Abstract
Following a screen for neuromuscular mouse mutants, we identified ostes, a novel N-ethyl N-nitrosourea-induced mouse mutant with muscle atrophy. Genetic and biochemical evidence shows that upregulation of the novel, uncharacterized transient receptor potential polycystic (TRPP) channel PKD1L2 (polycystic kidney disease gene 1-like 2) underlies this disease. Ostes mice suffer from chronic neuromuscular impairments including neuromuscular junction degeneration, polyneuronal innervation and myopathy. Ectopic expression of PKD1L2 in transgenic mice reproduced the ostes myopathic changes and, indeed, caused severe muscle atrophy in Tg(Pkd1l2)/Tg(Pkd1l2) mice. Moreover, double-heterozygous mice (ostes/+, Tg(Pkd1l2)/0) suffer from myopathic changes more profound than each heterozygote, indicating positive correlation between PKD1L2 levels and disease severity. We show that, in vivo, PKD1L2 primarily associates with endogenous fatty acid synthase in normal skeletal muscle, and these proteins co-localize to costameric regions of the muscle fibre. In diseased ostes/ostes muscle, both proteins are upregulated, and ostes/ostes mice show signs of abnormal lipid metabolism. This work shows the first role for a TRPP channel in neuromuscular integrity and disease.
INTRODUCTION
Discovering the molecular mechanisms underlying neuromuscular degenerative diseases is pivotal to the development of therapies for such heterogenous disorders. Model organisms have assisted in the identification of novel neuromuscular disease genes and genetic pathways (reviewed in 1–3). We have exploited mouse mutagenesis to identify new neuromuscular mutants. In phenotype-driven screens of mutagenized mice, simple measures of muscle strength such as wheel running or grip strength have failed to consistently identify mutants that show muscular defects at the histological level. In addition, measuring progressive motor neuron loss in mutagenized animals is not a suitable protocol for large-scale screening programmes. In contrast, overt phenotypes such as muscle atrophy, tremors, gait defects and growth retardation have been used to signpost abnormal neuromuscular function in mice (4,5). Using this simple paradigm, we identified ostes in an N-ethyl N-nitrosourea (ENU) screen (6). The aim of this work was to characterize this novel neuromuscular phenotype in the mouse and to provide insights into the underlying molecular mechanism. We present evidence that the product of the genetically linked Pkd1l2 (polycystic kidney disease gene 1-like 2) gene is grossly overexpressed in ostes/ostes mice, and that ectopic expression of PKD1L2 in transgenic mice leads to similar neuromuscular features. To date, no analysis of PKD1L2 protein expression, localization or in vivo function has been done. Here, we show that PKD1L2 is the first transient receptor potential polycystic (TRPP) channel to be associated with neuromuscular function and disease. Additionally, we show that abnormal lipid metabolism in PKD1L2-dysregulated mice is a key feature of this disease.
RESULTS
Ostes mice show a complex neuromuscular phenotype
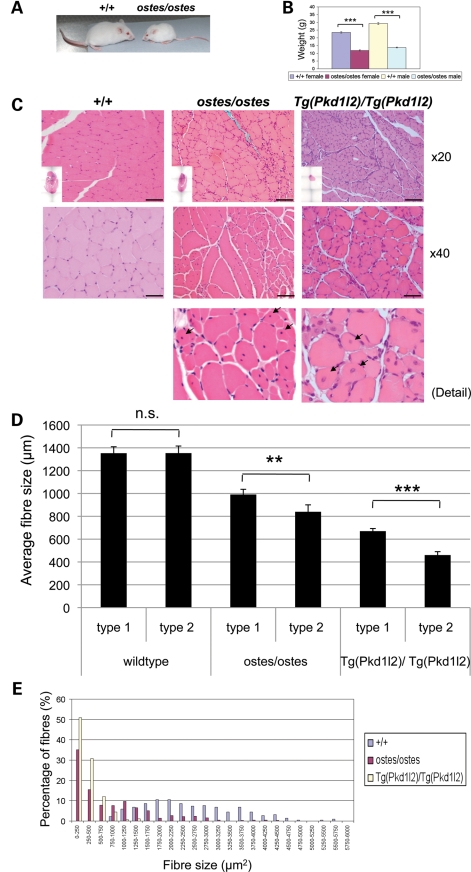
We assessed the neuromuscular system in ostes/ostes mice at three levels of analysis. Grossly, ostes/ostes mice become recognizable by their small size and reduced weight between 3 and 5 weeks of age (Fig. 1A) and remain small and light throughout life (see weights of 12-week-old mice, Fig. 1B). Ostes/ostes mice also show a moderate tremor of the hind limbs upon suspension by the tail and have a poor reaching response. Conversely, ostes/+ mice do not display any overt abnormal phenotype. Sections of 24 ostes/ostes tissues including heart did not reveal any pathology except for muscle at this level of examination (data not shown). Haematoxylin and eosin (H&E)-stained sections of hind limb muscles from ostes/ostes mice show striking fibre size asymmetry, including large compensatory fibres with internal nuclei (representative pictures and quantifications in Fig. 1C, D and E). In addition, clusters of fibres with pyknotic nuclei and scattered, small centrally nucleated fibres are present, indicating fibre atrophy and some regeneration, respectively (Fig. 1C and Supplementary Material, Fig. S1). There is no evidence of macrophage infiltration (Supplementary Material, Fig. S2). Additionally, ostes/ostes mice show widespread muscle atrophy, as indicated by the average cross-sectional area of the muscle fibres (Fig. 1D and E) and reflected in the overall muscle group size (see insets in Fig. 1C). Despite the lack of gross phenotype in ostes/+ mice, similar pathological hallmarks are evident in ostes/+ hind limb muscle, but the pathology is milder and there is no evidence of muscle atrophy (Fig. 1I and Supplementary Material, Fig. S3), indicating a semi-dominant action of the ostes mutation in muscle.
Figure 1.
