Abstract
The vinegar fly, Drosophila melanogaster, is a popular model for the study of invertebrate antiviral immune responses. Several picorna-like viruses are commonly found in both wild and laboratory populations of D. melanogaster. The best-studied and most pathogenic of these is the dicistrovirus Drosophila C virus. Among the uncharacterized small RNA viruses of D. melanogaster, Drosophila A virus (DAV) is the least pathogenic. Historically, DAV has been labelled as a picorna-like virus based on its particle size and the content of its RNA genome. Here, we describe the characterization of both the genome and the virion structure of DAV. Unexpectedly, the DAV genome was shown to encode a circular permutation in the palm-domain motifs of the RNA-dependent RNA polymerase. This arrangement has only been described previously for a subset of viruses from the double-stranded RNA virus family Birnaviridae and the T=4 single-stranded RNA virus family Tetraviridae. The 8 Å (0.8 nm) DAV virion structure computed from cryo-electron microscopy and image reconstruction indicates that the virus structural protein forms two discrete domains within the capsid. The inner domain is formed from a clear T=3 lattice with similarity to the β-sandwich domain of tomato bushy stunt virus, whilst the outer domain is not ordered icosahedrally, but forms a cage-like structure that surrounds the core domain. Taken together, this indicates that DAV is highly divergent from previously described viruses.
INTRODUCTION
As obligate intracellular pathogens, viruses have complex interactions with their hosts. Comparatively little is known about the specific responses to virus infection in insects. Recently, insect responses that are specific for virus infection have been investigated by using the genetically tractable Drosophila melanogaster (vinegar fly) model and the most pathogenic of its viruses, the dicistrovirus Drosophila C virus (DCV) (Cherry & Perrimon, 2004; Cherry et al., 2005; Deddouche et al., 2008; Ding & Voinnet, 2007; Dostert et al., 2005; Galiana-Arnoux et al., 2006; Hedges & Johnson, 2008; Hedges et al., 2008; Roxstrom-Lindquist et al., 2004; Sabatier et al., 2003; Teixeira et al., 2008; van Rij et al., 2006) and the birnavirus Drosophila X virus (Zambon et al., 2005). In future studies of the range and specificity of Drosophila responses to virus infection, it would be valuable to use viruses that naturally infect Drosophila with a range of pathogenicities.
During the 1970s, 11 viruses were described from Drosophila (Brun & Plus, 1980; Gateff et al., 1980; Jousset & Plus, 1975; Plus, 1978). Three of these, which were isolated from D. melanogaster, were described as picorna-like viruses based on morphological and biophysical characteristics. The three viruses, DCV, Drosophila A virus (DAV) and Drosophila P virus, were shown to be different viruses based on serology and sizes of the capsid proteins (Jousset et al., 1972; Plus et al., 1976). These viruses are natural pathogens of D. melanogaster, with about 40 % of wild populations being infected with one or more of these viruses (Plus et al., 1975). The three viruses have been shown to vary in their pathogenicity, with DCV being the most pathogenic and DAV the least. It is important to have an understanding of molecular characteristics of viruses to facilitate these studies (Huszar & Imler, 2008). Whilst DCV has been well-characterized and belongs to the genus Cripavirus of the family Dicistroviridae (Christian et al., 2005; Johnson & Christian, 1998), no detailed characterization of the DAV genome or virion structure has been reported.
There are many small RNA viruses of insects that have non-enveloped, 25–40 nm particles that encapsidate positive-sense RNA genomes (Christian & Scotti, 1998; Fauquet et al., 2005). Of these, the majority with sequenced genomes are included in four taxonomic groups: the families Nodaviridae, Dicistroviridae and Tetraviridae and the floating genus Iflavirus (Fauquet et al., 2005). However, in recent years, a number of viruses from insects have been characterized that do not fit clearly into one of these groups (Habayeb et al., 2006; Hartley et al., 2005; van der Wilk et al., 1997). Furthermore, within the family Tetraviridae, there are viruses with diverse genome organizations that are classified together because of the T=4 symmetry of their capsid particles (Agrawal & Johnson, 1992; Gorbalenya et al., 2002; Gordon et al., 1995, 1999; Hanzlik et al., 1995; Pringle et al., 2001, 2003). As more invertebrate RNA viruses are characterized fully, it is becoming clear that there is a previously unrecognized diversity of genome organization and expression strategies within this group of viruses. These findings give us insight both into virus evolution and into other biological processes (Koonin et al., 2008).
Here, we describe the molecular and structural characterization of DAV. The 30 nm particles encapsidate a positive-sense RNA that encodes two large open reading frames (ORFs). The first of these has significant sequence similarities to the RNA-dependent RNA polymerases (RdRPs) of the tetraviruses and birnaviruses that have permuted RdRPs, whilst the second ORF encodes the capsid protein. Analysis of the structure of DAV virions shows a unique capsid arrangement.
METHODS
Propagation of DAV in D. melanogaster.
A stock of Champetières flies was obtained from the Drosophila Genetic Resource Centre of the Kyoto Institute of Technology, Japan (stock no. 103403), and a virus-free population was established essentially as described by Brun & Plus (1980). The Australian isolate DAVHD was kindly provided by Dr Peter Christian (Christian, 1992). DAVHD was diluted in sterile insect saline (0.6 % NaCl, 0.04 % KCl, 0.024 % CaCl2 and 0.02 % NaHCO3) and injected into the haemocoel of virus-free D. melanogaster. Injected flies were maintained on standard Drosophila medium (Sigma) for 10 days at 25 °C and then frozen at −20 °C until purification.
Virus purification.
DAVHD-infected flies were homogenized in 50 mM Tris buffer, pH 7.4, and homogenates were centrifuged at 5000 r.p.m. for 5 min to pellet fly debris. The virus was purified from the supernatant by pelleting through a 3 ml 10 % sucrose cushion at 27 000 r.p.m. at 12 °C for 3 h in a SW41 swing-bucket rotor (Beckman). The resuspended virus was layered onto a continuous 10–40 % (w/v) sucrose gradient and centrifuged at 27 000 r.p.m. at 12 °C for 3 h. The virus-containing fractions were harvested, then diluted in 50 mM Tris (pH 7.4) and virus was pelleted by centrifugation at 27 000 r.p.m., at 12 °C for 3 h. The virus was resuspended in 50 mM Tris (pH 7.4) at 4 °C overnight, aliquotted and stored at −20 °C until use. DAVHD virions were also isolated from infected DL2 cells by using a similar method after cells were lysed by repetitive freeze–thawing at −20 °C.
