Abstract
Amyotrophic lateral sclerosis/parkinsonism–dementia complex (ALS/PDC) is a fatal neurodegenerative disease found in the Chamorro people of Guam and other Pacific Island populations. The etiology is unknown, although both genetic and environmental factors appear important. To identify loci for ALS/PDC, we conducted both genome-wide linkage and association analyses, using approximately 400 microsatellite markers, in the largest sample assembled to date, comprising a nearly complete sample of all living and previously sampled deceased cases. A single, large, complex pedigree was ascertained from a village on Guam, with smaller families and a case–control sample ascertained from the rest of Guam by population-based neurological screening and archival review. We found significant evidence for two regions with novel ALS/PDC loci on chromosome 12 and supportive evidence for the involvement of the MAPT region on chromosome 17. D12S1617 on 12p gave the strongest evidence of linkage (maximum LOD score, Zmax = 4.03) in our initial scan, with additional support in the complete case–control sample in the form of evidence of allelic association at this marker and another nearby marker. D12S79 on 12q also provided significant evidence of linkage (Zmax = 3.14) with support from flanking markers. Our results suggest that ALS/PDC may be influenced by as many as three loci, while illustrating challenges that are intrinsic in genetic analyses of isolated populations, as well as analytical strategies that are useful in this context. Elucidation of the genetic basis of ALS/PDC should improve our understanding of related neurodegenerative disorders including Alzheimer disease, Parkinson disease, frontotemporal dementia and ALS.
INTRODUCTION
Amyotrophic lateral sclerosis/parkinsonism-dementia complex [ALS/PDC (MIM 105500)] is a fatal neurodegenerative disease of unknown etiology found in the Chamorro people of Guam (1–3), a number of Japanese families located on the Kii peninsula of Japan (4,5) and the Auyu and Jakai people of New Guinea (6). The neuropathology in the Chamarro and Japanese subjects consists of neurofibrillary tangles (NFTs) in both the brain and spinal cord (7–10), with a higher NFT burden in regions corresponding to the respective clinical manifestations of ALS and/or PDC. Clinically, ALS in Chamorros is similar to ALS in other populations (11), and PDC shares features with Parkinson's disease (MIM 168600), but is also accompanied by early and progressive dementia (1,7). The frequency of Guam ALS and PDC was highest in the 1950s, with similar prevalences and annual incidences of about 140 and 50 per 100 000, respectively (2,12,13). In the past 40 years, the age-specific incidence of ALS has steadily declined, whereas the incidence of PDC appeared to decline more modestly until the 1980s and then to increase slightly (14,15). A recent survey identified a prevalence of PDC that is nearly the same as that of the 1950s, but with an increased age-at-onset and decreased annual incidence rate (16). The changing incidence rates and age-at-onset suggest the existence of a changing environmental factor. Cycad toxins, infectious agents and mineral deficiencies have been extensively investigated, but a causal role has not been established, suggesting a complex etiology (17–20).
A genetic basis for ALS/PDC is supported by evidence from both epidemiologic and genetic studies. Early reports noted familial clustering of ALS/PDC, with multiple cases of ALS, PDC, or both in the same individual, often occurring in relatives of probands and in successive generations (1–3). Segregation analyses supported a major gene model, consistent with autosomal dominant inheritance with reduced penetrance, possibly affected by environmental factors (21–23). A 40 year prospective study found significantly higher incidence rates of ALS/PDC in siblings and offspring of cases compared with the expected rates in the Chamorro population (24). Also, despite similar environments and lifestyles, the village of Umatac, which was relatively isolated until the 1950s, had a much higher annual incidence rate of ALS (approximately 250/100 000 in the 1950s) than any other Guam village (2,3,12,13,25). Finally, the population has the hallmarks of a genetic isolate: Chamorros are descendants of aboriginal Austronesian populations from Island Southeast Asia (26–28), with a population size of 40 000–100 000 in the Mariana Islands by the time of the Spanish explorations, a precipitous reduction in population size to approximately 1500 by the end of the 18th century as a consequence of this contact (29,30), and subsequent growth to a current population size on Guam of approximately 65 000.
Population isolates, such as the Chamorros of Guam, provide unique opportunities, but pose significant challenges for mapping complex traits (31–34). A population isolate arises from a small group of founders, and is consequently likely to be more genetically homogeneous than are outbred populations, with sometimes a single founder mutation identified in all affected individuals. The reduced genetic heterogeneity can improve the power to detect linkage (34–36) and diminish concerns about population stratification in case–control studies. Environmental factors also tend to be less heterogeneous among individuals because of shared environment and culture (32,34). Additionally, the availability of genealogical records sometimes enables the reconstruction of large pedigrees, which have higher power than small pedigrees for linkage analysis (37). However, a major disadvantage of population isolates is that the statistical methods designed for genetic analysis of outbred populations can be difficult to apply (31). The large and complex pedigrees found in population isolates often require pedigree simplification to make computations feasible, yet reducing pedigree complexity can result in loss of power (38,39), and breaking inbreeding loops can inflate the type I error rate (40,41). Furthermore, classic association tests, which rely on the independence of cases and controls, may not be valid in populations where randomly selected individuals are more likely to be related (42), particularly among cases (43). Finally, isolates often constitute unique populations with finite sample sizes, which can limit the power to study endemic diseases and the feasibility of replication studies (44).
Despite a few previous studies, a major locus for ALS/PDC has not been identified. A genome-wide association study of a small sample of 41 case and control subjects from Guam, followed by linkage analysis of interesting regions in five small families, identified 17 markers with moderate evidence for association with PDC, but no region with a maximum LOD score Zmax > 3 in linkage analysis (45). A single-marker Zmax = 1.8 was obtained on chromosome (chr) 20, with multipoint Zmax > 2 on chr 14 and 20. Case–control studies also demonstrated that variants in the microtubule-associated protein tau [MAPT (MIM 157140)] gene on chr 17 are associated with ALS, PDC and Guam dementia (46,47). These previous attempts to map ALS/PDC loci have been hindered by limited power because of small sample sizes and the lack of information from a genome linkage scan.
To identify loci for ALS/PDC of Guam, we conducted a genome scan in the largest sample of Chamorros studied to date, representing half a century of data collection (14,22,48) and a complete or nearly complete sample of all available cases, both living and deceased. We performed genome-wide linkage analysis with a large, complex pedigree from the village of Umatac, and with a number of small nuclear families from the rest of Guam. We also performed a genome-wide association study in a sample of cases and controls assembled from archival data and screening efforts targeting the entire current Chamorro population on the island of Guam, and the pedigree from Umatac. The late onset of the disease, the complexity of the Umatac pedigree structure and the population history of the Chamorro people all posed significant analytical challenges. We describe our approach to these challenges along with the results of our findings supporting significant evidence for the existence of one to two major genes for ALS/PDC on chr 12, and additional support for the involvement of the MAPT region on chr 17.
RESULTS
Genome-wide linkage analysis
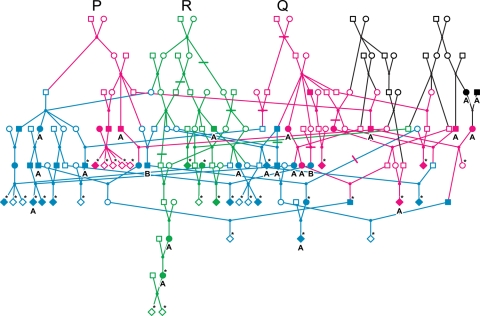
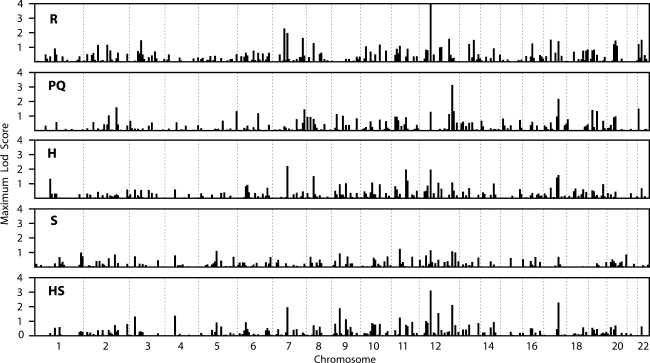
We carried out a genome-wide linkage scan of an extended and complex Umatac kindred (U pedigree; Fig. 1) divided into three subcomponents (R, PQ and H), and of 11 simple-structure non-Umatac kindreds (S families; Table 1). The R and PQ subpedigrees were defined by the descendents of particular founder couples, and overlap; the H subpedigrees were defined by identifying simpler pedigrees on the basis of sampled individuals in the most recent generations, and do not overlap. We obtained significant evidence for linkage (Zmax > 3) in some of these subcomponents in the initial genome scan for markers on two regions of chr 12, with suggestive evidence for linkage (Zmax > 2) on chr 7 and 17 (Fig. 2, Table 2).
Figure 1.
The U pedigree (n = 159). Solid symbols represent individuals affected by ALS (A), PDC, or both (B) ALS and PDC. Asterisks denote genotyped individuals. Other individuals were almost always deceased and unavailable for genotyping. The overlapping portions of the PQ (magenta) and R (green) subpedigrees are shown in blue, and the remainder of the U pedigree is shown in black.
Table 1.
Description of Umatac R, PQ and H subpedigrees and non-Umatac S families
| Subpedigree | Families | Individuals (genotyped) | Affected (genotyped) | Unaffected (genotyped) | Loops |
|---|---|---|---|---|---|
| R | 1 | 93 (26) | 32 (16) | 9 (9) | 7 |
| PQ | 1 | 107 (28) | 40 (16) | 10 (10) | 7 |
| H | 4 | 105 (32) | 44 (18) | 12 (12) | 4 |
| H1 | 1 | 66 (20) | 24 (11) | 7 (7) | 2 |
| H2 | 1 | 24 (5) | 13 (4) | 1 (1) | 2 |
| H3 | 1 | 8 (2) | 4 (2) | 0 (0) | 0 |
| H4 | 1 | 7 (5) | 3 (1) | 4 (4) | 0 |
| S | 11 | 53 (24) | 22 (22) | 1 (1) | 0 |
Figure 2.
Genome-wide single-marker linkage analysis results. Bar graph of maximum LOD score for each marker, plotted at the position of each marker, in the R and PQ founder subpedigrees, H non-founder subpedigrees, independent S families and combined HS data set.
Table 2.