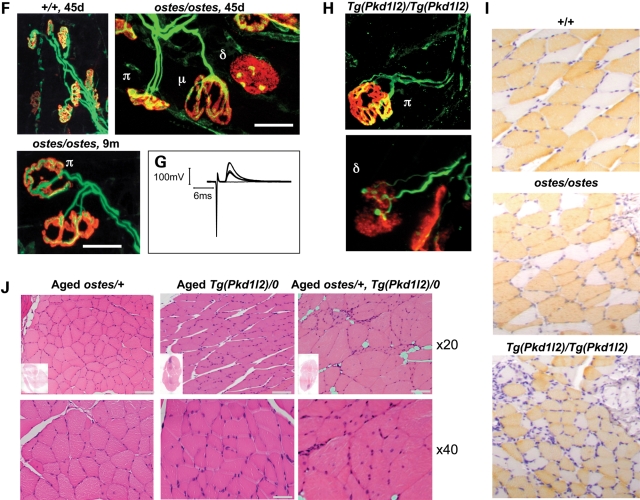
Neuromuscular phenotypes of ostes and Tg(Pkd1l2) mice. (A) Photograph of representative 5-week-old wild-type (+/+) and ostes/ostes mice. Ostes/ostes mice are small and weigh less than wild-type mice from 3–5 weeks of age. (B) Weight of 12-week-old wild-type (+/+) and ostes/ostes mice. Both male and female ostes/ostes mice weigh significantly less than wild-type mice at 12 weeks of age (females: P = 5.873E − 16, n = 9 ostes/ostes, 22 wild-type; males: P = 1.143E − 12, n = 11 ostes/ostes, 22 wild-type). Data are represented as mean ± SEM. (C) Representative sections of gastrocnemius skeletal muscle from 45-day-old wild-type (+/+), ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) mice, stained with H&E. Evidence of degenerative changes are present in ostes/ostes muscle, including muscle atrophy, small fibres, striking fibre size asymmetry, pyknotic nuclei and centrally nucleated fibres (arrows). Muscles from 45-day-old Tg(Pkd1l2)/Tg(Pkd1l2) mice show similar pathological hallmarks. Scale bar: ×20 magnification, 100 μm, ×40, 50 μm. Inset: H&E-stained levelled cross-sections of whole hind limbs from the same mice show the reduced size and muscle atrophy in ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) hind limbs. (D) Mean fibre size according to fibre type in soleus muscle from wild-type (+/+), ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) mice. Mean fibre size in both ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) skeletal muscle is reduced compared with wild-type. Type-2 fibres show a small but significant reduction in size in ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) muscle. Fibre size was analysed in MYH7-stained sections (slow twitch) of soleus muscle from three 45-day-old mice containing at least 80 fibres each. Data are represented as mean ± SEM. (E) Fibre size distribution in 45-day-old wild-type (+/+), ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) skeletal muscle. Fibre size was analysed in five H&E-stained sections of gastrocnemius from 45-day-old mice containing at least 80 fibres each. (F) Immunofluorescence photographs of NMJs in wholemount lumbrical muscle from wild-type (+/+) and ostes/ostes mice. Acetylcholine receptors are labelled in red, and presynaptic boutons and axons in green. Wild-type (+/+) endplates are mononeuronally innervated, with one axon leading to each endplate. Ostes/ostes endplates show both normal mononeuronal innervation (μ), alongside abnormal polyneuronal innervation (π) where two or more axon terminals innervate one endplate, in both 45-day-old and 9-month-old mice. In addition, some ostes/ostes endplates are denervated (δ), characterized by a plaque-like morphology of acetylcholine receptor staining and only partial or absent occupation by axon terminals. Scale bar, 20 µm. (G) Example of an intracellular recording of nerve-evoked EPPs from an ostes/ostes EDL muscle, showing functional polyneuronal innervation. (H) Immunofluorescence photographs of NMJs in wholemount hind limb muscle from 26-day-old Tg(Pkd1l2)/Tg(Pkd1l2) mice. Examples of an abnormal polyneuronal innervation (π) and a plaque-like partially denervated endplate (δ) are shown. (I) Fibre type analysis of levelled cross-sections of 45-day-old +/+, ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) hind limb skeletal muscle. There is no evidence of fibre type grouping, and fibre size asymmetry does not correlate with specific fibre type in either ostes/ostes or Tg(Pkd1l2)/Tg(Pkd1l2) muscle. (J) Representative sections of gastrocnemius skeletal muscle from aged (>6 months) ostes/+, Tg(Pkd1l2)/0 and ostes/+, Tg(Pkd1l2)/0 mice, stained with H&E. Ostes/+ skeletal muscle shows similar but much milder pathological hallmarks than ostes/ostes muscle, and no evidence of overall muscle atrophy. Skeletal muscle from hemizygous Tg(Pkd1l2)/0 mice does not show any obvious myopathic changes or atrophy. However, skeletal muscle from aged ostes/+, Tg(Pkd1l2)/0 mice shows marked fibre size asymmetry, very small fibres and centrally nucleated regenerating fibres. Scale bar: ×20, 100 μm, ×40, 50μm. Inset: H&E-stained cross-sections of whole hind limbs from the same mice show no significant differences in size.
Pyknotic nuclear clumps have been associated with denervation of muscle fibres in amyotrophic lateral sclerosis and bulbospinal muscular atrophy. Therefore, endplates, axons and presynaptic axon terminals in lumbrical muscle wholemounts from 45-day-old ostes/ostes, ostes/+ and wild-type mice were immunostained and examined for signs of denervation. Confocal microscopy revealed many denervated endplates in ostes/ostes muscles, characteristically showing a plaque-like morphology, with or without contact to a visible degenerating nerve terminal, or only partially occupied by a small number of nerve terminal boutons (Fig. 1F). Additionally, ∼40% of ostes/ostes motor endplates were polyneuronally innervated by collateral branches of two or three axons. All endplates in wild-type littermates had only one axon terminal per endplate. We assessed the function of polyinnervated junctions in ostes mice by intracellular recordings of nerve-evoked endplate potentials (EPPs) in extensor digitorum longus (EDL) and soleus muscle from ostes/ostes (n = 3) mice. Recordings from ostes/ostes muscle in response to graded stimulation of the nerve supply resolved two components to the EPP with distinct thresholds in 20–30% of muscle fibres (n = 15–18 fibres per muscle), indicating functional polyneuronal innervation (Fig. 1G). These recordings corroborated the morphological evidence of enduring polyneuronal innervation in ostes/ostes mice. Fibre type distribution in the soleus muscle region did not show any differences between ostes/ostes and wild-type staining. Fibre size asymmetry did not correlate with specific fibre type, and there was no evidence of fibre type grouping (Fig. 1I and Supplementary Material, Fig. S4).
To assess whether this polyinnervation was due to a delay in developmental synapse elimination at the neuromuscular junction (NMJ), we examined motor endplates in 9-month-old ostes/ostes mice (Fig. 1F). Polyinnervated endplates were also observed in these mice, suggesting that this phenotype was persistent and not due to prolongation of the period over which synapse elimination would normally occur (1–2 weeks postnatal). Ostes/+ mice also showed evidence of polyinnervation, but at lesser incidence than ostes/ostes mice, and polyinnervation was not unequivocally found to be functional (n = 2 mice, data not shown). In both young and old ostes/ostes and ostes/+ mice, normal mononeuronal innervation of endplates was also observed (Fig. 1F). Therefore, three distinctive types of endplates were characteristically observed in lumbrical muscles from ostes/ostes mice: monoinnervated (∼50%, n = 3), denervated (∼10%, n = 3) and polyinnervated (∼40%, n = 3).
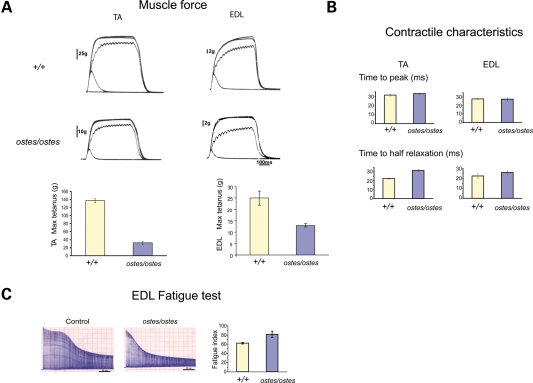
The specific effects of the morphological changes in ostes/ostes muscle on muscle function were also established. We found that the hind limb muscles of ostes/ostes mice were weaker than the corresponding muscles in age-matched wild-type controls. Whereas tibialis anterior (TA) muscles in wild-type mice produced a maximum tetanic force of 137g (±5.3 SEM, n = 6), this was reduced to only 32g (±2.1 SEM, n = 10) in ostes/ostes mice (Fig. 2A). Similarly, the maximum force in EDL muscles of wild-type mice was 25g (±3.1 SEM, n = 8) compared with only 13g (±0.8 SEM, n = 7) in ostes/ostes mice (Fig. 2A). The contractile characteristics of these muscles in ostes/ostes mice were also assessed. Although the time to peak force and the time taken to reach half relaxation force were the same in TA and EDL muscles of ostes/ostes and wild-type mice (Fig. 2B), there was a significant change in the fatigue characteristics of EDL muscles in ostes/ostes mice (Fig. 2C). EDL is normally a fast muscle that fatigues rapidly when repeatedly stimulated (Fig. 2C). However, EDL in ostes/ostes mice has a higher fatigue index than that of wild-type (Fig. 2C), indicating that it has become more fatigue resistant than normal.