Transmission electron microscopy (TEM) of purified virus.
Purified DAVHD was analysed by TEM as described previously (Johnson & Ball, 2003).
Nuclease treatment.
For nuclease treatment, 1.2 μg DAVHD double-stranded (ds) RNA, prepared in vitro as described previously (Hedges & Johnson, 2008), and Flock House virus (FHV) RNA (Johnson et al., 2001) was incubated at 37 °C with 10 U RNase ONE (Promega) for 45 min. Samples were then diluted in RNA loading buffer (Ambion), heated to 65 °C and separated on a non-denaturing 1 % agarose gel; products were visualized by staining with ethidium bromide.
cDNA synthesis, cloning and sequence determination.
RNA was extracted from sucrose gradient-purified DAVHD virions by using TriReagent (Sigma-Aldrich) as per the manufacturer's instructions, with the addition of 20 μg glycogen to increase RNA recovery. First- and second-strand cDNA was synthesized by using a SuperScript double-stranded cDNA synthesis kit (Invitrogen) with random hexamers as recommended by the manufacturer. The cDNA products were ligated into the EcoRV site of phosphatase-treated pBluescript SK+ (Stratagene) and sequenced. From these sequences, DAV-specific primers were designed and used for further cDNA synthesis using SuperScript reverse transcriptase III (Invitrogen) as per the manufacturer's instructions. 5′ RACE (random amplification of cDNA ends) was performed as indicated by the manufacturer (Invitrogen) to obtain sequence at the 5′ end of the virus genome. A poly(A) tail was added to the 3′ end of purified DAVHD RNA by using poly(A) polymerase (Ambion), and the 3′ RACE system (Invitrogen) was used as per the manufacturer's instructions to clone the 3′ end of the DAV RNA. Nucleotide sequences were assembled and analysed by using dnastar software.
Cells.
Drosophila DL2 cells (Schneider, 1972) were maintained at 26 °C in Schneider's medium (Invitrogen) supplemented with 10 % heat-inactivated fetal bovine serum and antibiotics. DL2 cells were seeded at 7×106 cells per well into six-well tissue-culture plates and left to adhere at 26 °C overnight. Cells were infected with purified DAVHD virions in 500 μl additive-free Schneider's medium or mock-infected and supplemented with 1.5 ml Schneider's complete medium following incubation at 26 °C for 2 h. Infected cells were harvested at 0, 1, 2, 3 and 5 days post-infection (p.i.), washed in PBS and lysed in either 1 ml TriReagent or 50 μl SDS-PAGE loading buffer.
SDS-PAGE and Western blot analysis.
Virus and cell lysate samples were heated at 99 °C in 1× sample buffer for 5 min before electrophoresis through SDS–10 % polyacrylamide gels at 170 V. Alternatively, samples were loaded with sample buffer and a reducing agent on a NuPAGE 4–12 % Bis–Tris gradient gel in MES running buffer (Invitrogen) and electrophoresed at 200 V. Proteins were visualized by staining with Coomassie brilliant blue.
For Western blot analysis, separated proteins were transferred from SDS-PAGE gels to a nitrocellulose membrane and, following blocking, were incubated with a 1/5000 dilution of an anti-DAV antiserum overnight. Polyclonal antisera raised against both the original French DAV isolate (Jousset et al., 1972) and the Australian isolate DAVHD (Christian, 1992) were kindly provided by Dr Peter Christian. The membrane was washed then incubated with a 1/5000 dilution of a monoclonal anti-rabbit IgG conjugated with alkaline phosphatase for 2 h. Following three washes of the membrane in TBST (Tris-buffered saline, Tween 20), bound antibody was visualized with 0.5 % nitro blue tetrazolium and 0.25 % 5-bromo-4-chloro-indolyl phosphate (BCIP) in 1 M Tris buffer (pH 9.0). SeeBlue II+ prestained markers (Invitrogen) were used to allow alignment of proteins between Western blot and SDS-PAGE analysis.
Mass spectrometry.
Peptide-mass fingerprinting and sequence analysis were conducted by using matrix-assisted laser desorption–ionization time-of-flight (MALDI-TOF) and electrospray ion trap (ESI-MS/MS) mass analysis, respectively. Samples were digested with modified trypsin (Promega) directly from solution or as gel pieces after SDS-PAGE. For MALDI-TOF, sinapinic acid was dissolved in 50 % acetonitrile, 0.1 % trifluoroacetic acid (TFA) at half-saturation concentration (saturated sinapinic acid solution was diluted with an equal volume of the solvent) as matrix. A 1 μl aliquot of the DAVHD sample was mixed with 5 μl matrix; 1 μl of the mixed solution was deposited onto a clean plate as one spot and the sample was analysed in a 4700 MALDI TOF/TOF system (Applied Biosystems) and a Bruker Biflex III mass spectrometer. Linear mode was used above 4000 m/z and reflectron mode below this threshold. Liquid chromatography–tandem mass spectrometry (LC-MS/MS) used an Agilent XCT-Ultra 6330 ion-trap mass spectrometer connected to an Agilent 1100 CapLC and ChipCube. Peptides were eluted and loaded onto the analytical capillary column (43 mm×75 μm inner diameter, also packed with 5 μm Zorbax 300SB-C18 particles) connected in line to the mass spectrometer with a flow of 600 nl min−1. Peptides were eluted from a Zorbax 300SB-C18 system (Agilent) with a 5–90 % acetonitrile gradient (0.1 % formic acid) over 10 min. Data-dependent acquisition of collision-induced dissociation MS/MS was utilized. Protein identifications from peptide MS and MS/MS data were based on mascot (Matrix Science) searches of both an in-house database containing all DAV ORFs and the GenBank non-redundant database. Intact protein analysis was performed on a Bruker micrOTOF ESI-TOF after a similar C18 reverse-phase separation. Bruker MaxEnt deconvolution software was used to determine the mass of the capsid protein.
Cryo-electron microscopy (cryoEM) analysis of particles.