Chromosome regions with suggestive (Zmax > 2) maximum LOD scores in pairwise genome-wide linkage analysis
| Marker | Locationa | R | PQ | H | S | HS |
|---|---|---|---|---|---|---|
| D7S516 | 41.69 | 0.00 | 0.00 | 0.09 | 0.00 | 0.08 |
| D7S484 | 53.50 | 0.08 | 0.00 | 0.00 | 0.00 | 0.00 |
| D7S510 | 59.93 | 2.29 | 0.30 | 0.01 | 0.00 | 0.00 |
| D7S519 | 69.03 | 0.09 | 0.20 | 0.07 | 0.25 | 0.23 |
| D7S502 | 78.65 | 1.98 | 0.00 | 2.22 | 0.06 | 1.96 |
| D7S669 | 90.42 | 0.18 | 0.14 | 0.00 | 0.00 | 0.00 |
| D7S630 | 98.44 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 |
| D12S364 | 30.60 | 0.21 | 0.05 | 0.46 | 0.00 | 0.20 |
| D12S310 | 36.06 | 0.83 | 0.00 | 0.50 | 0.80 | 1.01 |
| D12S1617 | 44.03 | 4.03 | 1.29 | 1.96 | 1.15 | 3.11 |
| D12S345 | 53.09 | 0.00 | 0.00 | 0.00 | 0.26 | 0.14 |
| D12S85 | 61.34 | 0.98 | 0.12 | 0.02 | 0.03 | 0.05 |
| D12S78 | 111.87 | 1.58 | 1.13 | 0.24 | 0.09 | 0.29 |
| D12S1646 | 121.84 | 0.47 | 0.00 | 0.02 | 0.00 | 0.01 |
| D12S79 | 125.31 | 0.24 | 3.14 | 1.08 | 1.07 | 2.12 |
| D12S86 | 134.54 | 0.27 | 1.33 | 0.51 | 0.00 | 0.03 |
| D12S324 | 147.17 | 0.01 | 0.02 | 0.04 | 0.99 | 0.73 |
| D17S798 | 53.41 | 0.62 | 0.00 | 0.00 | 0.10 | 0.00 |
| D17S1868 | 64.16 | 0.00 | 0.97 | 1.42 | 0.00 | 0.46 |
| D17S787 | 74.99 | 1.41 | 2.18 | 1.59 | 0.69 | 2.29 |
| D17S944 | 82.56 | 0.00 | 0.00 | 0.00 | 0.18 | 0.01 |
| D17S949 | 93.27 | 0.41 | 0.00 | 0.12 | 0.01 | 0.06 |
Boldface indicates markers with Zmax > 2 in any data set.
aKosambi map (cM) as reported on the Marshfield web site.
The strongest evidence for linkage with an individual marker was obtained for D12S1617 at 44 cM. In the R subpedigree, genome-wide significant evidence of linkage was obtained: Zmax = 4.03 at a recombination fraction, θ, of 0. The combined H + S (HS) data set also gave significant evidence of linkage to this same marker (Zmax = 3.1, θ = 0), with the H families contributing Zmax = 1.96 to this total; no other markers yielded Zmax > 3 in the HS data set. Empirical P-values for these LOD scores, including the effects of pedigree simplification, were <0.0001 for R (maximum LOD = 2.87 across 10 000 replicates) and 0.0004 for HS. The PQ subpedigree also provided suggestive evidence in favor of linkage to D12S1617 (Zmax = 1.3, θ = 0). Results for D12S1617 were sensitive to the choice of the sample used to specify marker allele frequencies, as would be expected if sampling is through related cases with a disease locus close to the marker: when frequencies were derived from external cases or the U pedigree, Zmax was reduced by 29 or 89%, respectively, compared with the use of frequencies from unrelated controls. Similar patterns of reduced LOD scores using case allele frequencies were found for the H and PQ subpedigrees. Further evaluation of the evidence for the association with this marker is presented later.
A second region on chr 12q at 125 cM also gave a significant LOD score for D12S79 in the genome scan (Table 2). The PQ component of the U pedigree was the major contributor to this signal, yielding a significant Zmax = 3.14 at θ = 0, with support (Zmax > 1) from markers 9–14 cM on either side (Supplementary Material, Table S1). The combined HS data set also gave supportive evidence of linkage to D12S79 (Zmax = 2.12, θ = 0), with equal contributions to the total from the H and S data sets. The R subpedigree was essentially uninformative for D12S79 (Zmax = 0.2, θ = 0.2), but provided modest support to this region from nearby marker D12S78 (Zmax = 1.58). Computations with the PQ component were too slow (Table 3) to allow practical computation of an empirical P-value.
Table 3.
Linkage analysis results for key markers in subpedigrees with additional loops retained
| Subpedigree/marker | Loops (n = 7) | Zmaxa | Run timeb (days) |
|---|---|---|---|
| R/D12S1617 | 2 | 4.03 | <0.0007 |
| 3 | 3.88 | 0.04 | |
| 4 | 3.81 | 2.6 | |
| 4 | 3.85 | 4.7 | |
| 5 | 3.80 | 130 | |
| 7c | 3.15 ± 0.04 | 48/6 scans | |
| PQ/D12S79 | 3 | 3.14 | 0.04 |
| 4 | 2.67 | 4.7 | |
| 5 | 3.05 | 186 | |
| 7c | 2.91 ± 0.14 | 40/6 scans |
aMaximum LOD score obtained at θ = 0 for exact analysis, or mean LOD score ± SD at θ = 0.001 in six MCMC replicate runs. MCMC estimates at the exact location of the marker can be unstable, so a recombination fraction of 0.001 was used instead of 0.
bTotal run time on AMD Opteron 1.8 GHz processor for a single exact analysis, or six replicate MCMC runs.
cMCMC-based LOD score analysis using lm_bayes in MORGAN.
Sensitivity analyses suggested that breaking loops did not substantially alter the interpretation of the results (Table 3). Exact single-marker computations with five loops were extremely slow, taking over four CPU-months, whereas analyses based on Markov chain Monte Carlo (MCMC) methods, while still demanding (Table 3), remained computationally feasible for primary analyses and produced stable estimates for as many as seven loops. For D12S1617 in the R subpedigree, breaking connections led to only slight inflation of the LOD score in all but the deepest portion of the pedigree. For D12S79 in the PQ subpedigree, breaking loops resulted in modest LOD score increase or decrease depending on the particular individual and number of loops involved. For example, reducing the number of loops from five to four decreased Zmax at D12S79 from 3.05 to 2.67, whereas breaking four loops increased Zmax from 2.91 to 3.14 compared with the intact seven-loop PQ subpedigree.
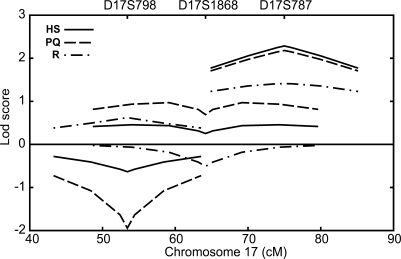
Suggestive LOD scores were also found near MAPT on chr 17 (Fig. 3), and for two markers on chr 7 (Table 2; Supplementary Material, Table S1). For D17S787 (75 cM), located ∼9 mb distal to MAPT, all data sets showed consistent but modest evidence of linkage, with Zmax at θ = 0 ranging from 1.4 in the R subpedigree to 2.3 in the HS data set. For D17S1868, located ∼3 mb distal to MAPT, the PQ subpedigree provided weak evidence in favor of linkage (Zmax = 1, θ = 0.05), while the R and HS data sets were largely uninformative. On chr 7, marker D7S502 (79 cM) showed evidence in favor of linkage in the H (Zmax = 2.2, θ = 0) and R (Zmax = 2.0, θ = 0) subpedigrees, while the PQ and S data sets were essentially uninformative at this marker. For D7S510 (60 cM), only the R subpedigree provided support for linkage (Zmax = 2.3, θ = 0).
Figure 3.
Chromosome 17 single-marker linkage analysis results. LOD score curves for HS data set (solid line), PQ subpedigree (dashes) and R subpedigree (dot-dashes). The MAPT gene is located between D17S798 and D17S1868, ∼3 Mb proximal to D17S1868.
Separate linkage analyses with ALS and PDC as distinct traits gave results that were consistent with a single disease. The two phenotypes mapped to the same regions on chr 12 and 17 and did not separately explain the two distinct peaks on chr 12. No Zmax > 3 was found in genome scans of ALS or PDC, separately, and higher Zmax values in the regions of interest were obtained by combining ALS and PDC in the trait model. Evidence in favor of linkage was obtained in all complex subpedigrees and in the S data set at markers D12S1617 (44 cM), D12S79 (125 cM) and D17S787 (∼10 cM distal to MAPT) for both ALS and PDC, analyzed as distinct traits.
Genome-wide association study
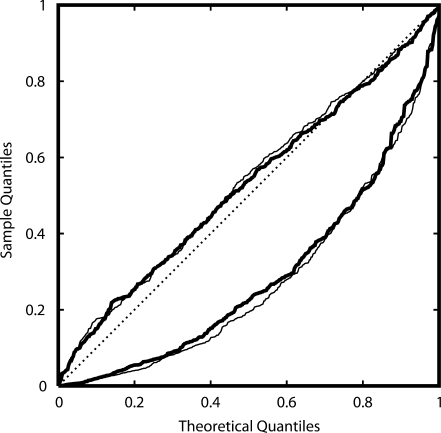
Initial genome-wide tests of association resulted in a markedly non-uniform P-value distribution from the analysis of both the full sample and the sample of all unrelated cases and controls (Fig. 4). While the existence of relatives in the Umatac sample was known, these results for the non-Umatac subjects suggested violation of analysis assumptions. Deviation from Hardy–Weinberg equilibrium (HWE) was unlikely to explain the aberrant distribution because there was little evidence of departure from HWE among cases or controls (not shown). Further investigation identified cryptic relatedness as the cause of the aberrant P-value distribution.
Figure 4.
Quantile–quantile plot of P-value distributions for χ2 test for allelic association in cases versus controls. Lines show P-value distributions before (lower lines) and after (upper lines) correcting for relationships in the sample, for the complete sample of affected and unaffected individuals in the population-based sample (thin lines) and the combined sample from the population-based sample plus the Umatac pedigree (thick lines). The uniform distribution expected under the null distribution of no association is shown along the diagonal (dotted line).