Figure 2.
Physiological assessment of ostes muscle. (A) Muscle force in ostes/ostes tibialis anterior (TA) and extensor digitorum longus (EDL) muscles. Examples of tension recordings of isometric contractions from wild-type (+/+) and ostes/ostes TA and EDL, showing twitch and tetanic contractions elicited by stimulating the motor nerve to both muscles. Both TA and EDL from ostes/ostes mice show decreased force, compared with wild-type (TA: wild-type n = 6, ostes/ostes n = 10; EDL: wild-type n = 8, ostes/ostes n = 7). Data shown as mean maximum tetanic force ± SEM. Max tetanus, maximum tetanic force. (B) Contractile characteristics of ostes/ostes TA and EDL muscles. Time to peak and time to half relaxation are not significantly different in wild-type and ostes/ostes muscle (TA: wild-type n = 6, ostes/ostes n = 10; EDL: wild-type n = 8, ostes/ostes n = 7). Data shown as mean time ± SEM. (C) Fatigue profile of ostes/ostes EDL muscle. Ostes/ostes EDL (n = 6) shows increased resistance to fatigue relative to wild-type EDL (n = 6). Data shown as mean fatigue index ± SEM.
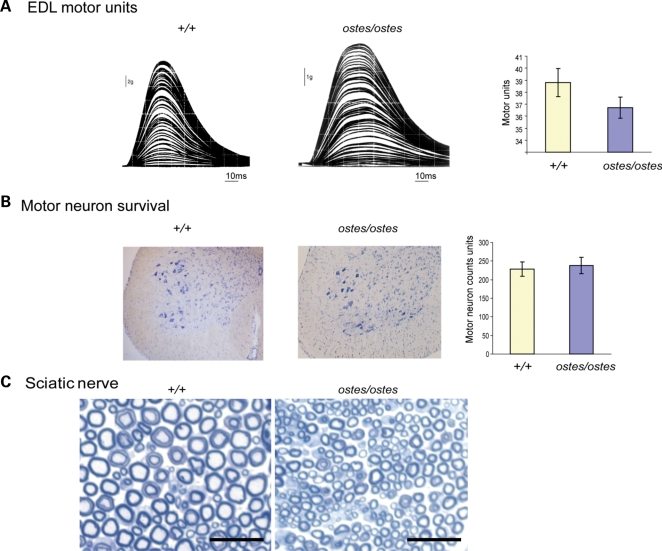
Despite the presence of some denervated NMJs, the loss of muscle force and change in fatigue characteristics observed in hind limb muscles of ostes/ostes mice were not due to a loss of motor units. Physiological assessment of motor unit survival showed that EDL muscles in wild-type and ostes/ostes mice were innervated by an average of 38.8 (±1.17 SEM, n = 6) and 36.7 (±0.88 SEM, n = 6) motor neurons, respectively (Fig. 3A). This finding, indicating that there was no loss of motor axons in ostes/ostes mice, was supported by morphological assessment of motor neuron survival (Fig. 3B). We found no difference in the number of motor neurons present in the lumbar spinal cord of ostes/ostes mice and wild-type littermates (Fig. 3B). This was supported by the lack of muscle paralysis in either ostes/+ or ostes/ostes mice at any age. Additionally, cross-sections of sciatic nerve showed no evidence of demyelination in ostes/ostes mice (n = 3) (Fig. 3C).
Figure 3.
Motor neuron survival in ostes. (A) Assessment of motor unit number in ostes/ostes EDL. Examples of stepwise increments in twitch force from wild-type (+/+) and ostes/ostes EDL in response to gradual intensity increases of stimulation of the motor nerve to the EDL. Motor unit number in ostes/ostes EDL is not significantly different to wild-type EDL (n = 6 per genotype). Data shown as mean motor unit number ± SEM. (B) Motor neuron survival in ostes/ostes mice. Shown are representative 20 μm-thick transverse sections of spinal cord from wild-type and ostes/ostes mice. Motor neurons are stained with gallocyanin (blue). Data shown as mean motor neuron counts ± SEM. The number of gallocyanin-stained motor neurons present in the sciatic motor pool of the ventral horn of each spinal cord was determined by counting every third serial section (n = 60 sections per mouse; n = 6 per genotype). (C) Representative cross-section of proximal regions of sciatic nerve from 6-week-old wild-type (+/+) (n = 3) and ostes/ostes mice (n = 3) fixed with osmium tetroxide and stained with methylene blue. No demyelination of ostes/ostes motor neurons as evidenced by reduced myelin-to-lumen staining or by ‘onion-bulb’ morphology is seen. Note that ostes/ostes motor neurons are markedly smaller than wild-type, reflecting the overall small size of ostes/ostes mice. Scale bar: 25 µm.
The ostes mutation maps to a 0.78 Mb region on distal chromosome 8
The ostes mutant was identified in a complex recessive mouse mutagenesis screen that involved ENU treatment of a hybrid (C3H/HeH × 101/H) strain (6). To facilitate genetic and functional analysis, the ostes line was crossed to wild-type BALB/c mice for 20 generations to generate a congenic ostes line. Genetic mapping on ostes/ostes progeny from these crosses established linkage to distal chromosome 8 between D8Mit213 and D8Mit13. Two further outcrosses, to the C57BL/6J and FVB inbred strains, also independently confirmed significant linkage to the same region on chromosome 8 (data not shown). Since the FVB strain showed the highest rate of polymorphism with the initial mutagenized background, this strain was selected for high-resolution mapping. We generated and genotyped approximately 500 intercross progeny including both affected (ostes/ostes) and non-affected (+/+ and ostes/+) offspring to utilize all recombination events. This analysis refined the non-recombinant region to 772 kb, between D8Mit89 and the in-house marker FM3 (see Materials and Methods). Pkd1l2, Gcsh, Bcmo1, Dynlrb2 and five additional predicted genes lie within this region, comprising 94 coding exons in total (Ensembl v49, May 2008; Supplementary Material, Table S1). All exons and splice acceptor/donor sites were amplified by PCR from genomic ostes/ostes DNA and screened for mutation by sequencing, but intriguingly, no mutation was revealed. To confirm this finding, we used a commercial next-generation sequencing approach (NGS) to sequence all exons in the interval. As no mutation in coding sequence was identified, this analysis confirmed our previous results (see Materials and Methods). To screen for cDNA variants, cDNA of all candidate genes except Pkd1l2 was successfully amplified and sequenced from primary cDNA libraries from wild-type and ostes/ostes tissues. This analysis revealed no ostes-specific splicing variants of these genes. Therefore, this raised the possibility that the ostes mutation could lie in a regulatory element and that misregulation of one of these genes, rather than a coding mutation, could underlie the ostes phenotype. Intriguingly, 30 sets of primers in a large number of combinations failed to amplify overlapping segments of the large Pkd1l2 cDNA from several tissues including the liver, kidney and muscle; therefore, a complete sequence scan for this particular cDNA was not achieved.
To evaluate expression, we performed microarray analysis on adult ostes/ostes and wild-type hind limb skeletal muscle using the RNG-MRC Mouse 25k array (MRC Harwell) which contained all of the nine genes from the non-recombinant region. No significant differences in the expression of these genes between ostes/ostes and wild-type samples were found (Supplementary Material, Fig. S5A; data deposited at ArrayExpress, accession number E-MEXP-1873). Pkd1l2 was the only gene in this region for which analysis of splicing variants or differences in expression could not be established despite its reported expression in skeletal muscle (7). This was not due to technical problems, because detection of Pkd1l2 to levels amenable for analysis failed in direct cDNA amplifications, Q-PCR (see what follows) and microarray experiments, whereas cDNAs from other genes in the non-recombinant region were efficiently detected using the same techniques and templates. However, we considered Pkd1l2 to be a good candidate on the basis of the association of the homologous PKD1 gene with disease and the fact that all genes in the non-recombinant region except Pkd1l2 had been effectively ruled out. Thus, for all these other genes, no coding or splicing mutations and no significant expression differences between the mutant and control were found. Therefore, we decided to assess expression of Pkd1l2 in ostes/ostes mice at the protein level.