DAV was prepared for cryoEM analysis by preservation in vitreous ice over holey carbon films from Protochips (Cflats). Grids were cleaned prior to freezing by using a plasma cleaner (Gatan) using a mixture of 75 % argon and 25 % oxygen for 10 s. A 3 μl aliquot of sample was applied to the grids, which were loaded into an FEI Vitrobot with settings at 4 °C and 100 % humidity. Grids were blotted for 4 s and stored at liquid nitrogen temperatures. An Oxford 3500CT side-entry cryo-stage was used for data collection.
Data were acquired by using a Tecnai F20 Twin transmission electron microscope operating at 120 keV, using a dose of approximately 19 e− Å−2 (0.19 e− nm−2) and a nominal underfocus ranging from 0.5 to 3.0 μm. In total, 2128 images were collected automatically at a nominal magnification of ×80 000 at a pixel size of 0.101 nm at the specimen level. All images were recorded with a Tietz F415 4K×4K pixel CCD camera (15 μm pixel) utilizing the Leginon data-collection software (Suloway et al., 2005). Experimental data were processed with the Appion software package (Lander et al., 2009), which interfaces with the Leginon database infrastructure. The contrast transfer function (CTF) for each micrograph was estimated by using the Automated CTF Estimation (ace) package (Mallick et al., 2005). In total, 29 940 particles were selected automatically from the micrographs by using a template-based particle picker (Roseman, 2003) and extracted at a box size of 512 pixels. Only those particles whose CTF estimation had an ace confidence of 80 % or better were extracted. Phase correction of the single particles was carried out during creation of the particle stack. Stacked particles were binned by a factor of two for the final reconstruction. The final stack contained 22 103 particles, with 17 063 contributing to the final density. The three-dimensional reconstruction was carried out by using the eman reconstruction package (Ludtke et al., 1999). Resolution was assessed by calculating the Fourier shell correlation (FSC) at a cut-off of 0.5, which provided a value of 8.32 Å (0.832 nm) resolution. Calculation of the resolution by measure (Sousa & Grigorieff, 2007) at a 0.5 cut-off yielded a resolution of 8.69 Å (0.869 nm).
RESULTS AND DISCUSSION
DAV is a non-enveloped, icosahedral virus
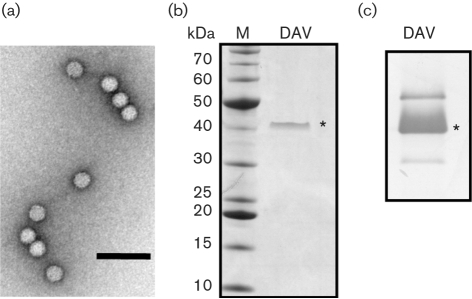
To visualize the morphology of DAV particles, sucrose gradient-purified virions were negatively stained and examined by using TEM. At ×100 000 magnification, icosahedral, non-enveloped particles were observed, with a mean diameter of approximately 30 nm (Fig. 1a). Small projections were visible on the virion surface in a regular array. The overall physical structure observed is consistent with that of other insect RNA viruses (Johnson & Reddy, 1998; Tate et al., 1999). Higher-resolution analysis is discussed in greater detail below.
Fig. 1.
Analysis of DAV virions and structural proteins. (a) Sucrose gradient-purified virions were negatively stained and visualized by TEM at ×100 000 magnification. Bar, 100 nm. (b) Proteins present in purified virus particles were separated by electrophoresis through a 4–12 % Bis–Tris gradient gel. Total protein was visualized by staining with Coomassie blue. The capsid protein is indicated by an asterisk. Sizes of molecular mass markers are shown on the left. (c) Virion proteins separated through an SDS–10 % polyacrylamide gel were transferred to a nitrocellulose membrane and detected with a rabbit polyclonal anti-DAV antibody. The band marked with an asterisk corresponds to the 42 kDa protein indicated in (b).
Structural protein analysis
To visualize DAV protein components, sucrose gradient-purified virions were analysed by using SDS-PAGE (Fig. 1b). A single major protein band was observed and the mass of this protein was estimated to be 42 kDa by comparison with protein molecular mass standards. This suggested that DAVHD has a single major capsid protein of approximately 42 kDa, which is similar to the estimated size of the major structural protein described previously for DAV (Plus et al., 1976). There was no evidence of the two minor proteins of 32 and 73 kDa that were observed when the original French DAV isolate was analysed by Plus et al. (1976). Western blot analysis was performed to analyse the protein components in DAVHD, using an aliquot of the original anti-DAV antiserum that was raised against the French DAV isolate (Jousset et al., 1972). Three distinct bands were detected by this analysis (Fig. 1c). The intense band indicated by the asterisk (Fig. 1c) migrates at a similar position to the 42 kDa protein observed in SDS-PAGE, showing that the 42 kDa protein is recognized by the original DAV antiserum. Two additional bands were detected at lower intensity and are observed consistently in Western blot analyses of DAVHD virions. The molecular masses of the minor proteins were estimated to be 50 and 36 kDa for the larger and smaller bands, respectively. Whilst these proteins were not observed in the SDS-PAGE gel shown in Fig. 1(b), they were seen by SDS-PAGE analysis when virus was overloaded (data not shown). It is possible that these additional proteins are analogous to the two minor proteins described by Plus et al. (1976), although the difference in size of the larger protein is difficult to reconcile. Unfortunately, no virus was recovered following passage of two different samples of the original French DAV isolate (which had been stored at −20 °C for several decades) either in flies or in cell culture, so it was not possible to make direct comparisons between the isolates.
DAV replicates in cell culture
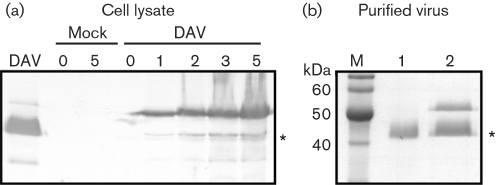
To investigate whether DAV replication and assembly occur in cell culture, Drosophila DL2 cells were infected with DAVHD and accumulation of capsid protein in cells was analysed by Western blotting (Fig. 2a). Proteins that co-migrated with each of the three proteins identified in purified virions were also detected in infected cells. No proteins were detected either in the mock-infected cells or in the sample taken at time zero, the latter confirming that the input virus was below the level of detection in this assay. All three DAV proteins were detected 1 day p.i. (Fig. 2a) and the quantity of the proteins increased over time, indicating that virus protein synthesis was occurring in cell culture. However, the ratio of the 50 and 42 kDa proteins was reversed compared with that observed in the purified virion sample (Fig. 1b). To compare the ratio of proteins in virus derived from cell culture with those in virus derived from flies, virions were purified from both DL2 cells and Drosophila by using the same method and the resulting samples were analysed by SDS-PAGE (Fig. 2b). The virus sample purified from DL2 cells contained two major proteins of similar abundance and the sizes corresponded to the 50 and 42 kDa proteins typically seen in Western blot analysis (Fig. 2b, lane 2). In contrast, virus purified from flies, as noted previously, contained only a single major 42 kDa protein (Fig. 2b, lane 1). The sucrose gradient-purified virus from cultured DL2 cells was shown to be infectious in naïve DL2 cells (data not shown).