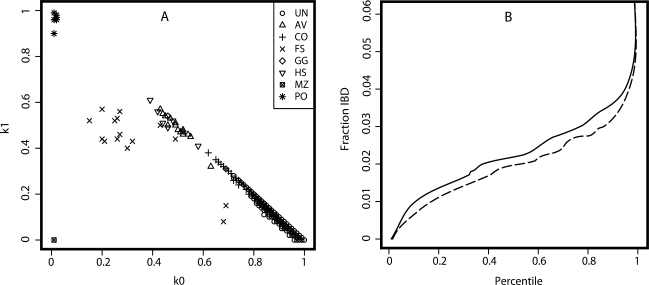
Using the genome-wide marker data, we identified clusters of related individuals (Fig. 5A) who had a strong effect on the performance of the association testing. Inferred pairwise relationships and estimated k-statistics used to estimate kinship coefficients demonstrate that a substantial majority (72.3%) of putatively unrelated pairs of individuals had estimated k1 > 5% and therefore showed evidence of cryptic relatedness. In addition, the proportion of the genome shared identical by descent was cumulatively higher between pairs of unrelated cases than pairs of unrelated controls, as would be expected for a genetic disease (Fig. 5B) (43), thereby providing further support for an underlying genetic basis.
Figure 5.
Relationships in the population-based sample. (A) Estimated k coefficients and relationships inferred from the genome-wide marker data in population-based Chamorro cases and controls. Maximum likelihood estimates of the multilocus probability of sharing 0 (k0) or 1 (k1) alleles identical by descent; k2 = 1 − k0 − k1. As estimated by Relpair: UN, unrelated; AV, avuncular; CO, first cousins; FS, full siblings; GG, grandparent-grandchild; HS, half-siblings; MZ, monozygotic twins (duplicate sample); PO, parent-offspring. (B) Cumulative distribution of estimated IBD sharing among pairs of individuals for cases (solid line) and controls (dashed line).
Three markers on chr 12 continued to show evidence for the association with ALS/PDC (Table 4) after we developed and applied a test that allowed the use of the entire sample for such analysis. The strongest evidence for association was obtained for D12S1617. Evidence for association in the entire sample was suggestive based on a relationship-corrected χ2 test (49) (P = 0.097, 5th percentile of tests), but became nominally significant (q = 0.0058) when results from the genome-wide linkage scan were incorporated as weights in the computation of the false-discovery rate (FDR). For D12S1617, alleles 252 and 258 had consistently higher frequencies in cases compared with controls across samples (Table 5), with allele 252 showing an increased allele frequency in the U pedigree and allele 258 showing an increased frequency in the non-Umatac samples. Subsequent follow-up genotyping (see what follows) also identified evidence for the association with nearby marker D12S1048, providing evidence for association from the relationship-corrected χ2 test (P = 0.055, 3rd percentile of tests), and a significant FDR (q = 0.015). Finally, D12S79 gave a nominally significant FDR of 0.05.
Table 4.
Association of chromosome 12 markers in affected versus unaffected subjects
| Markera | Position | Nominal P-valueb | Corrected P-valuec | Percentiled | FDR q-value |
|---|---|---|---|---|---|
| D12S352 | 0.00 | 0.837 | 0.951 | 95.6 | 0.99 |
| D12S99 | 12.60 | 0.076 | 0.384 | 33.3 | 0.51 |
| D12S336 | 19.68 | 0.486 | 0.753 | 75.2 | 0.95 |
| D12S364 | 30.60 | 0.519 | 0.863 | 88.5 | 0.94 |
| D12S310 | 36.06 | 0.185 | 0.435 | 39.2 | 0.21 |
| D12S1606 | 42.74 | 0.695 | 0.840 | 90.4 | 0.45 |
| D12S1057 | 43.85 | 0.045 | 0.193 | 13.0 | 0.26 |
| D12S1617 | 44.03 | 0.006 | 0.097 | 5.1 | 0.0058 |
| D12S1596 | 45.95 | 0.708 | 0.884 | 90.0 | 0.99 |
| D12S1640 | 48.70 | 0.245 | 0.482 | 43.4 | 0.51 |
| D12S345 | 53.09 | 0.120 | 0.429 | 38.4 | 0.49 |
| D12S2080 | 54.09 | 0.238 | 0.457 | 40.9 | 0.61 |
| D12S1048 | 56.38 | 0.004 | 0.055 | 2.7 | 0.015 |
| D12S85 | 61.34 | 0.064 | 0.316 | 26.5 | 0.40 |
| D12S368 | 66.03 | 0.443 | 0.705 | 70.3 | 0.63 |
| D12S83 | 75.17 | 0.020 | 0.167 | 11.3 | 0.21 |
| D12S326 | 86.40 | 0.025 | 0.150 | 9.8 | 0.16 |
| D12S351 | 95.56 | 0.972 | 0.991 | 99.8 | 0.99 |
| D12S346 | 104.65 | 0.291 | 0.635 | 61.3 | 0.65 |
| D12S78 | 111.87 | 0.792 | 0.947 | 95.3 | 0.94 |
| D12S1646 | 121.84 | 0.611 | 0.793 | 80.9 | 0.99 |
| D12S79 | 125.31 | 0.068 | 0.341 | 29.4 | 0.05 |
| D12S86 | 134.54 | 0.157 | 0.601 | 57.1 | 0.77 |
| D12S324 | 147.17 | 0.025 | 0.139 | 9.3 | 0.089 |
| D12S1659 | 155.94 | 0.962 | 0.991 | 100.0 | 0.99 |
| D12S1723 | 164.63 | 0.776 | 0.933 | 94.6 | 0.96 |
aFollow-up markers in italics.
bNo correction for relationship information.
cCorrected for relationship information.
dSample percentile of resulting relationship-corrected χ2 test statistic.
Table 5.
D12S1617 allele frequencies
| Alleles | Full sample |
Unrelated |
U Pedigree |
S Pedigree | |||
|---|---|---|---|---|---|---|---|
| Unaffected (206), n (%) | Affected (358), n (%) | Controls (176), n (%) | Cases (76), n (%) | Unaffected (30), n (%) | Affected (40), n (%) | Affected (42), n (%) | |
| 248 | 1 (0.5) | 0 (0) | 1 (1) | 0 (0) | |||
| 250 | 6 (3) | 4 (1) | 6 (3) | 3 (1) | 0 (0.0) | 1 (2.5) | |
| 252 | 17 (8) | 48 (13) | 11 (6) | 30 (11) | 6 (20.0) | 15 (37.5) | 3 (7.1) |
| 254 | 0 (0) | 1 (0.3) | 0 (0) | 1 (0) | |||
| 256 | 13 (6) | 8 (2) | 13 (7) | 7 (3) | 1 (2.4) | ||
| 258 | 58 (28) | 121 (34) | 51 (29) | 94 (34) | 7 (23.3) | 10 (25.0) | 17 (40.5) |
| 260 | 18 (9) | 46 (13) | 13 (7) | 38 (14) | 5 (16.7) | 4 (10.0) | 4 (9.5) |
| 262 | 38 (18) | 50 (14) | 35 (20) | 38 (14) | 3 (10.0) | 2 (5.0) | 10 (23.8) |
| 264 | 36 (18) | 64 (18) | 30 (17) | 51 (18) | 6 (20.0) | 7 (17.5) | 6 (14.3) |
| 266 | 7 (3) | 4 (1) | 7 (4) | 4 (1) | |||
| 268 | 9 (4) | 7 (2) | 6 (3) | 5 (2) | 3 (10.0) | 1 (2.5) | 1 (2.4) |
| 270 | 1 (0.5) | 4 (1) | 1 (1) | 4 (1) | |||
| 272 | 2 (1) | 1 (0.3) | 2 (1) | 1 (0) | |||
n = number of alleles.
Follow-up linkage analysis on chromosome 12
As is typical of follow-up markers, heterozygosity (h) was lower than that of the genome scan markers. For all of the six follow-up markers, h was at or below the median (h = 0.79) of a typical Caucasian sample. Although the overall heterozygosity of the genome scan markers in this sample was also less than in a Caucasian sample—the median h = 0.75 in this sample is the lowest quartile in a Caucasian sample—five of the six follow-up markers had h below the median in this sample, and four of the six markers had h < 0.69, the lowest quartile in the genome scan markers in this sample.
Multipoint linkage analyses with consecutive pairs of markers supported evidence of linkage to both regions on chr 12. Pairwise linkage analysis of markers near D12S1617 had little effect on evidence for linkage to this region (Table 6). LOD scores remained strong for two-marker linkage analyses on 12p that involved D12S1617 and the closest markers on either side: Zmax = 2.66 and 2.48 in the HS and R pedigrees, respectively, for D12S1617-D12S1596, and Zmax = 2.45 in the R pedigree for D12S1057-D12S1617, with the LOD score maximizing at the position of D12S1617 in both cases. These results are consistent with expectations from multipoint analysis with less informative flanking markers. Multipoint analysis also supported evidence of linkage to 12q, but localization of the linkage peak was slightly more variable: two-marker analysis with markers immediately surrounding D12S79 was strongest for markers D12S79-D12S86, with Zmax = 2.94 and 1.55 for the PQ and HS pedigrees, respectively, maximizing ∼3 cM distal to D12S79, whereas LOD score curves in analyses of D12S1646-D12S79 were lower and maximized >0.7 cM proximal to D12S79. The R subpedigree was essentially uninformative for two-marker analyses of 12q, whereas the PQ subpedigree was uninformative for two-marker analyses of 12p. Analysis of marker–marker recombination patterns identified some evidence for marker-map inconsistencies, consistent with possible distortion of the genetic map in the sample, which could be caused by biological phenomenon such as undetected genotyping error or an inversion segregating in the families (Supplementary Material).
Table 6.
Maximum LOD scores in pairwise analysis and association study results for follow-up markers on chromosome 12p
| Marker | Location | R | PQ | H | S | HS | Corrected P-value | FDR q-value |
|---|---|---|---|---|---|---|---|---|
| D12S1606 | 42.74 | 0.79 | 0.00 | 0.86 | 0.06 | 0.90 | 0.84 | 0.45 |
| D12S1057 | 43.85 | 0.02 | 0.03 | 0.00 | 0.22 | 0.00 | 0.19 | 0.26 |
| D12S1617 | 44.03 | 4.03 | 1.29 | 1.96 | 1.15 | 3.11 | 0.097 | 0.0057 |
| D12S1596 | 45.95 | 0.00 | 0.00 | 0.00 | 0.32 | 0.01 | 0.88 | 0.99 |
| D12S1640 | 48.70 | 0.50 | 0.00 | 0.34 | 0.01 | 0.22 | 0.48 | 0.51 |
| D12S345 | 53.09 | 0.00 | 0.00 | 0.00 | 0.26 | 0.14 | 0.43 | 0.45 |
| D12S2080 | 54.09 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.46 | 0.61 |
| D12S1048 | 56.38 | 0.06 | 0.00 | 1.06 | 0.50 | 1.56 | 0.055 | 0.0153 |
Italics indicate original genome scan markers. Boldface indicates markers with Zmax >1.5 in any data set.