PKD1L2 protein is upregulated in ostes mice
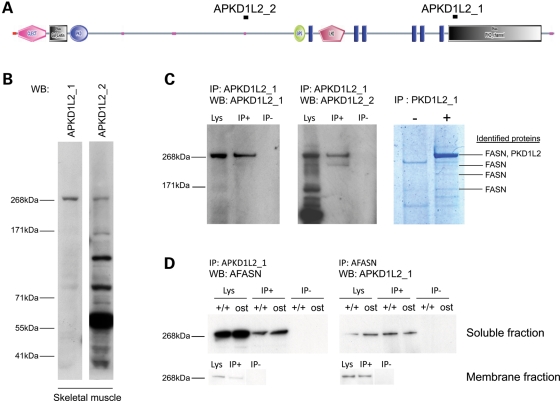
We assessed Pkd1l2 as a candidate gene for ostes by generating PKD1L2-specific antibodies and examining PKD1L2 protein in ostes/ostes muscle. Two rabbit polyclonal antibodies, APKD1L2_1 and APKD1L2_2, were generated and thoroughly characterized (Fig. 4). Western blot analysis of soluble skeletal muscle extracts from wild-type mice showed that both antibodies recognized a band migrating at ∼268 kDa (Fig. 4B), consistent with the PKD1L2 expected size (272 kDa). Western blot analysis of multiple tissues showed that this band was the largest protein recognized in several tissues (Supplementary Material, Fig. S6A), suggesting that this protein is likely to be full-length PKD1L2. We used immunoprecipitation to confirm the specificity of these antibodies. Antibody APKD1L2_1 specifically and efficiently immunoprecipitated a protein migrating at ∼268 kDa from wild-type skeletal muscle lysates (Fig. 4C). Analysis of the corresponding band by peptide mass fingerprinting identified PKD1L2 among other proteins (see below and Fig. 4C). In addition, the immunoprecipitated protein was also recognized by the second antibody, APKD1L2_2 (Fig. 4C). We then assessed PKD1L2 protein expression in ostes/ostes skeletal muscle. Since PKD1L2 is a predicted transmembrane protein, PKD1L2 expression was assessed in both soluble and membrane fractions. The results showed that PKD1L2 is strongly accumulated in ostes/ostes skeletal muscle compared with wild-type (n = 8, representative samples shown in Fig. 5). Accumulation was observed in both soluble and membrane fractions (Fig. 5). These results suggested that the upregulation of PKD1L2 underlies the ostes phenotype.
Figure 4.
Analysis of PKD1L2 protein in skeletal muscle. (A) Domain composition of PKD1L2 with the annotated position of peptides used to raise antibodies APKD1L2_1 and APKD1L2_2. Functional domains as predicted by SMART (http://smart.embl-heidelberg.de/). C-LECT, C-type lectin (CTL) or carbohydrate-recognition domain (CRD); Pfam Gal_Lectin, galactose-binding lectin domain; PKD, PKD domain; GPS, G-protein-coupled receptor proteolytic site domain; LH2, lipoxygenase homology 2 (beta barrel) domain; blue rectangles, transmembrane domains; pink dots, low-complexity region. (B) Western blot analysis using PKD1L2 antibodies. Both antibodies APKD1L2_1 and APKD1L2_2 recognize a ∼268 kDa protein in wild-type soluble skeletal muscle extracts. APKD1L2_2 recognizes other bands of smaller size. WB, antibody used for western blot. (C) Immunoprecipitation and peptide analysis using APKD1L2_1 antibody on wild-type skeletal muscle extract. APKD1L2_1 specifically immunoprecipitates a ∼268 kDa band (left panel) that is also recognized by APKD1L2_2 (middle panel). The right panel shows a Coomassie-stained gel in which the immunoprecipitated protein complex has been resolved. Proteins identified by mass spectrometry are indicated. IP, immunoprecipitation; WB, western blot; Lys, lysate; IP+, immunoprecipitated sample; IP−, negative control. (D) Reciprocal co-immunoprecipitation of PKD1L2 and FASN in skeletal muscle. Western blot analysis using AFASN antibody specifically recognizes a ∼268 kDa band from protein complexes immunoprecipitated using APKD1L2_1 antibody in skeletal muscle soluble fraction, from both wild-type and ostes/ostes mice. Western blot analysis using APKD1L2 antibody specifically recognizes a ∼268 kDa band from protein complexes immunoprecipitated using AFASN antibody in skeletal muscle soluble fraction, from both wild-type and ostes/ostes mice. PKD1L2 and FASN co-immunoprecipitation was also observed in membrane fractions (bottom panels). IP, immunoprecipitation; WB, western blot; Lys, lysate; IP+, immunoprecipitated sample; IP−, negative control.
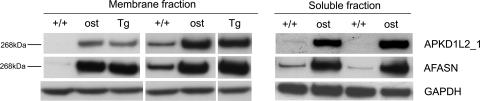
Figure 5.
Western blot analysis of skeletal muscle extracts from wild-type, ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) mice. (A) PKD1L2 and FASN show increased levels in the membrane fraction of ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) skeletal muscle. Samples shown are from individual mice (n = 2 per genotype). (B) PKD1L2 and FASN show increased levels in the soluble fraction of ostes/ostes skeletal muscle. Samples shown are from individual mice (n = 2 per genotype). PKD1L2 and FASN were detected, respectively, with antibodies APKD1L2_1 and mAFASN. +/+, wild-type; ost, ostes/ostes; Tg, Tg(Pkd1l2)/Tg(Pkd1l2).
In search of a putative regulatory mutation within the vicinity of the Pkd1l2 gene that would explain the upregulation of this gene in mutant mice, we used NGS to sequence unique non-coding sequences in a 250 kb genomic segment encompassing Pkd1l2 and flanking genes Bcmo1 and Gcsh, that is, all genomic DNA except repetitive elements in that interval. This analysis produced 149 kb of sequence from ostes/ostes DNA, but no change was revealed, suggesting that a regulatory mutation could exist in a non-conserved regulatory element somewhere else within the non-recombinant interval.