Fig. 2.
Detection of virus protein synthesis in infected Drosophila DL2 cells. (a) DL2 cells were either mock-infected or infected with DAV and incubated at 26 °C until harvesting. Cell samples were taken at 0, 1, 2, 3 and 5 days p.i. and lysed with sample buffer. Cellular proteins were separated by electrophoresis through an SDS–10 % polyacrylamide gel, transferred to a nitrocellulose membrane and incubated with an anti-DAV rabbit polyclonal antiserum to detect virus proteins. The asterisk indicates the 42 kDa capsid protein. Virus proteins were detected at 1 day p.i. and their levels increased over time. No DAV-specific bands were detected in the mock-infected samples. (b) Proteins from DAVHD virions purified from infected DL2 cells (lane 2) and adult Drosophila (lane 1) were electrophoresed through an SDS–10 % polyacrylamide gel stained with Coomassie blue. The asterisk indicates the 42 kDa capsid protein.
The relationship between the capsid proteins was not clear from these data. However, the disappearance of the 50 kDa protein and correlated increase in the 42 kDa protein is reminiscent of the cleavage of a capsid protein precursor. Furthermore, a small protein, estimated to be 6 kDa in mass, was observed on SDS-PAGE gels overloaded for the major structural protein (Fig. 3). Many viruses encode their capsid proteins in a precursor polyprotein. Within the insect single-stranded (ss) RNA viruses, both nodaviruses and tetraviruses produce a capsid precursor protein that is cleaved maturationally toward the C terminus to produce the two mature capsid proteins (Johnson & Reddy, 1998). One interpretation of the DAV capsid protein profile is that the 50 kDa protein is a capsid precursor protein, which is cleaved during maturation of the virion to produce the major 42 kDa capsid protein. If this is the case for DAV, then the higher abundance of the 50 kDa protein in virus from DL2 cells may be due to incomplete cleavage of the precursor protein during maturation. However, in contrast to nodaviruses and tetraviruses, no asparagine residue was identified in the DAV sequence as the potential cleavage site. Further analysis will be required to determine the relationship between these proteins. The 36 kDa protein may initiate from the second AUG of the capsid ORF, as is observed in viruses of the family Nodaviridae (Johnson & Ball, 2003).
Fig. 3.
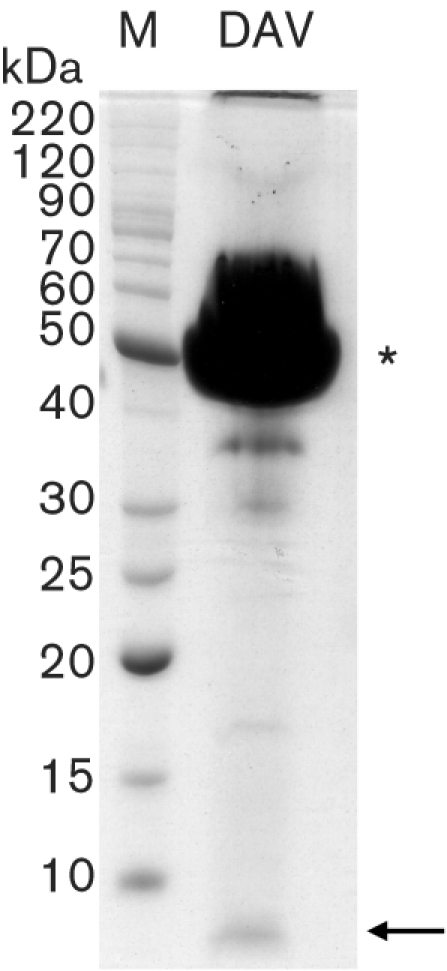
Identification of a small structural protein. Virion proteins were overloaded and separated by electrophoresis through an SDS–10 % polyacrylamide gel. Total protein was stained with Coomassie blue. The asterisk indicates the 42 kDa capsid protein band that is consistently observed in SDS-PAGE analyses. The arrow indicates the small protein band that is estimated to be approximately 6 kDa.
Genome analysis
To analyse the nucleic acid encapsidated in virions, RNA extracted from sucrose gradient-purified DAV was resolved on a 1 % agarose gel and nucleic acids were visualized by staining with ethidium bromide (Fig. 4). This showed a smear of RNA, which spanned sizes of approximately 1000–5000 nt compared with ssRNA markers; this size range was confirmed by analysis on a denaturing formaldehyde agarose gel (data not shown). This suggested that the RNA was degraded during the extraction and electrophoresis processes, although the distinct edges of the smear and intact markers indicate that there was no RNase contamination in the electrophoresis process. The RNA smear was observed repeatedly, even when FHV RNA extracted concurrently with the DAV sample was shown to be intact (Fig. 4). An alternative RNA-extraction kit (Invitrogen) also did not improve the integrity of the RNA (data not shown). Degradation of RNA purified from tetravirus particles is common (Gordon & Hanzlik, 1998; Gordon et al., 1999; Pringle et al., 2003) and it is possible that removal of the genomic RNA from the virion results in RNA degradation. We cannot discount other possible explanations for the apparent diversity in size of the encapsidated RNA. However, despite the smearing, an approximation of the maximum RNA size was inferred from the distinct upper size cut-off of 5000 nt.
Fig. 4.