DISCUSSION
ALS/PDC of Guam is a complex neurodegenerative trait that disproportionately affects the Chamorro people. We report significant evidence for two distinct regions with novel ALS/PDC loci on chr 12 on the basis of both genome-wide linkage analysis of a large complex pedigree and a series of smaller families, and association analysis of a population-based sample of cases and controls. We also obtained suggestive evidence of linkage to a region on chr 17 that includes MAPT. This signal on chr 17 is consistent with our previous work that showed that two sites, one in MAPT and one immediately upstream of MAPT, have independent high-risk alleles that are associated with Guam disease (47). Our study shows that ALS/PDC has a complex etiology, possibly involving multiple genes, even in this isolated population, and illustrates both the power of such samples and some of the challenges of genome scans in such ethnic isolates.
Statistically significant evidence for a novel locus for ALS/PDC on chr 12p was supported by both linkage and association analyses. Evidence for linkage to D12S1617, located at 44 cM on the Marshfield map, is robust, with evidence for linkage in all subpedigrees extracted from the large, complex U pedigree, from an independent collection of smaller families from non-Umatac villages, and under a variety of loop-breaking and intact pedigree computations. Support for a risk locus on chr 12p also derives from the combination of insensitivity of the results to most of the compromises needed to analyze the complex U pedigree coupled with positive results from information provided by both linkage and association analyses. The one exception to the robust results is the sensitivity to marker allele frequency, which was present in the genome scan for this marker only. We also found evidence of multiple disease mutation events or, more likely, either mutation at the marker or historical recombination events between the D12S1617 marker and the trait locus, since the Umatac and non-Umatac pedigrees appeared to be segregating two different alleles for D12S1617, with these alleles having inflated frequencies in the pedigrees relative to a population-based sample of controls. The introduction, through recombination, of a second associated marker allele is plausible in a rapidly growing population when the disease mutation is a few centuries old. Such situations have been observed for other disorders with <1 cM between the marker and trait locus (50). However, the existence of multiple haplotypes harboring disease mutations can substantially reduce the power of genetic studies, even in genetically homogeneous populations, when the joining connections between the alternative disease haplotypes are missing, as may be the case in this sample (38,39).
Evidence for linkage to 12q was also statistically significant. The support for linkage to D12S79, located at 125.3 cM, was modestly more sensitive to analysis conditions than for 12p. Results for 12q were less consistent across different components of the large complex pedigree than were the equivalent analyses for the 12p region, although the independent series of small families provided nearly equivalent support for linkage to both regions. In addition, there was modest evidence of linkage disequilibrium with markers in this region as measured by the FDR, and there was little sensitivity to the choice of marker allele frequency. The lack of consistency among overlapping complex pedigree components can be explained by missing key ancestral joining components, chance differences in marker information in the separate components or the effects of loop-breaking, which could either have caused inflated or deflated LOD scores.
There are several possible explanations for the presence of two regions with evidence of linkage on chr 12. The first hypothesis is the existence of two risk loci. This would be consistent with the complex etiology of ALS/PDC previously suggested by traditional segregation analyses (21–23). A second hypothesis is that there is only a single risk locus on chr 12, with the second signal representing a false-positive result caused by misspecification of some aspect of the analysis model or the data. This includes marker allele frequency misspecification because of stochastic events such as genetic drift, or the possibility of an undetected chromosomal rearrangement, which would distort the map. Inversions exist, including one that was recently reported in a single individual (51) with breakpoints located near the two markers with strongest evidence for linkage in our sample. However, we were unable to obtain a high-quality karyotype required to determine whether there was cytological evidence for an inversion. Our marker–marker linkage analyses gave some evidence of map discrepancies that could indicate such an event in our data, although again the complexity of the pedigree limited our ability to carry out the computations needed to fully use the available information. Finally, the need for pedigree simplification could have had different effects at different markers, perhaps producing two apparent peaks. Although analysis of the U pedigree in its entirety could help resolve localization of the trait locus, such an analysis is currently infeasible with any existing software.
Two of the chromosomal regions implicated harbor-known loci for related neurodegenerative diseases. Mutations in LRRK2 [leucine-rich repeat kinase 2 (MIM 609007)] on 12q12 cause autosomal dominant parkinsonism (52,53) and sporadic Parkinson disease (54,55). D12S1617, the marker in our initial analyses for which we found the strongest evidence of linkage, is located only ∼12 cM away from LRRK2. D12S1048, which showed significant evidence of association and weak evidence of linkage is located ∼263 kb closer to LRRK2, making LRRK2 a possible candidate gene for ALS/PDC of Guam. The peripherin gene [PRPH (MIM 170710)] in this region is also implicated as a gene for ALS in humans (56,57) and in mice (58). Finally, there are reports of late-onset familial Alzheimer disease mapping to chr 12p (59,60) in a region that spans that implicated here, suggesting that this region may be important for taupathies. To our knowledge, there are no known loci for disorders related to ALS/PDC in the 12q region of interest. Mutations in MAPT and nearby progranulin [PGRN (MIM 138945)] on chr 17q21 have been associated with frontotemporal dementia and parkinsonism (61–63), and SNPs in this region show association with ALS/PDC of Guam (47). However, we sequenced the PGRN gene in Guam ALS and PDC subjects and the results were negative (data not shown). Although our results on 17q place the putative risk locus several centimorgans from MAPT and our results on 12p are several centimorgans from LRRK2, localization of the position of the Zmax in these data may be complicated by misspecification of unknown aspects of the data or analysis model. Therefore, both signals, on 12p and chr 17, are consistent with previous studies and lend further support to a role for LRRK2/PRPH and MAPT in mediating genetic susceptibility to ALS/PDC.
Two of our results underscore the challenges that isolated populations pose in the search for the genetic basis of complex traits. First, the highly complex pedigree structures that are typical in such populations introduce computational limitations into the analysis. Despite the intrinsic information in such samples, necessary pedigree reduction may result in either a loss of power or inflated evidence for linkage, complicating the detection and interpretation of linkage signals, as we have shown. Similarly, the number of markers that can be incorporated into multipoint analysis is limited by computational feasibility, thereby restricting potential gains in linkage information and resolution from follow-up markers, which also tend to be less informative than are the genome scan markers. This computational problem is particularly challenging in studies that use SNPs instead of multiallelic markers, because of the importance, then, of multipoint analysis for obtaining high-linkage information. Multiallelic markers, therefore, remain most useful in such contexts, although emerging sequencing technologies may eventually provide alternatives through phased haplotypes that avoid the computational burden of unphased SNPs. Second, association studies in isolated populations are complicated by the difficulty of obtaining a truly unrelated sample of cases and controls, so that traditional association tests that assume independence among observations are invalid in this context. While the use of genomic controls (42,64) has been proposed to account for such cryptic relatedness, evaluation of this approach has been limited to simulated genome-wide marker data with uniform numbers of alleles and allele frequencies. We found that quantile–quantile (qq) plots of genome-wide P-values were highly useful for diagnosing cryptic relatedness in our sample, and that the estimation of k coefficients along with statistical inference of relationships provided a practical method of identifying and correcting for cryptic relationships (49). We feel that adjustment for cryptic relationships, rather than removal of samples, is likely to be preferable in the quest to maintain high power, while controlling for type I error, in studies like this one with a finite available sample size. Finally, to maximize the use of scarce samples, there may be advantages, in such populations, to combining information from both linkage and association analyses, with methods such as the weighted false discovery rate (65).
Our results suggest that ALS/PDC of Guam may be influenced by as many as three loci: two on chr 12 and one on chr 17. Other previously implicated genes or regions such as variants in TRPM7 on chr 15 (66) and regions on chr 14 and chr 20 (45) are not supported by our current results. A genome scan also cannot address the effects of possible somatic or mitochondrial mutations (67) which therefore remain possible risk modifiers. Given the Umatac pedigree structure, the evidence is most consistent with additive genetic effects on the risk scale, since the need for joint genotypes would not have the multigenerational patterns seen in the Umatac pedigree, nor would it be likely that significant evidence for linkage would be obtained with a single-locus model-based approach used here. Additional work is needed to elucidate this genetic basis, including using dense markers in the regions of interest. Ultimately, although the use of a genetic isolate such as the population here is not a panacea for dissecting the genetic basis of complex traits, the eventual elucidation of ALS/PDC mutation(s) in this population could improve our understanding of not only ALS/PDC, but also related neurodegenerative diseases such as Alzheimer disease, Parkinson disease, frontotemporal dementia, ALS and other dementias.
MATERIALS AND METHODS
Sample ascertainment and diagnostic assessment
Cases, controls and small families were ascertained from the entire Chamorro population of Guam, whereas a single, large, complex pedigree (described in what follows) was ascertained and followed over a half century from the village of Umatac. We believe that virtually all living Chamarro cases were identified and recruited into the study, with as many additional archival samples tracked down and obtained as was possible to augment the sample size, including samples that had been collected as long as 50 years ago. Subject recruitment and diagnostic assessment procedures have previously been described (16,47). Briefly, archival clinical data and DNA samples were obtained from the National Institutes of Neurological and Communicative Disorders and Stroke Intramural Research program in Guam (1956–1983), and additional subjects were recruited through a neurological screening program (1995–2006) targeting all Chamorros ≥65 years on Guam (48) and through referral from or collaboration with additional investigators elsewhere (14,16). Cases who reported a family history of ALS or PDC were later recruited into the family study. All subjects (or proxies in the case of demented patients) provided written informed consent to participate in the study, and the study was approved by Institutional Review Boards at the University of Guam, University of California-San Diego and University of Washington. The diagnosis of PDC required insidious onset and gradual progression of primary parkinsonism and dementia, either of which could be the initial feature. Controls were cognitively and neurologically normal. For analysis purposes, individuals were considered to have an affected, unaffected or unknown phenotype. Affected individuals had a diagnosis of ALS and/or PDC. Unaffected individuals did not have a neurological or neuropsychological disease at last follow-up. Individuals with unknown phenotype had no disease information or had a diagnosis of a condition with unknown relationship to ALS/PDC, i.e. Alzheimer disease (MIM 104300) or dementia.