Transgenic overexpression of PKD1L2 causes ostes-like myopathic and innervation changes
We tested the hypothesis that the overexpression of PKD1L2 found in ostes/ostes mice causes neuromuscular dysfunction in these mice by generating a transgenic mouse overexpressing PKD1L2. Since a full-length Pkd1l2 cDNA could not be amplified (discussed earlier), we opted to use a bacterial artificial chromosome (BAC) to generate a transgenic line. A BAC (RP23-269J16) was selected which contained the full-length genomic sequence of Pkd1l2 and the small flanking genes Bcmo1 and Gcsh. These flanking sequences were required to include the promoter and other possible regulatory elements in the vicinity of the Pkd1l2 gene. Following microinjection of the BAC into fertilized oocytes, a transgenic founder was identified (Tg(Pkd1l2)_#1). Subsequent crosses showed that the BAC was transmitted to 57.5% (57 of 99) of offspring from this mouse, thus indicating a single integration site. We examined hemizygous Tg(Pkd1l2)/0 mice for evidence of neuromuscular dysfunction. Tg(Pkd1l2)/0 mice were indistinguishable from wild-type littermates and did not show any overt phenotype, and histological analysis did not reveal any anomalies on H&E-stained hind limb skeletal muscle sections (Fig. 1J). However, on intercrossing Tg(Pkd1l2)/0 mice, we found that homozygous Tg(Pkd1l2)/Tg(Pkd1l2) mice showed extreme growth retardation and developed progressive hind limb dragging and paralysis between 4 and 6 weeks of age. Nineteen per cent of intercross offspring displayed this phenotype (13/64), and 3% were found dead around this age (2/64), indicating that some post-natal loss may be occurring. H&E-stained skeletal muscle sections from these mice showed extreme fibre size asymmetry, pyknotic nuclei, small centrally nucleated fibres and no evidence of macrophage infiltration, strongly recapitulating the pathological hallmarks seen in ostes/ostes mice (Fig. 1C, D and E and Supplementary Material, Figs S1 and S2). Similar pathology was seen in pharyngeal muscle and diaphragm, but not in any other 23 tissues analysed including the heart (not shown). Moreover, fibre type analysis revealed a similar distribution of fast and slow fibres to ostes/ostes muscle, again showing a lack of both fibre-specific hypertrophy and clustering (Fig. 1I and Supplementary Material, Fig. S4). However, both ostes and Tg(Pkd1l2) homozygotes showed a small but statistically significant reduction of type-2 fibre size in soleus muscle (Fig. 1D). Additionally, polyinnervated and denervated endplates were also evident in homozygous Tg(Pkd1l2)/Tg(Pkd1l2) mice (n = 4, representative examples shown in Fig. 1H).
Given the similarities between the ostes and Tg(Pkd1l2) myopathies, these results theoretically reduced the genetic cause of the ostes disease to the upregulation of any of the three genes in the BAC: Bcmo1, Pkd1l2 or Gcsh. We had already established that PKD1L2 protein was elevated in ostes mice. To unequivocally establish how expression levels of these three candidate genes correlated with the ostes neuromuscular disease, further expression analysis was carried out. Real-time quantitative PCR was performed using skeletal muscle from 10 ostes/ostes mice and 10 age- and sex-matched wild-type controls. The results were consistent with the microarray results described earlier: expression of Bcmo1 or Gcsh was not significantly different in ostes/ostes mice (Supplementary Material, Fig. S5B), whereas Pkd1l2 was not detected at sufficient levels for a reliable analysis. Therefore, Bcmo1 and Gcsh have no coding or regulatory mutations in ostes/ostes mice. Western blot analysis of skeletal muscle from Tg(Pkd1l2)/Tg(Pkd1l2) mice showed that full-length PKD1L2 was indeed overexpressed in this line (Fig. 5). Further proof that the myopathies described in ostes and Tg(Pkd1l2) homozygous mice share a common pathogenic mechanism could be obtained from making double heterozygotes. Thus, if ectopic expression of PKD1L2 as a result of an ENU mutation or expression from a BAC clone was respectively underlying myopathic changes in ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) mice, we could expect double-heterozygous mice (ostes/+, Tg(Pkd1l2)/0) to show a more severe pathology than the mild changes observed in ostes/+ mice. In young mice, approximately 45 days old, the presence of the BAC clearly exacerbated the mild myopathic changes of ostes heterozygotes without adding any new distinctive feature (Supplementary Material, Fig. S3). This result suggests that increasing the copy number of the Pkd1l2 gene results in worsening of the pathology caused by the ENU mutation. Moreover, ostes/+, Tg(Pkd1l2)/0 mice (n = 3) showed a late-onset muscular atrophy and weakness of the hind limbs between 6 and 10 months old, with foot-curling, unstable gait and similar myopathic changes to those observed in ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) muscles (Fig. 1J). These features were not found in aged ostes/+ (n = 6) or Tg(Pkd1l2)/0 mice (n = 3) (Fig. 1J). These results indicated that ostes and Tg(Pkd1l2) interact genetically and strongly supported the hypothesis that the upregulation of PKD1L2 causes neuromuscular disease. These results did not strictly exclude the possibility that a new mutation caused by the insertion of the BAC and not the BAC itself was responsible for the phenotype observed in the transgenic line. However, the specificity of the pathological changes in the ENU-induced and BAC-induced mutant lines and the genetic interaction between the underlying genetic loci made that possibility unlikely.
PKD1L2 interacts with fatty acid synthase in vivo
Our data showed that overexpression of PKD1L2 was associated with neuromuscular dysfunction in two mouse mutants. However, neither PKD1L2 nor other TRPP channels have previously been shown to have roles in muscle. To explore a molecular role for PKD1L2 in muscle, we examined PKD1L2 protein complexes by immunoprecipitation from adult mouse skeletal muscle lysates. Immunocomplexes precipitated with antibody APKD1L2_1 were analysed by SDS–PAGE and mass spectrometry to identify PKD1L2-interacting proteins. Upon gel staining, a very prominent band was observed migrating at ∼268 kDa, which was absent in the negative control lane (Fig. 4C). A peptide mass fingerprint was generated from this band by mass spectrometry. Database search using ProFound identified PKD1L2, as expected, but additionally the enzyme fatty acid synthase (FASN) was identified as the top scorer. FASN was also identified from bands of smaller molecular weight, probably reflecting partial degradation (Fig. 4C). The identity of this protein was confirmed by western blot with a monoclonal antibody against FASN (mAFASN) following immunoprecipitation with APKD1L2_1 antibody, from both wild-type and ostes/ostes skeletal muscle soluble fractions (Fig. 4D). To further confirm the association between PKD1L2 and FASN, we carried out immunoprecipitation using mAFASN on wild-type and ostes/ostes skeletal muscle soluble fractions. FASN was efficiently immunoprecipitated, and subsequent western blot analysis confirmed that PKD1L2 was present in the immunoprecipitated complex. The same results were obtained using wild-type skeletal muscle membrane fraction (Fig. 4D). Since the molecular weight of FASN is equivalent to that of PKD1L2 (272 kDa), and the two proteins co-migrate, this analysis did not exclude the possibility that the APKD1L2_1 antibody might be cross-reacting with FASN protein. To test for such cross-reactivity, we examined PKD1L2 and FASN protein expression in different mouse tissues. This analysis demonstrated that whereas FASN was strongly expressed in every tissue examined, expression of PKD1L2 was more restricted, thus indicating that the APKD1L2_1 antibody does not recognize FASN (Supplementary Material, Fig. S6A). Analysis of the same samples using non-denaturing conditions produced a different pattern for both antibodies on western blots (data not shown), suggesting that PKD1L2_1 antibodies did not cross-react against the FASN native form either. Moreover, affinity-purified antibody APKD1L2_2, raised against a different PKD1L2 epitope, also immunoprecipitated FASN (Supplementary Material, Fig. S6B), and the reciprocal immunoprecipitation using mAFASN antibody also produced a ∼268 kDa protein recognized by antibody APKD1L2_2, confirming the co-immunoprecipitation of FASN and PKD1L2 (Supplementary Material, Fig. S6B).