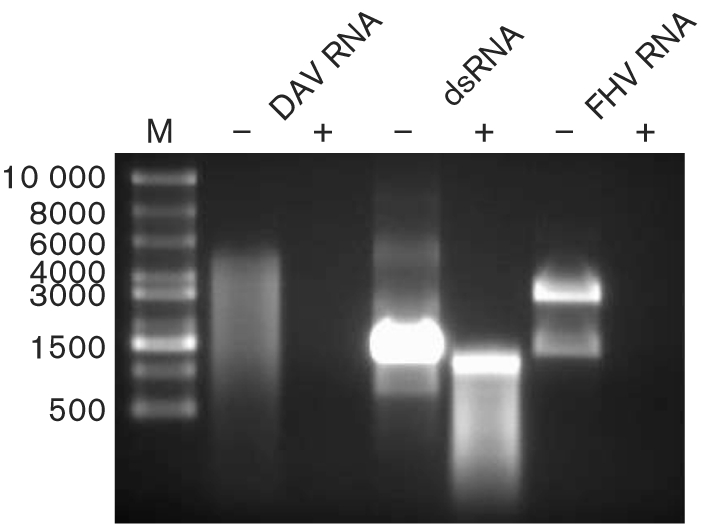
Analysis of DAV genomic nucleic acid. RNA was extracted from purified DAVHD virions and mock-treated (−) or treated with RNase ONE (Promega) (+), which degrades ssRNA only. As controls, a synthetic dsRNA product from DCV and the ssRNA genome from FHV were treated similarly. Following nuclease treatment, products were separated on a 1 % agarose gel and visualized by ethidium bromide staining. Sizes of the ssRNA markers (M; in nt) are indicated on the left.
Nucleic acid was extracted from sucrose gradient-purified DAV and treated with RNase, resulting in degradation of the DAV genome (Fig. 4). Under the same conditions, the genome of the ssRNA virus FHV was also degraded, whereas a synthetic dsRNA transcribed in vitro remained intact (Fig. 4). This suggests that the genome of DAV is most likely to be ssRNA.
By using a combination of approaches, clones corresponding to genomic RNA were synthesized and 4806 nt of genome sequence was determined. The sequences of all of the recovered clones formed a contiguous sequence, suggesting that DAV has a single genomic RNA. The 5′ and 3′ ends of the genomic RNA were targeted by using RACE. Two independent clones were identified with the same 5′ end; however, it is possible that the genome includes additional nucleotides at the 5′ terminus. Interestingly, 3′ RACE was only successful when a synthetic poly(A) tail was added to the DAVHD RNA, indicating that the DAV genome is not polyadenylated.
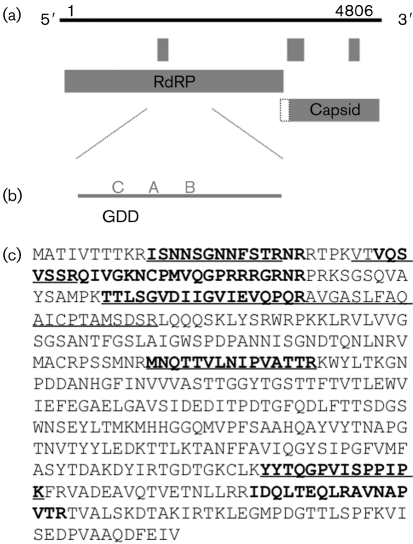
The genomic sequence was analysed for ORFs. The plus-sense strand encoded two major ORFs and there were an additional three small ORFs encoding potential proteins containing >50 aa (Fig. 5). The 5′-proximal ORF was the largest, encoding a putative protein of 1075 aa with a predicted molecular mass of 121 kDa. The 3′-proximal ORF encoded 442 codons, which are predicted to comprise a protein of 48.4 kDa. The second ORF initiates from a methionine that is 43 nt downstream of the first ORF stop codon and is in the +1 reading frame compared with the first. The 3′ ORF overlaps the 5′ coding region upstream of the initiating methionine (Fig. 5a). It is therefore possible that a +1 frameshift would allow synthesis of a polyprotein including both large ORFs. However, +1 frameshifts are uncommon and it seems unlikely that the capsid proteins would be translated in this inefficient manner.
Fig. 5.
Organization of the DAV genome. (a) The 4806 nt DAV genome encodes two major ORFs and three additional small ORFs that contain >50 aa. The ORFs as shown start with an AUG codon; the extension (dotted line) on the capsid ORF indicates that this ORF overlaps the RdRP ORF, but the first methionine is downstream of the RdRP ORF. (b) Analysis of the 5′-proximal ORF showed high similarity of DAV RdRP regions to those of members of the families Tetraviridae and Birnaviridae. These viruses have a permuted RdRP, with the GDD region upstream of other palm subdomains. DAV RdRP also showed this permuted arrangement, as indicated by C–A–B. (c) MS analysis indicates that the 3′-proximal large ORF encodes the capsid protein. The deduced sequence of the capsid protein is shown and the peptides identified by MALDI-TOF (in bold type) and LC-MS/MS (underlined) are indicated.
To identify proteins with sequence similarity to the deduced sequence of the 5′-proximal ORF, the translated sequence was used to search the non-redundant sequence databases by using blast. Significant matches were found to a large region of the RdRP proteins from two members of the family Tetraviridae and several members of the family Birnaviridae. The highest similarity was found to the RdRPs of Thosea asigna virus (TaV) and Euprosterna elaeasa virus (EeV), which are closely related viruses from the family Tetraviridae. TaV and EeV both have an unusual permutation in the palm domain of the RdRP (Gorbalenya et al., 2002). The nearly ubiquitous arrangement that is observed in RNA viruses is that the GDD box (motif C) of the palm subdomain follows the conserved A and B motifs (A–B–C) (Poch et al., 1989). However, in both TAV and EeV RdRPs, the C motif precedes the others in a C–A–B arrangement (Fig. 5b). This permuted RdRP is also observed in some dsRNA viruses from the family Birnaviridae, and structural analysis of the permuted RdRP has shown a similar arrangement to the canonical RdRP of other viruses (Garriga et al., 2007; Gorbalenya et al., 2002). Interestingly, the RdRP of DAV also contains this C–A–B permutation (see Supplementary Fig. S1, available in JGV Online).