Pedigree assembly
Large and complex pedigree
In 1962, all adults over age 20 living in Umatac had a neurologic examination, and pedigrees were assembled (25). The village of Umatac was chosen because it had the highest prevalence of ALS/PDC and its population was descended from a small number of families; thus ALS/PDC in this village could be the result of a founder mutation. Most Chamorros in Umatac were part of a single extended pedigree of more than 1100 individuals with 262 sibships, reaching back eight generations to seven founder couples. More recently, all identifiable additional cases of ALS/PDC related to this pedigree were ascertained (46) and DNA samples were obtained from both affected and unaffected consenting individuals in the most recent generations.
Simple pedigrees
We identified 11 small families, referred to as the S data set, from a population-based sample of cases not living in Umatac. These families were identified from ongoing family recruitment efforts, follow-up of decendents of an original case–control study carried out >30 years ago (13,24), and as a result of quality control analyses utilizing the available genome scan data on the case–control sample, described in what follows. The 11 families consisted of 6 affected sibships, 3 affected half-sibships and 2 three-generation families. Genealogical records confirmed the accuracy of the inferred relationships where such information was available.
Linkage analysis
Strategy and rationale
The complete Umatac pedigree has many inbreeding loops and cannot be analyzed intact by any currently available software. A necessary and relatively efficient strategy is therefore to perform an initial genome scan on simplified versions of the pedigree. Although there have been multiple suggestions for heuristic approaches to pedigree simplification (68–70), none has been shown to produce optimal results by comparison with analysis that uses the complete pedigree structure. Our goal was therefore to eliminate sufficient loops such that each resulting pedigree component could be analyzed for a full genome scan, one marker at a time, in <2 weeks of CPU time, while retaining as much pedigree information as possible to both maximize power (38,39,70) and minimize type I errors (40,41) induced by simplification. Follow-up analyses were then performed in regions of interest, using more complex pedigree structures and multiple markers to more fully utilize the available linkage information, as well as to evaluate sensitivity to the choice of model parameters. The computational demands limited the number of such follow-up studies that were feasible, especially on more complex pedigrees and with multiple markers, and prevented the use of simulation-based approaches for obtaining empirical significance levels or estimates of power across the full range of pedigree simplifications used.
Pedigree simplification
The most informative subset of the complete complex pedigree was extracted by selecting all sampled affected individuals, their sampled siblings and descendents and all ancestors needed to join these individuals. This resulted in a single pedigree, referred to as the U pedigree, with 159 members including 61 phenotyped individuals (48 affected and 13 unaffected) and 35 genotyped individuals (20 affected, 13 unaffected, 2 unknown phenotype). The structure of the U pedigree was highly complex, consisting of 5 of the original 7 founder couples in the complete Umatac pedigree, 8 generations and 24 loops (Fig. 1). We took two pragmatic approaches towards pedigree simplification. Both approaches resulted in pedigrees that had higher mean pairwise kinship coefficients within versus between simplified pedigrees: a feature that is key in such simplifications for retaining mapping power while avoiding false-positive results (69,70).
Our first approach was to eliminate the deepest ancestral generations because they introduce the greatest analytic complexity and have the smallest effect when removed (38,39,70). The advantage of this approach was that nearly all the genotyped individuals, in the most recent generations, could be preserved for analysis. Eliminating the founders and one successive generation, and pruning additional uninformative individuals, resulted in the H data set, comprised of 4 disjoint pedigree components (H1–H4) ranging in size and complexity from 7 members and no loops to 66 members and 2 loops (Table 1). The independent H and S data sets were also combined (HS data set) for analysis. The HS data set included all but three of the available genotyped affected family members. Mean kinship coefficients within the four H pedigrees ranged from 0.038 to 0.238, whereas mean kinship coefficients between members of each H pedigree and members of the other three H pedigrees were much lower, ranging from 0.004 to 0.011.
The second approach was to select a founder pair and all of their descendents in order to preserve information regarding a potential shared ancestral mutation. We eliminated a few additional connections, primarily in the most ancestral portion of the pedigree, to reduce the number of loops as needed to achieve computationally practical pedigrees. The R, P and Q subpedigrees are the three largest resulting subpedigrees (Fig. 1), but are not disjoint components so that the sum of their membership exceeds the total size of the U pedigree. The R subpedigree is the largest subpedigree, descended from a single pair of founders with 93 members and 7 loops. We broke five loops and retained two loops in the R subpedigree to make genome-wide linkage analysis feasible. The P and Q subpedigrees are individually smaller than the R subpedigree and were combined (PQ subpedigree) for analysis, yielding a pedigree with 107 members and 7 loops. Four loops were broken, and three were retained for the genome scan with the PQ subpedigree. Mean kinship coefficients within the R and PQ pedigrees were 0.035 and 0.027, respectively, whereas they were 0.009 and 0.013, respectively, for kinship coefficients between members of each of these pedigrees and the rest of the U pedigree.
Trait and marker model in linkage analysis
We assumed an autosomal dominant mode of inheritance, disease allele frequency of 1%, age-dependent penetrances and a constant sporadic disease rate of 0.005 for the low-risk genotype in parametric linkage analysis. This model was supported by previous segregation analyses (21,25), by the transmission across multiple generations for what is ordinarily a rare disease and by confinement of affected individuals to a relatively small component of the entire extended Umatac pedigree. Penetrances for the high-risk genotypes in six liability classes (≤30, 31–40, 41–50, 51–60, 61–70 and >70 years) were calculated using a cumulative normal distribution in unaffected individuals and normal probability density function in affected individuals (71), with a mean of 49.8 years and standard deviation of 10.1 years. These parameters were obtained from the 61 affected individuals with available age-of-onset data in the full 1169-member Umatac pedigree. Individuals with missing age and unknown disease status were assigned to the lowest liability class, with a penetrance of 0.007 for disease-allele carriers. All 13 unaffected individuals had age data; there were 20/48 affected individuals with missing age at onset who were assigned to the 41–50 liability class with a penetrance of 0.247.
Marker allele frequencies were estimated from an external sample of 88 Chamorro controls for use in linkage analyses of U pedigree components, and from the full population-based sample of 236 Chamorros (see case–control sample) in linkage analyses of the S families. Marker alleles present in the U pedigree but not the controls were assigned the U pedigree allele frequency, and frequencies were scaled to sum to 1. For multipoint analysis, the Haldane map function was used, but all results are presented on a Kosambi map obtained from the Marshfield map (http://research.marshfieldclinic.org/genetics/home/index.asp) (72).
Linkage analysis methods
The initial genome scan was performed using pairwise parametric LOD score analysis with FASTLINK version 4.1P (73). Single-marker analysis has the important advantages of being robust to marker map misspecification and trait model misspecification for linkage detection (74,75) even when the disease is influenced by more than one locus (76). It was also computationally much more feasible on a genome-wide scale in these data.
Regions with suggestive (Zmax > 2) single-marker results were followed up with multipoint LOD score linkage analysis. Although multipoint analysis can potentially increase the available mapping information, it has the disadvantage of being sensitive to both model (77) and map misspecification (78–80). In order to increase robustness to map misspecification and to decrease computational burden, we performed multipoint analysis using only pairs of sequential markers across chromosomal regions of interest, which allows model misspecification to be absorbed into maximization of the LOD score in the flanking intervals (77).
To allow additional practical multipoint computation in key regions on these complex pedigrees, we used the MCMC parametric linkage analysis program lm_bayes (81) from the MORGAN v.2.8.1 software package (http://www.stat.washington.edu/thompson/Genepi/pangaea.shtml). MCMC expands the situations where linkage analysis is computationally practical, and the implementation in MORGAN works well under a wide variety of conditions (82). The program lm_bayes utilizes both the phenotype and marker data in MCMC sampling, which can result in better LOD score estimates for traits with high penetrance compared with MCMC procedures such as lm_markers in the MORGAN package and Simwalk2 (83,84) that only utilize the marker data (81). An MCMC-based approach was necessary because of the computational challenges: exact computation with a single two-marker analysis took approximately 60 CPU-days on the R pedigree with two loops, and was therefore not further pursued. For two-marker analyses in the PQ and H pedigrees with up to three loops, we performed five replicate scans using different random number seeds, a main run length of 1 million iterations, Rao-Blackwellized estimates of the LOD score and the default settings for other parameters; this produced reliable LOD score estimates (SD <0.12 LOD units) and took <1 CPU-day for each two-marker scan on a 1.8 GHz AMD Opteron processor. Because of the computational challenges even with MCMC methods, we limited multipoint analysis only to regions with an initial LOD score >3.
For single-marker analysis in pedigrees with seven loops, we took additional steps to shorten the MCMC analysis to ∼1 week on a single processor. We set the locus:meiosis sampling probability ratio to perform an average of only 50 locus samples in the main run, and we eliminated the preliminary run, used to set the ‘pseudopriors’ or sampling probabilities at each recombination fraction, by instead using the exact LOD scores in the same pedigrees with four to five loops to set the pseudopriors. Pseudopriors were calculated by inverting the exact likelihood ratios at each position and scaling the resulting probabilities to sum to one. To evaluate MCMC performance, we performed six replicate scans using different seeds and a main run length of either 500 000 or 1 million iterations.
Empirical significance and sensitivity analyses
We wrote a customized Perl script to carry out simulations, when possible, in order to evaluate statistical significance of LOD scores, including the effects of simplifying and partitioning the large and complex U pedigree. For computational reasons, this was possible only for the H and S (or HS) and two-loop version of the R pedigree components. Even the simplest three-loop version of the PQ component would have required >400 CPU-days to complete an equivalent simulation, as would more complex versions of the PQ and R pedigrees (Table 3), with more complex versions of these pedigree components requiring up to several CPU-years for such analyses. We used Genedrop from the MORGAN v.2.8.1 software package to generate simulated marker data sets under the null hypothesis of no linkage. We simulated marker data on the complete, complex, Umatac pedigree, basing the simulation on the sample allele frequencies and assumed marker map. We followed this with linkage analysis, using the real trait data and simplified pedigree structures as used in the original data analysis, and computed the empirical P-value as the fraction of data sets, out of 10 000 replicates, in which the LOD score was at least as high as that observed for the real data.
We evaluated the sensitivity of our analysis in key regions to several factors. A higher disease allele frequency of 5% was used in the ALS/PDC analysis to accommodate the possibilities of multiple disease alleles at the trait locus or that a key pedigree loop might have been broken. Results using Chamorro marker allele frequencies derived from 140 cases versus 88 controls were compared. The impact of breaking loops was evaluated by carrying out computations with additional loops, to the extent possible.