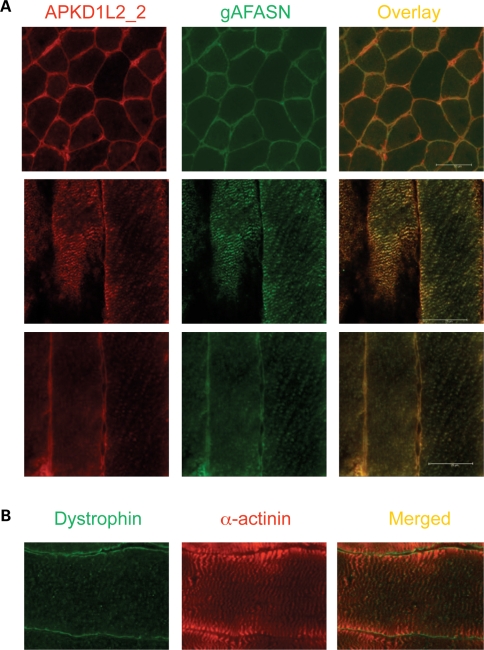
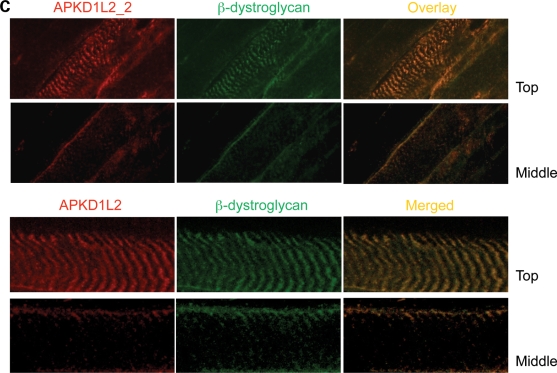
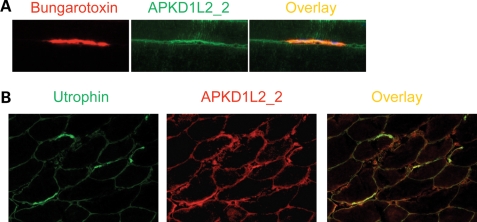
Despite PKD1L2 being a predicted transmembrane protein and FASN a soluble cytoplasmic enzyme, their association as revealed by immunoprecipitation of endogenous proteins suggested that these proteins must have overlapping subcellular localizations. To assess this, immunostainings on cross- and longitudinal sections of fresh-frozen skeletal muscle samples were performed with APKD1L2_1 and a goat polyclonal antibody against FASN (gAFASN). PKD1L2 and FASN were found localized to the membrane on cross-sections (Fig. 6A). On longitudinal sections, both antibodies produced overlapping localizations at striations overlying the Z-discs on the surface of the fibre (Fig. 6A, middle panel) but not at the centre (Fig. 6A, bottom panel). A similar result was obtained with mAFASN antibody (Supplementary Material, Fig. S7), suggesting a costameric localization of both proteins. Costameric distribution was supported by co-localization of PKD1L2 with the costameric transmembrane protein β-dystroglycan (8) on serial views of longitudinal sections (Fig. 6C). An identical pattern was observed with antibody APKD1L2_2, and for both PKD1L2 antibodies, the fluorescent signal was effectively blocked upon pre-incubation with their respective antigenic peptides (data not shown). On the other hand, expression of PKD1L2 was also detected at slightly higher levels at the postsynaptic membrane of the NMJs (Fig. 7A and B).
Figure 6.
Co-localization of PKD1L2 and FASN in skeletal muscle. (A) Confocal microscopic analysis of fresh-frozen skeletal muscle sections from wild-type mice immunostained with APKD1L2_1 and gAFASN antibodies. Top panel, a cross-section of muscle showing PKD1L2 and FASN co-localization at the membrane. Middle panel: a view of the surface of longitudinally sectioned fibres showing PKD1L2 and FASN with overlapping localizations to striations. Bottom panel: a deeper level of the same fibres showing PKD1L2 and FASN with overlapping localizations at the membrane. (B) For comparison, a view of the mid-level of a longitudinal fibre stained with dystrophin and α-actinin using the same conditions. Note that, in contrast to the bottom panel shown in (A), the Z-discs are stained here. (C) Top panel: superficial (‘Top’) and middle views of longitudinal fibres showing PKD1L2 co-localization with the costameric protein β-dystroglycan. Bottom panel: a close-up view of top and middle levels of a single fibre. Scale bar: 25 μm.
Figure 7.
PKD1L2 localizes to NMJs. (A) A longitudinal section showing the NMJ stained with bungarotoxin and overlapping localization with PKD1L2 (using APKD1L2_2 antibodies). Note that PKD1L2 is slightly increased at the postsynaptic membrane. (B) NMJs identified on cross-section with antibodies against utrophin. Note that PKD1L2 is slightly increased at the NMJs.
FASN is overexpressed in ostes mice
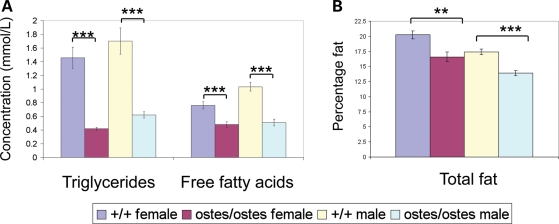
We speculated that the overexpression of PKD1L2 in ostes/ostes skeletal muscle could affect the expression or activity of FASN in this tissue. Surprisingly, we found that FASN was overexpressed in ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) skeletal muscle by western blot analysis, mirroring the overexpression of PKD1L2 (n = 12, representative samples shown in Fig. 5). Overexpression of FASN was observed in both soluble and membrane fractions (Fig. 5) and was also confirmed to occur at the transcriptional level (Supplementary Material, Fig. S8A). The growth defect exhibited by the ostes mouse is counterintuitive to an overexpression of FASN, as the latter is generally found in conditions of metabolic overload such as obesity (reviewed in 9). We therefore hypothesized that PKD1L2 acts as a negative regulator of FASN and that the upregulation of PKD1L2 in the ENU and transgenic lines results in decreased FASN activity. In this hypothesis, the increased expression of FASN in skeletal muscle of ostes mice would result from a positive feedback regulatory mechanism. Relative activity of FASN to total amount of FASN protein was measured in soluble fractions of ostes/ostes and wild-type muscle by spectrophotometric analysis of NADPH > NADP+ oxidation. Although there is a clear trend showing reduced relative activity in ostes/ostes mice, FASN activity varied widely in individual samples of the same genotype, making the difference between ostes/ostes and control statistically not significant (P = 0.236) (Supplementary Material, Fig. S8B). Therefore, we looked for other biochemical markers of lipid metabolism to determine whether fatty acid biosynthesis was abnormal in ostes/ostes mice. We assessed the blood plasma lipid profile of ostes/ostes mice (Fig. 8A). Both triglyceride (TG) and free fatty acid (FFA) levels were significantly and markedly decreased in plasma from both male and female ostes/ostes mice, compared with wild-types (Fig. 8A). Furthermore, total body fat content of ostes/ostes mice was analysed by dual-energy X-ray absorptiometry (DEXA) analysis (Fig. 8B). The total fat percentage of body mass was significantly decreased in ostes/ostes mice from a mean wild-type percentage of ∼20 to ∼17% in female ostes/ostes mice and from ∼17 to ∼14% in male ostes/ostes mice (Fig. 8B). These results indicated a reduction of lipogenesis in ostes mice.
Figure 8.
Lipid metabolism in ostes mice. (A) Lipid profiling in ostes/ostes mice. Blood lipids were analysed from fasted ostes/ostes (n = 9 female, 11 male) and wild-type (n = 12 female, 12 male) mice for triglyceride (TG) and free fatty acid (FFA). Both male and female ostes/ostes mice showed significantly decreased levels of both TG and FFA (TG: female: P = 3.107E − 05; male: P = 0.0001; FFA: female: P = 0.0005; male: P = 2.221E − 06). Data represented are mean blood plasma levels ± SEM. (B) Total percentage body fat in ostes/ostes mice. Total body fat was analysed by DEXA analysis in ostes/ostes (n = 9 female, 11 male) and wild-type (n = 22 female, 22 male) mice. Total percentage body fat was significantly decreased in both female and male ostes/ostes mice, compared with wild-type (female: P = 1.922E − 03, male: P = 5.023E − 06). Data represented are mean percentage body fat ± SEM.