To identify the structural protein-coding region, the sequence of the virion proteins was analysed. N-terminal sequencing of the structural protein was unsuccessful both from gel-purified protein and from whole virus, suggesting that the protein is blocked (data not shown). Protease mass mapping of virions in solution and of the 42 kDa capsid protein band after SDS-PAGE was tried next. By using a combination of MALDI-TOF and LC-MS/MS, seven and six peptides, respectively, spanning 29 % of the 3′ ORF were identified, indicating that this ORF encodes the capsid protein (Fig. 5c). The predicted mass of 48.4 kDa is also consistent with the precursor protein band visualized on the protein gels (50 kDa). LC-MS/MS of purified particles produced a signal for the mature capsid protein of 40 480±3 Da. This does not match with a single cleavage event leading to capsid protein maturation as in insect nodaviruses and tetraviruses, both of which occur autocatalytically at an asparagine residue (Johnson & Reddy, 1998). The fact that the mature protein mass does not match the amino acid sequence is, however, consistent with the N terminus being modified as discussed above; this will be investigated in future experiments.
Sequence-based fold recognition of the capsid protein
A blast search of the capsid protein's amino acid sequence against known insect virus genomes revealed a single homologue in the family Nodaviridae: 27 % identity was exhibited to the Wuhan nodavirus between residues 330 and 442. For further sequence-based analyses of the capsid structure, various fold-recognition methods were utilized via the structure prediction meta server (Ginalski et al., 2003), revealing a folding motif consistent with several members of the families Tombusviridae [carnation mottle virus (CMV), tobacco necrosis virus, tomato bushy stunt virus (TBSV)] and Sobemoviridae (cocksfoot mottle virus, rice yellow mottle virus, southern bean mosaic virus) (see Supplementary Fig. S2, available in JGV Online). The results point to a conserved canonical β-sandwich fold beginning around residue 74 and ending around residue 236. Two viruses from the family Tombusviridae, CMV and TBSV, whose structures contain a C-terminal domain that protrudes from the capsid surface, additionally exhibited a fold similarity to the DAV sequence between residues 270 and 315. Conceivably, the N-terminal region of DAV forms an evolutionarily conserved β-sandwich domain while the C-terminal domain forms a surface extension, much like the structures of CMV and TBSV. Indeed, cryoEM analysis of the Malabaricus grouper nervous necrosis virus (MGNNV), a fish nodavirus that appears to display a similar domain organization upon sequence-based fold recognition (Tang et al., 2002), revealed a C-terminal domain that was separated from the β-sandwich domain that formed the core capsid of the virus particle. The N-terminal portion of the sequence is highly charged, including many arginines, which indicates that it may interact with the RNA and have a role in particle assembly (Tang et al., 2001; Venter et al., 2009). A closer look at the structure of DAV particles via cryoEM methods was necessary to elucidate the overall structural morphology.
CryoEM analysis of DAV particles
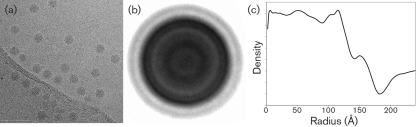
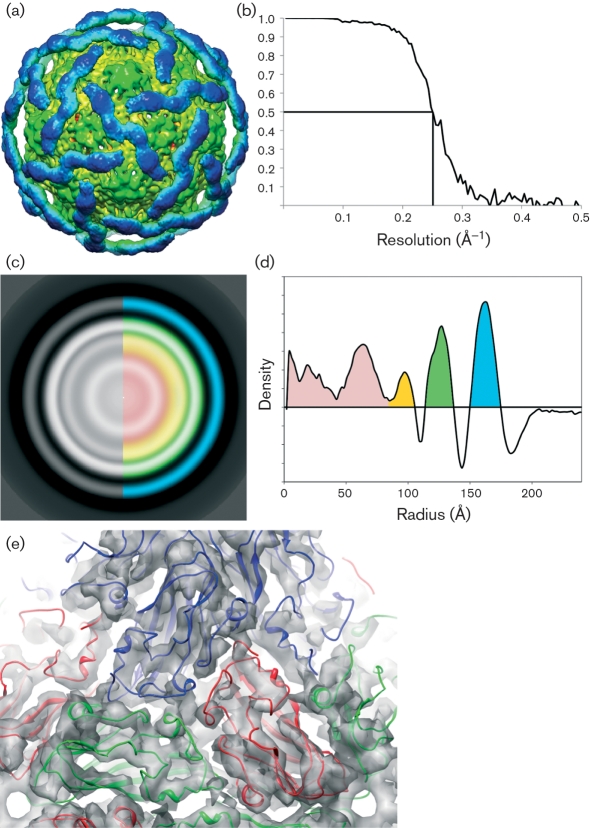
The structure of DAV was examined in detail by cryoEM image-reconstruction techniques. Interestingly, unprocessed, high-contrast cryoEM images of the virus particles (Fig. 6a) exhibited a structural morphology that was very reminiscent of MGNNV TEM images, whose radial density distribution shows two distinct protein shells (Tang et al., 2002). Averaging of the computationally extracted and centred DAV particles from the micrographs confirms that the virus indeed appears to have a divided capsid shell (Fig. 6b). From the selected particles, a three-dimensional structure was computed to 8 Å (0.8 nm) resolution as estimated by the FSC method, using a value of 0.5 (Fig. 7a, b).
Fig. 6.
CryoEM analysis of DAV virions. (a) Representative electron micrograph of sucrose gradient-purified DAV in the frozen–hydrated state. (b) Averaged two-dimensional density corresponding to 22 000 centred raw particles. (c) One-dimensional radial plot of the averaged two-dimensional particles, showing discrete peaks for the capsid layers.
Fig. 7.
Analysis of three-dimensional reconstruction of DAV particles. (a) Surface representation of the DaV cryoEM reconstruction contoured at 2σ. The inner N-terminal domain (green) is well-ordered with visible secondary-structural features, whereas the outer C-terminal domain (blue) contains only very-low-resolution features. (b) Estimated resolution of the reconstruction is 8 Å (0.8 nm) according to FSC criteria (y-axis) at 0.5. (c) Radially averaged slice through the centre of reconstruction. Blue, C-terminal domain; green, N-terminal domain; orange and red, RNA. (d) Plot showing density by radius [colours as in (c)]. (e) Docking of the β-sandwich domain of the TBSV crystal structure into the N-terminal domain of the DaV cryoEM density.