We separately analyzed ALS and PDC as distinct traits to determine whether separate linkage peaks were explained by distinct loci for the two phenotypes. Individuals with a diagnosis of PDC were considered to have unknown phenotype in the ALS analysis, and vice versa; two individuals with ALS and PDC were considered to be affected in both phenotype-specific analyses. Trait model parameters were the same as for ALS/PDC, except that phenotype-specific penetrances were calculated on the basis of the age of onset distribution in 29 individuals with ALS (mean = 45.1 years, SD = 10.4 years) and 33 individuals with PDC (mean = 53.8 years, SD = 7.7 years), respectively.
Case–control analysis
We carried out a case–control analysis of the genome scan data as an alternative strategy. The justification for this was that: (i) it provides complementary information; (ii) the genome scan data were available with the analysis being computationally rapid; (iii) the small population at the end of the 18th century followed by rapid growth over the next two centuries (29,85) predicts long tracts of linkage disequilibrium complete with power >80% to detect association with genome scan STRPs at a nominal significance level of 0.001 in the available sample (86); and (iv) STRPs can have considerable power to detect association in the context of recent founder events at moderate genetic distances (86). These analyses were initially carried out on a sample of 137 putatively unrelated cases (mean age 57.9 ± 11.4 years) and 88 age-matched controls (mean age 51.87 ± 15.8 years at examination), representing the entire available sample of population-based non-Umatac cases. To achieve the maximum possible number of affected individuals that could be included, we later also included all sampled subjects from Umatac, providing, in total, 157 cases and 101 controls in the full sample, while accounting for relationships among individuals with a newly developed method of analysis as described in what follows. Finally, to achieve maximum power to detect association with this analysis, given the modest total available sample size, in addition to a standard case–control analysis, we also carried out analysis with a weighted false-discovery rate (wFDR), which increases power to detect association by incorporating weights on the basis of genomewide linkage analysis results to provide an estimate of the FDR (65).
We investigated the existence of unidentified relationships in the sample by several methods that used the genome-wide marker data. We used Relpair v.2.0.1 (http://csg.sph.umich.edu/boehnke/relpair.php) (87), with an assumption of a 0.5% genotyping error rate and estimates of marker allele frequencies from the full case–control sample. We also computed maximum likelihood estimates (88,89) of k coefficients (90), which are the multilocus probability of sharing 0, 1 or 2 (k0, k1, k2) alleles identical by descent with no restrictions on the types of relationships allowed. Finally, genealogical and clinical records were reviewed for all individuals found to be related on the basis of genome-wide marker data.
Evidence for association between case status and marker allele frequencies was evaluated with χ2 tests. Provided that the assumptions of independence among observations and HWE are satisfied, P-values are expected to have a uniform (0,1) distribution except for a few markers that are near to and in linkage disequilibrium with the disease locus. We used qq plots to assess departures of the genome-wide P-value distribution from uniformity because such departures can indicate violation of the analysis assumptions, for example, caused by the presence of cryptic relatedness or population stratification. Marked deviation from the expected null distribution of P-values was obtained even when the Umatac subjects were excluded, and led to the identification of cryptic relatedness in this sample. As a result, and also to allow the use of the maximal available sample, we developed a corrected χ2 test on the basis of estimating relatedness from genotype data (89) and correcting the test for this relatedness (91). Details of this extension and its properties are described elsewhere (49), including the information that the corrected test is slightly conservative. Finally, to maximize power to detect association and to use the joint information from both the linkage and association analyses in the interpretation of the association results, we computed q-values using the weighted false discovery rate (FDR) as described elsewhere (65). For the estimation of the FDR, we used the genome scan LOD scores for the HS pedigree configuration, since this was the only pedigree configuration that used the full sample, an exponential model for each marker, and an empirical estimate of the proportion of tests representing the null distribution (92). To evaluate evidence for Hardy–Weinberg disequilibrium (HWD) and population structure, we performed an exact test for HWD for multiallelic markers using the MCMC program hwe in the Hardy software package (http://www.stat.washington.edu/thompson/Genepi/pangaea.shtml) (93), and we used Structure (http://pritch.bsd.uchicago.edu/software.html) (94,95) to evaluate evidence for the existence of admixture or distinct subpopulations. However, the results from Structure were difficult to interpret because of sensitivity to the choice of model assumptions and prior distributions, and this approach was therefore not pursued further.
Markers and laboratory procedures
Genomic DNA was isolated from available frozen brain tissue or fresh blood samples. Genotypes were determined by PCR amplification of polymorphic loci using primers labeled with fluorescent tags. We genotyped 408 microsatellite markers, primarily from the ABI Prism Linkage mapping set, Version 2.5, HD5 (http://home.appliedbiosystems.com/): 402 genome scan markers on all subjects, and 6 additional markers on chromosome 12p on the pedigree samples. The six additional markers on 12p, only, were chosen because the markers in the genome scan panel did not provide strong additional support for a locus on 12p, beyond the initial marker providing evidence for linkage. DNA fragments were analyzed using an ABI377 or an ABI3100 DNA sequencing instrument and GeneScan and Genotyper software. Quality control of the resulting data involved validating pedigree structures, identifying and eliminating genotyping errors, mapping of markers back to somatic cell hybrids to verify chromosomal location and evaluation of consistency of the data with published maps, as described in the Supplementary Material.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was funded by the National Institutes of Health (AG14382; AG11762; AG05136; and GM46255) and by the Department of Veterans Affairs.
ACKNOWLEDGEMENTS
We wish to thank Dr Chris C. Plato, who passed away in March 2008, and whose meticulous work and generosity provided the initial Umatac pedigree, without which the current study would not have been possible. We also wish to acknowledge Hiep Nguyen for computer assistance, as well as the efforts of research assistants and clinicians who have assisted with the laboratory work and in identifying, evaluating and referring patients in Guam and elsewhere over the years.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hirano A., Krooth R.S., Lessell S., Kurland L.T. Parkinsonism-dementia complex, an endemic disease on the island of Guam. 1. Clinical features. Brain. 1961;84:642–661. doi: 10.1093/brain/84.4.642. [DOI] [PubMed] [Google Scholar]
- 2.Koerner D.R. Amyotrophic lateral sclerosis on Guam—a clinical study and review of the literature. Ann. Intern. Med. 1952;37:1204–1220. doi: 10.7326/0003-4819-37-6-1204. [DOI] [PubMed] [Google Scholar]
- 3.Kurland L.T., Mulder D.W. Epidemiologic investigations of amyotrophic lateral sclerosis. 1. Preliminary report on geographic distribution, with special reference to the Mariana Islands, including clinical and pathologic observations. Neurology. 1954;4:438–448. doi: 10.1212/wnl.4.6.438. [DOI] [PubMed] [Google Scholar]
- 4.Kuzuhara S., Kokubo Y. Atypical parkinsonism of Japan: amyotrophic lateral sclerosis-parkinsonism-dementia complex of the Kii peninsula of Japan (Muro disease): an update. Mov. Disord. 2005;20:S108–S113. doi: 10.1002/mds.20548. [DOI] [PubMed] [Google Scholar]
- 5.Mimuro M., Kokubo Y., Kuzuhara S. Similar topographical distribution of neurofibrillary tangles in amyotrophic lateral sclerosis and parkinsonism-dementia complex in people living in the Kii peninsula of Japan suggests a single tauopathy. Acta Neuropathol. (Berl.) 2007;113:653–658. doi: 10.1007/s00401-007-0197-6. [DOI] [PubMed] [Google Scholar]
- 6.Gajdusek D.C., Salazar A.M. Amyotrophic lateral sclerosis and parkinsonian syndromes in high-incidence among the Auyu and Jakai people of west New-Guinea. Neurology. 1982;32:107–126. doi: 10.1212/wnl.32.2.107. [DOI] [PubMed] [Google Scholar]
- 7.Hirano A., Malamud N., Kurland L.T. Parkinsonism-dementia complex, an endemic disease on the island of Guam. 2. Pathological features. Brain. 1961;84:662–679. doi: 10.1093/brain/84.4.662. [DOI] [PubMed] [Google Scholar]
- 8.Hirano A., Malamud N., Elizan T.S., Kurland L.T. Amyotrophic lateral sclerosis and parkinsonism-dementia complex on Guam—further pathologic studies. Arch. Neurol. 1966;15:35–51. doi: 10.1001/archneur.1966.00470130039004. [DOI] [PubMed] [Google Scholar]
- 9.Hof P.R., Nimchinsky E.A., Bueescherrer V., Buee L., Nasrallah J., Hottinger A.F., Purohit D.P., Loerzel A.J., Steele J.C., Delacourte A., et al. Amyotrophic-lateral-sclerosis parkinsonism-dementia complex of Guam—quantitative neuropathology, immunohistochemical analysis of neuronal vulnerability, and comparison with related neurodegenerative disorders. Acta Neuropathol. (Berl.) 1994;88:397–404. doi: 10.1007/BF00389490. [DOI] [PubMed] [Google Scholar]