DISCUSSION
In this work, we have characterized the neuromuscular phenotype of ostes mice and explored the underlying molecular mechanism. Indeed, the genetic mechanism leading to the upregulation of the linked Pkd1l2 gene in ostes mice remains elusive despite meticulous sequencing efforts. However, as indicated by the total protein levels, Pkd1l2 is the only gene in the non-recombinant region that is upregulated in ostes/ostes mice, and there are identical pathological findings in ostes/ostes and Tg(Pkd1l2)/Tg(Pkd1l2) mice. Common histological hallmarks include muscle fibre asymmetry, scattered regenerating fibres, a lack of fibre type grouping, whole-muscle atrophy and an irregular innervation pattern. Conversely, typical dystrophic changes such as excessive connective tissue or macrophage infiltration are not present in either mutant. Disease manifestation is more severe in Tg(Pkd1l2)/Tg(Pkd1l2) probably because the BAC transgene does not exactly replicate the changes in Pkd1l2 gene expression caused by the ostes ENU mutation. Intriguingly, neither ostes/+ nor Tg(Pkd1l2)/0 mice showed any overt phenotype at young or old age (>9 months), but the double heterozygote suffered from a more severe myopathy that resulted in late-onset gait defects. Hind limb muscle histology of the latter showed changes qualitatively similar to those found in young ostes/ostes mice and much more profound than those of age-matched single heterozygotes. Altogether, these results suggested that overexpression of Pkd1l2, which was demonstrated at the protein level in both mutants, provokes neuromuscular disease in mice. Pathological changes were not found in any other tissue, including sciatic nerve and spinal cord, suggesting that skeletal muscle is the primary target tissue of this disease. The polyinnervation defects found in ostes mice could thus be secondary to the myopathic changes, but since PKD1L2 accumulates at NMJs on the postsynaptic membrane, a role for this protein in regulating the stability of NMJs is possible and merits further investigation.
PKD1L2 belongs to the TRPP family of proteins. The transient receptor potential (TRP) superfamily comprises a large group of cation channel proteins usually containing six transmembrane domains, which, despite having diverse functions and expression, appear to function as critical sensors of environmental stimuli, at both organism and cellular levels (reviewed in (10 and 11). The TRPP family includes two genes that are associated with human disease: mutation of TRPP1/PKD1 or TRPP2/PKD2 cause polycystic kidney disease in humans (12,13) and mice (14,15). Other TRPP1-like genes have been shown to have roles in taste reception (Pkd1l3 (16,17)) and reproductive success (Pkdrej (18)). Thus far, no TRPP genes have been associated with neuromuscular function or disease. PKD1L2 is a large, complex protein of unknown function. It is predicted to act as an ion channel (7) and has also been reported to bind in vitro to two G-proteins (19). The presence of FASN as the major component of the PKD1L2-immunoprecipitated complex in skeletal muscle suggests a functional relationship between these proteins in this tissue. In mammals, FASN synthesizes de novo long-chain, saturated fatty acids from malonyl-CoA and acetyl-CoA, and is therefore a critical enzyme in lipogenesis (20). In muscle, both imported and de novo-synthesized fatty acids are used as an energy source via beta oxidation. The increased levels of FASN protein in the skeletal muscle of ostes mice are due to the upregulation at the transcriptional level. This is not a non-specific response of skeletal muscle to a disease condition because the actively regenerating muscles of the muscular dystrophy ky mutant (21) do not show FASN upregulation (Supplementary Material, Fig. S9). However, the finding that FASN is overexpressed in ostes/ostes mice is intriguing because ostes/ostes mice are small and lean. Intriguingly, the dramatic accumulation of FASN in ostes/ostes skeletal muscle did not result in an increase of FASN activity. In fact, blood measurements of TGs and FFAs as well as total fat deposition would suggest that lipid metabolism is indeed compromised in the ostes homozygote. Given that muscle has a particularly high metabolic need for fatty acids driven by its high-energy demand, a reduction in FASN activity would predictably compromise muscle function. Mice homozygous for a null allele of FASN show embryonic lethality (22), but skeletal muscle was not examined in surviving Fasn heterozygous mice. A direct involvement of FASN activity in muscle disease or indeed the pathogenic mechanism of ostes cannot be tested at the present time, as a Fasn skeletal-muscle conditional knockout mouse has not been reported. However, a rare case of de novo fatty acid synthesis deficiency resulting in persistent myopathy and poor growth has been reported in humans. This defect was due to the deficiency of the acetyl-CoA carboxylase enzyme, which catalyses the formation of the FASN substrate malonyl-CoA (23), indicating that insufficient fatty acid synthesis can cause pathological changes in skeletal muscle. Moreover, the microarray data generated in this work identified lipid metabolism as the major biological pathway altered in ostes/ostes skeletal muscle: eight out of the top nine genes upregulated in ostes/ostes muscle have functions in lipid metabolism (Supplementary Material, Table S2; see also deposited data ArrayExpress, accession number E-MEXP-1873). Therefore, lipogenesis is altered at skeletal muscle and systemic levels in ostes/ostes mice. The precise reasons for the tissue specificity of this disease remain to be established. Concomitant upregulation of FASN and PKD1L2 was also detected in the kidney, but here the increase in FASN is smaller than that observed in muscle; overexpression of both PKD1L2 and FASN was not detected in other tissues from ostes/ostes mice, including the brain, spinal cord and liver (Supplementary Material, Fig. S10A). In conclusion, our data suggest that PKD1L2-induced dysregulation of lipid metabolism is a cause of complex neuromuscular defects in mice. On that basis, we propose that PKD1L2 and defective fatty acid synthesis should be assessed, respectively, as a genetic risk factor and as a precipitating cause of neuromuscular disorders in humans.
MATERIALS AND METHODS
Additional methods are detailed in Supplementary Material.
Antibodies
Polyclonal antibodies to mouse PKD1L2 were generated in rabbit using two PKD1L2 peptides (APKD1L2_1: LVASTKVRPEDQRRQEATRA; APKD1L2_2: GNSKLVGSAHIRQVRVRE) and independently affinity-purified (NeoMPS Inc., San Diego, CA, USA).
Genetic mapping
The in-house-generated marker used was a microsatellite marker FM3 (forward primer: 5′-TCTGGGTTGGATGGACATTT-3′; reverse primer: 5′-CCCAGATGATGGAAGGAGAA-3′). The ostes locus was genotyped on an FVB genetic background using D8Mit89 and D8Mit120 (MGI), and rs37591588 (NCBI).
BAC transgenesis
A BAC (identifier RP23-269J16) containing full-length genomic sequence of Pkd1l2 was obtained from BACPAC Resource Center, Children's Hospital Oakland Research Institute (CHORI), Oakland, CA 94609, USA. DNA was prepared from culture using the QIAGEN Large-Construct Kit, diluted to 1 ng/μl and microinjected into fertilized FVB oocytes. Resulting mice were screened for BAC integration by PCR with primers designed against the vector: forward 5′-ataaatggatgccctgcgta-3′; reverse 5′-ttcaacccagtcagctcctt-3′. One positive founder for the BAC was identified and bred to the FVB strain to generate a colony. The line was submitted to the Mouse Genomic Nomenclature Committee (The Jackson Laboratory) and designated the identifier Tg(Pkd1l2)1Blac (accession number MGI:3811813). For brevity, we refer to this line in this publication by the shortened identifier Tg(Pkd1l2).
