2006;26:7331–7341. doi: 10.1128/MCB.00581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen S, Simons SS., Jr A second pathway for the modulation of glucocorticoid receptor transactivation properties that involves hSur2. Mol Cell Endo. 2003;199:129–142. doi: 10.1016/s0303-7207(02)00333-7. [DOI] [PubMed] [Google Scholar]
- 85.Weigel NL, Moore NL. Steroid receptor phosphorylation: a key modulator of multiple receptor functions. Mol Endocrinol. 2007;21:2311–2319. doi: 10.1210/me.2007-0101. [DOI] [PubMed] [Google Scholar]
- 86.Kino T, Ichijo T, Amin ND, Kesavapany S, Wang Y, Kim N, Rao S, Player A, Zheng YL, Garabedian MJ, Kawasaki E, Pant HC, Chrousos GP. Cyclin-dependent kinase 5 differentially regulates the transcriptional activity of the glucocorticoid receptor through phosphorylation: clinical implications for the nervous system response to glucocorticoids and stress. Mol Endocrinol. 2007;21:1552–1568. doi: 10.1210/me.2006-0345. [DOI] [PubMed] [Google Scholar]
- 87.Huang CY, Beliakoff J, Li X, Lee J, Li X, Sharma M, Lim B, Sun Z. hZimp7, a novel PIAS-like protein, enhances androgen receptor-mediated transcription and interacts with SWI/SNF-like BAF complexes. Mol Endocrinol. 2005;19:2915–2929. doi: 10.1210/me.2005-0097. [DOI] [PubMed] [Google Scholar]
- 88.Sun J, Blair AL, Aiyar SE, Li R. Cofactor of BRCA1 modulates androgen-dependent transcription and alternative splicing. J Steroid Biochem Mol Biol. 2007;107:131–139. doi: 10.1016/j.jsbmb.2007.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pascual-Le Tallec L, Simone F, Viengchareun S, Meduri G, Thirman MJ, Lombes M. The elongation factor ELL (eleven-nineteen lysine-rich leukemia) is a selective coregulator for steroid receptor functions. Mol Endocrinol. 2005;19:1158–1169. doi: 10.1210/me.2004-0331. [DOI] [PubMed] [Google Scholar]
- 90.Chen S, Sarlis NJ, Simons SS., Jr Evidence for a common step in three different processes for modulating the kinetic properties of glucocorticoid receptor-induced gene transcription. J Biol Chem. 2000;275:30106–30117. doi: 10.1074/jbc.M005418200. [DOI] [PubMed] [Google Scholar]
- 91.Kaul S, Blackford JA, Jr, Cho S, Simons SS., Jr Ubc9 is a novel modulator of the induction properties of glucocorticoid receptors. J Biol Chem. 2002;277:12541–12549. doi: 10.1074/jbc.M112330200. [DOI] [PubMed] [Google Scholar]
- 92.Bai W, Tullos S, Weigel NL. Phosphorylation of Ser-530 facilitates hormone-dependent transcriptional activation of the chicken progesterone receptor. Mol Endo. 1994;8:1465–1473. doi: 10.1210/mend.8.11.7877616. [DOI] [PubMed] [Google Scholar]
- 93.McInerney EM, Katzenellenbogen BS. Different regions in activation function-1 of the human estrogen receptor required for antiestrogen- and estradiol-dependent transcriptional activation. J Biol Chem. 1996;271:24172–24178. doi: 10.1074/jbc.271.39.24172. [DOI] [PubMed] [Google Scholar]
- 94.Russcher H, Smit P, van den Akker EL, van Rossum EF, Brinkmann AO, de Jong FH, Lamberts SW, Koper JW. Two polymorphisms in the glucocorticoid receptor gene directly affect glucocorticoid-regulated gene expression. J Clin Endocrinol Metab. 2005;90:5804–5810. doi: 10.1210/jc.2005-0646. [DOI] [PubMed] [Google Scholar]
- 95.He Y, Szapary D, Simons SS., Jr Modulation of induction properties of glucocorticoid receptor-agonist and -antagonist complexes by coactivators involves binding to receptors but is independent of ability of coactivators to augment transactivation. J Biol Chem. 2002;277:49256–49266. doi: 10.1074/jbc.M205536200. [DOI] [PubMed] [Google Scholar]
- 96.Cheng J, Zhang C, Shapiro DJ. A functional serine 118 phosphorylation site in estrogen receptor-alpha is required for down-regulation of gene expression by 17beta-estradiol and 4-hydroxytamoxifen. Endocrinology. 2007;148:4634–4641. doi: 10.1210/en.2007-0148. [DOI] [PubMed] [Google Scholar]
- 97.Mercier L, Miller PA, Simons SS., Jr Antiglucocorticoid steroids have increased agonist activity in those hepatoma cell lines that are more sensitive to glucocorticoids. J Steroid Biochem. 1986;25:11–20. doi: 10.1016/0022-4731(86)90275-x. [DOI] [PubMed] [Google Scholar]
- 98.Bohl CE, Wu Z, Miller DD, Bell CE, Dalton JT. Crystal structure of the T877A human androgen receptor ligand-binding domain complexed to cyproterone acetate provides insight for ligand-induced conformational changes and structure-based drug design. J Biol Chem. 2007;282:13648–13655. doi: 10.1074/jbc.M611711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim Y, Sun Y, Chow C, Pommier YG, Simons SS., Jr Effects of acetylation, polymerase phosphorylation, and DNA unwinding in glucocorticoid receptor transactivation. J Steroid Biochem Molec Biol. 2006;100:3–17. doi: 10.1016/j.jsbmb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 100.Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O’Malley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 101.Aiyar SE, Sun JL, Blair AL, Moskaluk CA, Lu YZ, Ye QN, Yamaguchi Y, Mukherjee A, Ren DM, Handa H, Li R. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Auboeuf D, Honig A, Berget SM, O’Malley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- 103.Dong X, Shylnova O, Challis JR, Lye SJ. Identification and characterization of the protein-associated splicing factor as a negative co-regulator of the progesterone receptor. J Biol Chem. 2005;280:13329–13340. doi: 10.1074/jbc.M409187200. [DOI] [PubMed] [Google Scholar]
- 104.Auboeuf D, Dowhan DH, Kang YK, Larkin K, Lee JW, Berget SM, O’Malley BW. Differential recruitment of nuclear receptor coactivators may determine alternative RNA splice site choice in target genes. Proc Natl Acad Sci U S A. 2004;101:2270–2274. doi: 10.1073/pnas.0308133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 106.Zorio DA, Bentley DL. The link between mRNA processing and transcription: communication works both ways. Exp Cell Res. 2004;296:91–97. doi: 10.1016/j.yexcr.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 107.Reichardt HM, Umland T, Bauer A, Kretz O, Schutz G. Mice with an increased glucocorticoid receptor gene dosage show enhanced resistance to stress and endotoxic shock. Mol Cell Biol. 2000;20:9009–9017. doi: 10.1128/mcb.20.23.9009-9017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 109.Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinanen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi OP. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9:401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 110.Kasbohm EA, Guo R, Yowell CW, Bagchi G, Kelly P, Arora P, Casey PJ, Daaka Y. Androgen receptor activation by G(s) signaling in prostate cancer cells. J Biol Chem. 2005;280:11583–11589. doi: 10.1074/jbc.M414423200. [DOI] [PubMed] [Google Scholar]
- 111.Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 112.Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24:5157–51571. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.http://www.nursa.org/template.cfm?threadId=10222&dataType=Q-PCR&dataset=Tissue-specific%20expression%20patterns%20of%20nuclear%20receptors
- 114.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 115.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]