- 10.Hof P.R., Perl D.P. Neurofibrillary tangles in the primary motor cortex in Guamanian amyotrophic lateral sclerosis/parkinsonism-dementia complex. Neurosci. Lett. 2002;328:294–298. doi: 10.1016/s0304-3940(02)00523-2. [DOI] [PubMed] [Google Scholar]
- 11.Hirano A., Arumugas N., Zimmerma H.M. Amyotrophic lateral sclerosis—a comparison of Guam and classical cases. Arch. Neurol. 1967;16:357–363. doi: 10.1001/archneur.1967.00470220021003. [DOI] [PubMed] [Google Scholar]
- 12.Lessell S., Torres J., Kurland L.T., Hirano A. Parkinsonism-dementia complex—epidemiological considerations in Chamorros of Mariana Islands and California. Arch. Neurol. 1962;7:377–385. doi: 10.1001/archneur.1962.04210050013003. [DOI] [PubMed] [Google Scholar]
- 13.Reed D.M., Brody J.A. Amyotrophic lateral sclerosis and parkinsonism-dementia on Guam, 1945–1972. 1. Descriptive epidemiology. Am. J. Epidemiol. 1975;101:287–301. doi: 10.1093/oxfordjournals.aje.a112097. [DOI] [PubMed] [Google Scholar]
- 14.Plato C.C., Garruto R.M., Galasko D., Craig U.K., Plato M., Gamst A., Torres J.M., Wiederholt W. Amyotrophic lateral sclerosis and parkinsonism-dementia complex of Guam: changing incidence rates during the past 60 years. Am. J. Epidemiol. 2003;157:149–157. doi: 10.1093/aje/kwf175. [DOI] [PubMed] [Google Scholar]
- 15.Garruto R.M., Yanagihara R., Gajdusek D.C. Disappearance of high-incidence amyotrophic lateral sclerosis and parkinsonism-dementia on Guam. Neurology. 1985;35:193–198. doi: 10.1212/wnl.35.2.193. [DOI] [PubMed] [Google Scholar]
- 16.Galasko D., Salmon D.P., Craig U.K., Thal L.J., Schellenberg G., Wiederholt W. Clinical features and changing patterns of neurodegenerative disorders on Guam, 1997–2000. Neurology. 2002;58:90–97. doi: 10.1212/wnl.58.1.90. [DOI] [PubMed] [Google Scholar]
- 17.Gibbs C.J., Jr, Gajdusek D.C. Amyotrophic lateral sclerosis, Parkinson's disease, and the amyotrophic lateral sclerosis-Parkinsonism-dementia complex on Guam: a review and summary of attempts to demonstrate infection as the aetiology. J. Clin. Pathol. Suppl. (R Coll Pathol) 1972;6:132–140. [PMC free article] [PubMed] [Google Scholar]
- 18.Spencer P.S., Nunn P.B., Hugon J., Ludolph A.C., Ross S.M., Roy D.N., Robertson R.C. Guam amyotrophic-lateral-sclerosis parkinsonism dementia linked to a plant excitant neurotoxin. Science. 1987;237:517–522. doi: 10.1126/science.3603037. [DOI] [PubMed] [Google Scholar]
- 19.Garruto R.M., Fukatsu R., Yanagihara R., Gajdusek D.C., Hook G., Fiori C.E. Imaging of calcium and aluminum in neurofibrillary tangle-bearing neurons in parkinsonism-dementia of Guam. Proc. Natl Acad. Sci. USA. 1984;81:1875–1879. doi: 10.1073/pnas.81.6.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanagihara R., Garruto R.M., Gajdusek D.C., Tomita A., Uchikawa T., Konagaya Y., Chen K.M., Sobue I., Plato C.C., Gibbs C.J. Calcium and vitamin-D metabolism in Guamanian Chamorros with amyotrophic lateral sclerosis and parkinsonism-dementia. Ann. Neurol. 1984;15:42–48. doi: 10.1002/ana.410150108. [DOI] [PubMed] [Google Scholar]
- 21.Baileywilson J.E., Plato C.C., Elston R.C., Garruto R.M. Potential role of an additive genetic component in the cause of amyotrophic-lateral-sclerosis and parkinsonism-dementia in the Western Pacific. Am. J. Med. Genet. 1993;45:68–76. doi: 10.1002/ajmg.1320450118. [DOI] [PubMed] [Google Scholar]
- 22.Plato C.C., Reed D.M., Elizan T.S., Kurland L.T. Amyotrophic lateral sclerosis/parkinsonism-dementia complex of Guam. 4. Familial and genetic investigations. Am. J. Hum. Genet. 1967;19:617–632. [PMC free article] [PubMed] [Google Scholar]
- 23.Reed D.M., Torres J.M., Brody J.A. Amyotrophic lateral sclerosis and parkinsonism-dementia on Guam, 1945–1972. 2. Familial and genetic studies. Am. J. Epidemiol. 1975;101:302–310. doi: 10.1093/oxfordjournals.aje.a112098. [DOI] [PubMed] [Google Scholar]
- 24.Plato C.C., Galasko D., Garruto R.M., Plato M., Gamst A., Craig U.K., Torres J.M., Wiederholt W. ALS and PDC of Guam—forty-year follow-up. Neurology. 2002;58:765–773. doi: 10.1212/wnl.58.5.765. [DOI] [PubMed] [Google Scholar]
- 25.Plato C.C., Cruz M.T., Kurland L.T. Amyotrophic lateral sclerosis-parkinsonism dementia complex of Guam: further genetic investigations. Am. J. Hum. Genet. 1969;21:133–141. [PMC free article] [PubMed] [Google Scholar]
- 26.Vilar M., Lynch D., Chan C., Reiff D., Garruto R.M., Lum J.K. Peopling of the Marianas Islands: a mtDNA perspective. Am. J. Hum. Biol. 2008;20:237–237. doi: 10.1002/ajhb.22349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lum J.K., Cann R.L. mtDNA lineage analyses: origins and migrations of Micronesians and Polynesians. Am. J. Phys. Anthropol. 2000;113:151–168. doi: 10.1002/1096-8644(200010)113:2<151::AID-AJPA2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 28.Rogers R.F. Destiny's Landfall: A History of Guam. Honolulu: University of Hawaii Press; 1995. [Google Scholar]
- 29.Underwood J.H. The native origins of the neo-Chamorros of the Mariana Islands. Micronesia. 1976;12:203–209. [Google Scholar]
- 30.Yanagihara R.T., Garruto R.M., Gajdusek D.C. Epidemiological surveillance of amyotrophic lateral sclerosis and parkinsonism-dementia in the Commonwealth of the Northern Mariana Islands. Ann. Neurol. 1983;13:79–86. doi: 10.1002/ana.410130117. [DOI] [PubMed] [Google Scholar]
- 31.Bourgain C., Genin E. Complex trait mapping in isolated populations: are specific statistical methods required? Eur. J. Hum. Genet. 2005;13:698–706. doi: 10.1038/sj.ejhg.5201400. [DOI] [PubMed] [Google Scholar]
- 32.Peltonen L., Palotie A., Lange K. Use of population isolates for mapping complex traits. Nat. Rev. Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 33.Service S., DeYoung J., Karayiorgou M., Roos J.L., Pretorious H., Bedoya G., Ospina J., Ruiz-Linares A., Macedo A., Palha J.A., et al. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat. Genet. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- 34.Venken T., Del-Favero J. Chasing genes for mood disorders and schizophrenia in genetically isolated populations. Hum. Mutat. 2007;28:1156–1170. doi: 10.1002/humu.20582. [DOI] [PubMed] [Google Scholar]
- 35.Greenberg D.A., Abreu P., Hodge S.E. The power to detect linkage in complex disease by means of simple LOD-score analyses. Am. J. Hum. Genet. 1998;63:870–879. doi: 10.1086/301997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu F., Arias-Vasquez A., Sleegers K., Aulchenko Y.S., Kayser M., Sanchez-Juan P., Feng B.J., Bertoli-Avella A.M., van Swieten J., Axenovich T.I., et al. A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am. J. Hum. Genet. 2007;81:17–31. doi: 10.1086/518720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wijsman E.M., Amos C.I. Genetic analysis of simulated oligogenic traits in nuclear and extended pedigrees: summary of GAW10 contributions. Genet. Epidemiol. 1997;14:719–735. doi: 10.1002/(SICI)1098-2272(1997)14:6<719::AID-GEPI28>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 38.Chapman N., Leutenegger A.-L., Badzioch M., Bogdan M., Conlon E., Daw E., Gagnon F., Li N., Maia J., Wijsman E., et al. The importance of connections: joining components of the Hutterite pedigree. Genet. Epidemiol. 2001;21:S230–S235. doi: 10.1002/gepi.2001.21.s1.s230. [DOI] [PubMed] [Google Scholar]
- 39.Dyer T., Blangero J., Williams J., Göring H., Mahaney M. The effect of pedigree complexity on quantitative trait linkage analysis. Genet. Epidemiol. 2001;21:S236–S243. doi: 10.1002/gepi.2001.21.s1.s236. [DOI] [PubMed] [Google Scholar]
- 40.Leutenegger A.L., Genin E., Thompson E.A., Clerget-Darpoux F. Impact of parental relationships in maximum LOD score affected sib-pair method. Genet. Epidemiol. 2002;23:413–425. doi: 10.1002/gepi.10190. [DOI] [PubMed] [Google Scholar]
- 41.Genin E., ClergetDarpoux F. Consanguinity and the sib-pair method: an approach using identity by descent between and within individuals. Am. J. Hum. Genet. 1996;59:1149–1162. [PMC free article] [PubMed] [Google Scholar]
- 42.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 43.Voight B.F., Pritchard J.K. Confounding from cryptic relatedness in case-control association studies. PLoS Genet. 2005;1:302–311. doi: 10.1371/journal.pgen.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chanock S.J., Manolio T., Boehnke M., Boerwinkle E., Hunter D.J., Thomas G., Hirschhorn J.N., Abecasis G., Altshuler D., Bailey-Wilson J.E., et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 45.Morris H.R., Steele J.C., Crook R., Wavrant-De Vrieze F., Onstead-Cardinale L., Gwinn-Hardy K., Wood N.W., Farrer M., Lees A.J., McGeer P.L., et al. Genome-wide analysis of the Parkinsonism-dementia complex of Guam. Arch. Neurol. 2004;61:1889–1897. doi: 10.1001/archneur.61.12.1889. [DOI] [PubMed] [Google Scholar]
- 46.Poorkaj P., Tsuang D., Wijsman E., Steinbart E., Garruto R.M., Craig U.K., Chapman N.H., Anderson L., Bird T.D., Plato C.C., et al. TAU as a susceptibility gene for amyotropic lateral sclerosis-parkinsonism dementia complex of Guam. Arch. Neurol. 2001;58:1871–1878. doi: 10.1001/archneur.58.11.1871. [DOI] [PubMed] [Google Scholar]
- 47.Sundar P.D., Yu C.E., Sieh W., Steinbart E., Garruto R.M., Oyanagi K., Craig U.K., Bird T.D., Wijsman E.M., Galasko D.R., et al. Two sites in the MAPT region confer genetic risk for Guam ALS/PDC and dementia. Hum. Mol. Genet. 2007;16:295–306. doi: 10.1093/hmg/ddl463. [DOI] [PubMed] [Google Scholar]
- 48.Galasko D., Salmon D., Gamst A., Olichney J., Thal L.J., Silbert L., Kaye J., Brooks P., Adonay R., Craig U.K., et al. Prevalence of dementia in Chamorros on Guam—relationship to age, gender, education, and APOE. Neurology. 2007;68:1772–1781. doi: 10.1212/01.wnl.0000262028.16738.64. [DOI] [PubMed] [Google Scholar]