At these resolutions, the distribution of the capsid protein into two discrete and separate domains is plainly evident. Viewed down the fivefold axis, the density distribution of the reconstructed particle shows the core domain extending from a radius of 115 to 138 Å (11.5–13.8 nM) with a maximum at 127 Å (12.7 nm), whilst the outer domain has a radius of 150–175 Å (15.0–17.5 nm) with a maximum at 163 Å (16.3 nm) (Fig. 7c, d). These dimensions are similar to those of the MGNNV cryoEM structure, with two additional layers of density corresponding to RNA, the outermost of which appears to interact directly with the capsid shell. Examination of the inner domain of the DAV electron density reveals an unmistakable T=3 quasi-equivalent lattice, with many of the secondary-structural elements visible. Comparison of the core domain with the β-sandwich domain of TBSV shows a very similar structural organization, and was docked into place by using an automated real-space rigid body refinement (Fig. 7e). The docking shows that the network of subunit–subunit interactions that make up the capsid is highly conserved, and that capsid assembly in DAV is likely to be similar to that of TBSV and other tombusviruses. Although tombusviruses contain two distinct domains, it is noteworthy that they are not separated by the 10 Å (1.0 nm) gap present in the DAV structure. TBSV has a short polypeptide hinge connecting the N- and C-terminal domains, and it is probable that a similar, although much longer, hinge is seen in DAV.
An unanticipated finding is the lack of highly ordered structure corresponding to the outer domain. Whilst the inner domain shows a detailed T=3 organization, the outer domain exhibits no such lattice, but only ambiguous densities that appear to be an artefact of applying icosahedral symmetry to a disordered or dynamic protein domain. Despite a lack of quasi-symmetry, however, the outer domain appears to be somewhat ordered, forming a cage-like icosahedral lattice that surrounds the core domain. The densities attributed to this dynamic C-terminal domain appear to converge above the icosahedral and quasi-twofold axes. In this regard, DAV diverges from the structure of MGNNV, whose outer domain consists of large protrusions of density at the quasi-threefold axis.
Attempts to focus on this outer domain by masking out the interior region during the refinement did not provide any further insight into its organization, as the resulting densities were equally ambiguous, with little deviation from the map containing the N-terminal portion. Due to the disorganization and lack of quasi-equivalent symmetry present in the C-terminal region of density, fitting of the protruding domain (residues 272–387) of TBSV was not modelled into the map.
The β-sandwich that makes up the inner core of many viruses sets up a solid structural template from which other domains protrude. These protrusions are generally involved in biologically relevant processes, such as host-cell recognition, and it has been shown that in some viruses, such as rotavirus (Dormitzer et al., 2004), these domain extensions are dynamic and can undergo rearrangements during the virus life cycle. The cryoEM work shown here provides evidence that DAV may be one such virus, whose dynamic outer domains, with possible biological function, are anchored to a well-ordered, β-sandwich icosahedral core.
Conclusions
DAV was first described as a picorna-like virus. The data presented here show that DAV is related evolutionarily to the viruses with permuted RdRP core motifs in the families Tetraviridae and Birnaviridae. However, the capsid protein shows strongest sequence similarity to those of viruses in the family Nodaviridae and conserved protein-folding motifs with the members of the family Tombusviridae. In addition, the structure of DAV particles shows an inner T=3 core surrounded by a less-well-defined outer protein domain. Taken together, these features show that DAV is an unusual RNA virus with evolutionary relationships across a diverse range of invertebrate, fish and plant viruses.
Supplementary Material
Acknowledgments
We thank Peter Christian for providing virus and antisera and for discussions, Darren Obbard (University of Edinburgh, UK) and Peter Christian for sharing unpublished data and Lopson Kebapetswe and Ivy Mui Tan for fly injections. This research was supported by a University of Queensland development grant to K. N. J. Electron microscopic imaging and reconstruction were conducted at the National Resource for Automated Molecular Microscopy, which is supported by the NIH through the National Center for Research Resources P41 Program (RR17573). B. B. acknowledges support from the National Science Foundation, MCB 0646499 and NIH grant no. P20 RR-16455-06 from the Centers of Biomedical Research Excellence (COBRE) Program of the National Center For Research Resources. B. B. also receives support from the Center for Bio-Inspired Nanomaterials (Office of Naval Research grant no. N00014-06-01-1016). We also thank the Murdock Charitable Trust for support of the Mass Spectrometry Facility at Montana State University.
Footnotes
The GenBank/EMBL/DDBJ accession number for the DAV genome sequence reported in this paper is FJ150422.
Two supplementary figures, showing alignments of the RdRP and the deduced capsid protein sequences of DAV with those of other RNA viruses, are available with the online version of this paper.
References
- Agrawal, D. K. & Johnson, J. E. (1992). Sequence and analysis of the capsid protein of Nudaurelia capensis ω virus, an insect virus with T=4 icosahedral symmetry. Virology 190, 806–814. [DOI] [PubMed] [Google Scholar]
- Brun, G. & Plus, N. (1980). The viruses of Drosophila. In The Genetics and Biology of Drosophila, pp. 625–702. Edited by M. Ashburner & T. F. R. Wright. New York: Academic Press.
- Cherry, S. & Perrimon, N. (2004). Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat Immunol 5, 81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, S., Doukas, T., Armknecht, S., Whelan, S., Wang, H., Sarnow, P. & Perrimon, N. (2005). Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev 19, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian, P. D. (1992). A simple vacuum dot-blot hybridisation assay for the detection of Drosophila A and C viruses in single Drosophila. J Virol Methods 38, 153–165. [DOI] [PubMed] [Google Scholar]
- Christian, P. D. & Scotti, P. D. (1998). The picorna-like viruses of insects. In The Viruses: Insect Viruses II, pp. 301–336. Edited by L. K. Miller & L. A. Ball. New York: Plenum.
- Christian, P. D., Carstens, E. B., Domier, L., Johnson, J. E., Johnson, K. N., Nakashima, N., Scotti, P. D. & van der Wilk, F. (2005). Dicistroviridae. In Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses, pp. 783–788. Edited by C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger & L. A. Ball. San Diego, CA: Elsevier Academic Press.
- Deddouche, S., Matt, N., Budd, A., Mueller, S., Kemp, C., Galiana-Arnoux, D., Dostert, C., Antoniewski, C., Hoffmann, J. A. & Imler, J. L. (2008). The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in Drosophila. Nat Immunol 9, 1425–1432. [DOI] [PubMed] [Google Scholar]
- Ding, S. W. & Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormitzer, P. R., Nason, E. B., Prasad, B. V. & Harrison, S. C. (2004). Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 430, 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert, C., Jouanguy, E., Irving, P., Troxler, L., Galiana-Arnoux, D., Hetru, C., Hoffmann, J. A. & Imler, J. L. (2005). The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat Immunol 6, 946–953. [DOI] [PubMed] [Google Scholar]
- Fauquet, C. M., Mayo, M. A., Maniloff, J., Desselberger, U. & Ball, L. A. (editors) (2005). Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Academic Press.