FASN activity assay and HeLa cell culture
FASN-specific activity in soluble fraction of skeletal muscle from 3-month-old female wild-type (n = 4) and ostes/ostes (n = 4) mice was measured as described in Nepokroeff et al. (24), except that mice were fed ad libitum before sacrificing. Untreated, and serum-deprived and insulin-treated, HeLa cells were used as positive control. HeLa cell culture, treatment and protein isolation were performed as described in Alberts et al. (25). FASN protein amounts were quantified by densitometric analysis using QuantityOne software (Bio-Rad) from western blot of equally loaded cytosolic fractions and were normalized to GAPDH. FASN relative activity was calculated using the following formula: FASN activity = FASN-specific activity (units/mg/min)/FASN total protein (arbitrary units). Statistical testing was performed using a two-tailed Student's t-test.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
Funding for this work and to pay the Open Access publication charges for this article was provided by the UK Medical Research Council.
ACKNOWLEDGEMENTS
We thank Richard Gale, Rama Kurapati, James Green and MGU core facilities for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Allamand V., Campbell K.P. Animal models for muscular dystrophy: valuable tools for the development of therapies. Hum. Mol. Genet. 2000;9:2459–2467. doi: 10.1093/hmg/9.16.2459. [DOI] [PubMed] [Google Scholar]
- 2.Hafezparast M., Ahmad-Annuar A., Hummerich H., Shah P., Ford M., Baker C., Bowen S., Martin J.E., Fisher E.M. Paradigms for the identification of new genes in motor neuron degeneration. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 2003;4:249–257. doi: 10.1080/14660820310016084. [DOI] [PubMed] [Google Scholar]
- 3.Vainzof M., Ayub-Guerrieri D., Onofre P.C., Martins P.C., Lopes V.F., Zilberztajn D., Maia L.S., Sell K., Yamamoto L.U. Animal models for genetic neuromuscular diseases. J. Mol. Neurosci. 2008;34:241–248. doi: 10.1007/s12031-007-9023-9. [DOI] [PubMed] [Google Scholar]
- 4.Bittner R.E., Anderson L.V., Burkhardt E., Bashir R., Vafiadaki E., Ivanova S., Raffelsberger T., Maerk I., Hoger H., Jung M., et al. Dysferlin deletion in SJL mice (SJL-Dysf) defines a natural model for limb girdle muscular dystrophy 2B. Nat. Genet. 1999;23:141–142. doi: 10.1038/13770. [DOI] [PubMed] [Google Scholar]
- 5.Bridges L.R., Coulton G.R., Howard G., Moss J., Mason R.M. The neuromuscular basis of hereditary kyphoscoliosis in the mouse. Muscle Nerve. 1992;15:172–179. doi: 10.1002/mus.880150208. [DOI] [PubMed] [Google Scholar]
- 6.Bogani D., Willoughby C., Davies J., Kaur K., Mirza G., Paudyal A., Haines H., McKeone R., Cadman M., Pieles G., et al. Dissecting the genetic complexity of human 6p deletion syndromes by using a region-specific, phenotype-driven mouse screen. Proc. Natl Acad. Sci. USA. 2005;102:12477–12482. doi: 10.1073/pnas.0500584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li A., Tian X., Sung S.W., Somlo S. Identification of two novel polycystic kidney disease-1-like genes in human and mouse genomes. Genomics. 2003;81:596–608. doi: 10.1016/s0888-7543(03)00048-x. [DOI] [PubMed] [Google Scholar]
- 8.Rybakova I.N., Patel J.R., Ervasti J.M. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J. Cell Biol. 2000;150:1209–1214. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berndt J., Kovacs P., Ruschke K., Kloting N., Fasshauer M., Schon M.R., Korner A., Stumvoll M., Bluher M. Fatty acid synthase gene expression in human adipose tissue: association with obesity and type 2 diabetes. Diabetologia. 2007;50:1472–1480. doi: 10.1007/s00125-007-0689-x. [DOI] [PubMed] [Google Scholar]
- 10.Nilius B. TRP channels in disease. Biochim. Biophys. Acta. 2007;1772:805–812. doi: 10.1016/j.bbadis.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Venkatachalam K., Montell C. TRP channels. Annu. Rev. Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The European Polycystic Kidney Disease Consortium. The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell. 1994;77:881–894. doi: 10.1016/0092-8674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 13.Mochizuki T., Wu G., Hayashi T., Xenophontos S.L., Veldhuisen B., Saris J.J., Reynolds D.M., Cai Y., Gabow P.A., Pierides A., et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 1996;272:1339–1342. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- 14.Lu W., Peissel B., Babakhanlou H., Pavlova A., Geng L., Fan X., Larson C., Brent G., Zhou J. Perinatal lethality with kidney and pancreas defects in mice with a targetted Pkd1 mutation. Nat. Genet. 1997;17:179–181. doi: 10.1038/ng1097-179. [DOI] [PubMed] [Google Scholar]
- 15.Wu G., D’Agati V., Cai Y., Markowitz G., Park J.H., Reynolds D.M., Maeda Y., Le T.C., Hou H., Jr, Kucherlapati R., et al. Somatic inactivation of Pkd2 results in polycystic kidney disease. Cell. 1998;93:177–188. doi: 10.1016/s0092-8674(00)81570-6. [DOI] [PubMed] [Google Scholar]
- 16.Ishimaru Y., Inada H., Kubota M., Zhuang H., Tominaga M., Matsunami H. Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc. Natl Acad. Sci. USA. 2006;103:12569–12574. doi: 10.1073/pnas.0602702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LopezJimenez N.D., Cavenagh M.M., Sainz E., Cruz-Ithier M.A., Battey J.F., Sullivan S.L. Two members of the TRPP family of ion channels, Pkd1l3 and Pkd2l1, are co-expressed in a subset of taste receptor cells. J. Neurochem. 2006;98:68–77. doi: 10.1111/j.1471-4159.2006.03842.x. [DOI] [PubMed] [Google Scholar]
- 18.Sutton K.A., Jungnickel M.K., Florman H.M. A polycystin-1 controls postcopulatory reproductive selection in mice. Proc. Natl Acad. Sci. USA. 2008;105:8661–8666. doi: 10.1073/pnas.0800603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuasa T., Takakura A., Denker B.M., Venugopal B., Zhou J. Polycystin-1L2 is a novel G-protein-binding protein. Genomics. 2004;84:126–138. doi: 10.1016/j.ygeno.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Wakil S.J. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry. 1989;28:4523–4530. doi: 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- 21.Blanco G., Coulton G.R., Biggin A., Grainge C., Moss J., Barrett M., Berquin A., Marechal G., Skynner M., van Mier P., et al. The kyphoscoliosis (ky) mouse is deficient in hypertrophic responses and is caused by a mutation in a novel muscle-specific protein. Hum. Mol. Genet. 2001;10:9–16. doi: 10.1093/hmg/10.1.9. [DOI] [PubMed] [Google Scholar]
- 22.Chirala S.S., Chang H., Matzuk M., Abu-Elheiga L., Mao J., Mahon K., Finegold M., Wakil S.J. Fatty acid synthesis is essential in embryonic development: fatty acid synthase null mutants and most of the heterozygotes die in utero. Proc. Natl Acad. Sci. USA. 2003;100:6358–6363. doi: 10.1073/pnas.0931394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blom W., de Muinck Keizer S.M., Scholte H.R. Acetyl-CoA carboxylase deficiency: an inborn error of de novo fatty acid synthesis. N. Engl. J. Med. 1981;305:465–466. doi: 10.1056/NEJM198108203050820. [DOI] [PubMed] [Google Scholar]
- 24.Nepokroeff C.M., Lakshmanan M.R., Porter J.W. Fatty-acid synthase from rat liver. Methods Enzymol. 1975;35:37–44. doi: 10.1016/0076-6879(75)35136-7. [DOI] [PubMed] [Google Scholar]
- 25.Alberts A.W., Ferguson K., Hennessy S., Vagelos P.R. Regulation of lipid synthesis in cultured animal cells. J. Biol. Chem. 1974;249:5241–5249. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.