- 49.Choi Y., Wijsman E.M., Weir B.S. Case-control association testing in the presence of unknown relationships. Genet. Epidemiol. 2009 doi: 10.1002/gepi.20418. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goddard K.A.B., Yu C.E., Oshima J., Miki T., Nakura J., Piussan C., Martin G.M., Schellenberg G.D., Wijsman E.M. Toward localization of the Werner Syndrome gene by linkage disequilibrium and ancestral haplotyping: lessons learned from analysis of 35 chromosome 8p11:1–21.1 markers. Am. J. Hum. Genet. 1996;58:1286–1302. [PMC free article] [PubMed] [Google Scholar]
- 51.Korbel J.O., Urban A.E., Affourtit J.P., Godwin B., Grubert F., Simons J.F., Kim P.M., Palejev D., Carriero N.J., Du L., et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paisan-Ruiz C., Jain S., Evans E.W., Gilks W.P., Simon J., van der Brug M., de Munain A.L., Aparicio S., Gil A.M., Khan N., et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 53.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R.J., Calne D.B., et al. Mutations in LRRK2 cause autosomal-dominant Parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 54.Farrer L.A., Cupples L.A., Haines J.L., Hyman B., Kukull W.A., Mayeux R., Myers R.H., Pericak-Vance M.A., Risch N., van Duijn C.M. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. J. Am. Med. Assoc. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 55.Lesage S., Durr A., Tazir M., Lohmann E., Leutenegger A.L., Janin S., Pollak P., Brice A. LRRK2 G2019S as a cause of Parkinson's disease in North African Arabs. N. Engl. J. Med. 2006;354:422–423. doi: 10.1056/NEJMc055540. [DOI] [PubMed] [Google Scholar]
- 56.Gros-Louis F., Lariviere R., Gowing G., Laurent S., Camu W., Bouchard J.P., Meininger V., Rouleau G.A., Julien J.P. A frameshift deletion in peripherin gene associated with amyotrophic lateral sclerosis. J. Biol. Chem. 2004;279:45951–45956. doi: 10.1074/jbc.M408139200. [DOI] [PubMed] [Google Scholar]
- 57.Leung C.L., He C.Z., Kaufmann P., Chin S.S., Naini A., Liem R.K.H., Mitsumoto H., Hays A.P. A pathogenic peripherin gene mutation in a patient with amyotrophic lateral sclerosis. Brain Pathol. 2004;14:290–296. doi: 10.1111/j.1750-3639.2004.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robertson J., Doroudchi M.M., Nguyen M.D., Durham H.D., Strong M.J., Shaw G., Julien J.P., Mushynski W.E. A neurotoxic peripherin splice variant in a mouse model of ALS. J. Cell Biol. 2003;160:939–949. doi: 10.1083/jcb.200205027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pericak-Vance M.A., Bass M.P., Yamaoka L.H., Gaskell P.C., Scott W.K., Terwedow H.A., Menold M.M., Conneally P.M., Small G.W., Saunders A.M., et al. Complete genomic screen in late-onset familial Alzheimer's disease. Neurobiol. Aging. 1998;19:S39–S42. doi: 10.1016/s0197-4580(98)00037-2. [DOI] [PubMed] [Google Scholar]
- 60.Ertekin-Taner N. Genetics of Alzheimer's disease: a centennial review. Neurol. Clin. 2007;25:611–667. doi: 10.1016/j.ncl.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clark L.N., Poorkaj P., Wszolek Z., Geschwind D.H., Nasreddine Z.S., Miller B., Li D., Payami H., Awert F., Markopoulou K., et al. Pathogenic implications of mutations in the tau gene in pallido-ponto-nigral degeneration and related neurodegenerative disorders linked to chromosome 17. Proc. Natl Acad. Sci. USA. 1998;95:13103–13107. doi: 10.1073/pnas.95.22.13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baker M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A.D., Rollinson S., et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 63.Hutton M., Lendon C.L., Rizzu P., Baker M., Froelich S., Houlden H., Pickering-Brown S., Chakraverty S., Isaacs A., Grover A., et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 64.Bacanu S.A., Devlin B., Roeder K. Association studies for quantitative traits in structured populations. Genet. Epidemiol. 2002;22:78–93. doi: 10.1002/gepi.1045. [DOI] [PubMed] [Google Scholar]
- 65.Roeder K., Bacanu S.A., Wasserman L., Devlin B. Using linkage genome scans to improve power of association in genome scans. Am. J. Hum. Genet. 2006;78:243–252. doi: 10.1086/500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hermosura M.C., Nayakanti H., Dorovkov M.V., Calderon F.R., Ryazanov A.G., Haymer D.S., Garruto R.M. A TRPM7 variant shows altered sensitivity to magnesium that may contribute to the pathogenesis of two Guamanian neurodegenerative disorders. Proc. Natl Acad. Sci. USA. 2005;102:11510–11515. doi: 10.1073/pnas.0505149102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lynch D., Wanglund C., Spathis R., Chan C.W., Reiff D.M., Lum J.K., Garruto R.M. The contribution of mitochondrial dysfunction to a gene-environment model of Guamanian ALS and PD. Mitochondrion. 2008;8:109–116. doi: 10.1016/j.mito.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 68.Falchi M., Forabosco P., Mocci E., Borlino C.C., Picciau A., Virdis E., Persico I., Parracciani D., Angius A., Pirastu M. A genomewide search using an original pairwise sampling approach for large genealogies identifies a new locus for total and low-density lipoprotein cholesterol in two genetically differentiated isolates of Sardinia. Am. J. Hum. Genet. 2004;75:1015–1031. doi: 10.1086/426155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Axenovich T.I., Zorkoltseva I.V., Liu F., Kirichenko A.V., Aulchenko Y.S. Breaking loops in large complex pedigrees. Hum. Hered. 2008;65:57–65. doi: 10.1159/000108937. [DOI] [PubMed] [Google Scholar]
- 70.Liu F., Kirichenko A., Axenovich T.I., van Duijn C.M., Aulchenko Y.S. An approach for cutting large and complex pedigrees for linkage analysis. Eur. J. Hum. Genet. 2008;16:854–860. doi: 10.1038/ejhg.2008.24. [DOI] [PubMed] [Google Scholar]
- 71.Margaritte P., Bonaiti-Pellie C., King M.C., Clerget-Darpoux F. Linkage of familial breast cancer to chromosome 17q21 may not be restricted to early-onset disease. Am. J. Hum. Genet. 1992;50:1231–1234. [PMC free article] [PubMed] [Google Scholar]
- 72.Broman K.W., Murray J.C., Sheffield V.C., White R.L., Weber J.L. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cottingham R.W., Idury R.M., Schaffer A.A. Faster sequential genetic linkage computations. Am. J. Hum. Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 74.Clerget-Darpoux F., Bonaiti-Pellie C., Hochez J. Effects of misspecifying genetic parameters in LOD score analysis. Biometrics. 1986;42:393–399. [PubMed] [Google Scholar]
- 75.Ott J. Linkage analysis with misclassification at one locus. Clin. Genet. 1977;12:119–124. doi: 10.1111/j.1399-0004.1977.tb00913.x. [DOI] [PubMed] [Google Scholar]
- 76.Greenberg D.A., Hodge S.E. Linkage analysis under ‘random’ and ‘genetic’ reduced penetrance. Genet. Epidemiol. 1989;6:259–264. doi: 10.1002/gepi.1370060145. [DOI] [PubMed] [Google Scholar]
- 77.Risch N., Giuffra L. Model misspecification and multipoint linkage analysis. Hum. Hered. 1992;42:77–92. doi: 10.1159/000154047. [DOI] [PubMed] [Google Scholar]
- 78.Daw E.W., Thompson E.A., Wijsman E.M. Bias in multipoint linkage analysis arising from map misspecification. Genet. Epidemiol. 2000;19:336–380. doi: 10.1002/1098-2272(200012)19:4<366::AID-GEPI8>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 79.Fingerlin T.E., Abecasis G.R., Boehnke M. Using sex-averaged genetic maps in multipoint linkage analysis when identity-by-descent status is incompletely known. Genet. Epidemiol. 2006;30:384–396. doi: 10.1002/gepi.20151. [DOI] [PubMed] [Google Scholar]
- 80.Halpern J., Whittemore A.S. Multipoint linkage analysis. A cautionary note. Hum. Hered. 1999;49:194–196. doi: 10.1159/000022874. [DOI] [PubMed] [Google Scholar]
- 81.George A.W., Wijsman E.M., Thompson E.A. MCMC multilocus LOD scores: application of a new approach. Hum. Hered. 2005;59:98–108. doi: 10.1159/000085224. [DOI] [PubMed] [Google Scholar]
- 82.Wijsman E.M., Rothstein J.H., Thompson E.A. Multipoint linkage analysis with many multiallelic or dense diallelic markers: MCMC provides practical approaches for genome scans on general pedigrees. Am. J. Hum. Genet. 2006;79:846–858. doi: 10.1086/508472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lange K., Sobel E. A random walk method for computing genetic location scores. Am. J. Hum. Genet. 1991;49:1320–1334. [PMC free article] [PubMed] [Google Scholar]
- 84.Sobel E., Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am. J. Hum. Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 85.Garruto R.M., Gajdusek D.C., Chen K.M. Amyotrophic lateral sclerosis among Chamorro migrants from Guam. Ann. Neurol. 1980;8:612–619. doi: 10.1002/ana.410080612. [DOI] [PubMed] [Google Scholar]
- 86.Chapman N.H., Wijsman E.W. Genome screens using linkage disequilibrium tests: optimal marker characteristics and feasibility. Am. J. Hum. Genet. 1998;63:1872–1885. doi: 10.1086/302139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Epstein M.P., Duren W.L., Boehnke M. Improved inference of relationship for pairs of individuals. Am. J. Hum. Genet. 2000;67:1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Milligan B.G. Maximum-likelihood estimation of relatedness. Genetics. 2003;163:1153–1167. doi: 10.1093/genetics/163.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thompson E.A. Estimation of pairwise relationships. Ann. Hum. Genet. 1975;39:173–188. doi: 10.1111/j.1469-1809.1975.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 90.Cotterman C.W. Columbia: Ohio State University; 1940. A calculus for statistico-genetics. PhD thesis. [Google Scholar]
- 91.Bourgain C., Hoffjan S., Nicolae R., Newman D., Steiner L., Walker K., Reynolds R., Ober C., McPeek M.S. Novel case-control test in a founder population identifies P-selectin as an atopy-susceptibility locus. Am. J. Hum. Genet. 2003;73:612–626. doi: 10.1086/378208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guo S.W., Thompson E.A. Technical Report. Seattle: Department of Statistics, University of Washington; 1989. Analysis of sparse contingency tables: Monte Carlo estimation of exact P-values. [Google Scholar]
- 94.Falush D., Stephens M., Pritchard J.K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.