- Galiana-Arnoux, D., Dostert, C., Schneemann, A., Hoffmann, J. A. & Imler, J. L. (2006). Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat Immunol 7, 590–597. [DOI] [PubMed] [Google Scholar]
- Garriga, D., Navarro, A., Querol-Audi, J., Abaitua, F., Rodriguez, J. F. & Verdaguer, N. (2007). Activation mechanism of a noncanonical RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A 104, 20540–20545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gateff, E., Gissmann, L., Shrestha, R., Plus, N., Pfister, H., Schröder, J. & Zur Hausen, H. (1980). Characterization of two tumorous blood cell lines of Drosophila melanogaster and the viruses they contain. In Invertebrate Systems In Vitro, pp. 517–533. Edited by E. Kurstak, K. Maramorosch & F. Dübendorfer. Amsterdam: Elsevier/North-Holland Biomedical Press.
- Ginalski, K., Elofsson, A., Fischer, D. & Rychlewski, L. (2003). 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics 19, 1015–1018. [DOI] [PubMed] [Google Scholar]
- Gorbalenya, A. E., Pringle, F. M., Zeddam, J. L., Luke, B. T., Cameron, C. E., Kalmakoff, J., Hanzlik, T. N., Gordon, K. H. & Ward, V. K. (2002). The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol 324, 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, K. H. J. & Hanzlik, T. N. (1998). Tetraviruses. In The Viruses: Insect Viruses II, pp. 269–299. Edited by L. K. Miller & L. A. Ball. New York: Plenum.
- Gordon, K. H. J., Johnson, K. N. & Hanzlik, T. N. (1995). The larger genomic RNA of Helicoverpa armigera stunt tetravirus encodes the viral RNA polymerase and has a novel 3′-terminal tRNA-like structure. Virology 208, 84–98. [DOI] [PubMed] [Google Scholar]
- Gordon, K. H., Williams, M. R., Hendry, D. A. & Hanzlik, T. N. (1999). Sequence of the genomic RNA of Nudaurelia beta virus (Tetraviridae) defines a novel virus genome organization. Virology 258, 42–53. [DOI] [PubMed] [Google Scholar]
- Habayeb, M. S., Ekengren, S. K. & Hultmark, D. (2006). Nora virus, a persistent virus in Drosophila, defines a new picorna-like virus family. J Gen Virol 87, 3045–3051. [DOI] [PubMed] [Google Scholar]
- Hanzlik, T. N., Dorrian, S. J., Johnson, K. N., Brooks, E. M. & Gordon, K. H. J. (1995). Sequence of RNA2 of the Helicoverpa armigera stunt virus (Tetraviridae) and bacterial expression of its genes. J Gen Virol 76, 799–811. [DOI] [PubMed] [Google Scholar]
- Hartley, C. J., Greenwood, D. R., Gilbert, R. J., Masoumi, A., Gordon, K. H., Hanzlik, T. N., Fry, E. E., Stuart, D. I. & Scotti, P. D. (2005). Kelp fly virus: a novel group of insect picorna-like viruses as defined by genome sequence analysis and a distinctive virion structure. J Virol 79, 13385–13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges, L. M. & Johnson, K. N. (2008). The induction of host defence responses by Drosophila C virus. J Gen Virol 89, 1497–1501. [DOI] [PubMed] [Google Scholar]
- Hedges, L. M., Brownlie, J. C., O'Neill, S. L. & Johnson, K. N. (2008). Wolbachia and virus protection in insects. Science 322, 702. [DOI] [PubMed] [Google Scholar]
- Huszar, T. & Imler, J. L. (2008). Drosophila viruses and the study of antiviral host-defense. Adv Virus Res 72, 227–265. [DOI] [PubMed] [Google Scholar]
- Johnson, K. N. & Ball, L. A. (2003). Virions of Pariacoto virus contain a minor protein translated from the second AUG codon of the capsid protein open reading frame. J Gen Virol 84, 2847–2852. [DOI] [PubMed] [Google Scholar]
- Johnson, K. N. & Christian, P. D. (1998). The novel genome organization of the insect picorna-like virus Drosophila C virus suggests this virus belongs to a previously undescribed virus family. J Gen Virol 79, 191–203. [DOI] [PubMed] [Google Scholar]
- Johnson, J. E. & Reddy, V. (1998). Structural studies of nodaviruses and tetraviruses. In The Insect Viruses, pp. 171–223. Edited by L. K. Miller & L. A. Ball. New York: Plenum.
- Johnson, K. N., Johnson, K. L., Dasgupta, R., Gratsch, T. & Ball, L. A. (2001). Comparisons among the larger genome segments of six nodaviruses and their encoded RNA replicases. J Gen Virol 82, 1855–1866. [DOI] [PubMed] [Google Scholar]
- Jousset, F. X. & Plus, N. (1975). Étude de la transmission horizontale et de la transmission verticale des picornavirus de Drosophila melanogaster et de Drosophila immigrans. Ann Microbiol (Paris) 126, 231–249 (in French). [PubMed] [Google Scholar]
- Jousset, F. X., Plus, N., Croizier, G. & Thomas, M. (1972). Existence chez Drosophila de deux groupes de picornavirus de propriétés sérologiques et biologiques différentes. C R Acad Sci Hebd Seances Acad Sci D 275, 3043–3046 (in French). [PubMed] [Google Scholar]
- Koonin, E. V., Wolf, Y. I., Nagasaki, K. & Dolja, V. V. (2008). The big bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat Rev Microbiol 6, 925–939. [DOI] [PubMed] [Google Scholar]
- Lander, G. C., Stagg, S., Voss, N. R., Cheng, A., Fellmann, D., Pulokas, J., Yoshioka, C., Irving, C., Mulder, A. & other authors (2009). Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke, S. J., Baldwin, P. R. & Chiu, W. (1999). eman: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128, 82–97. [DOI] [PubMed] [Google Scholar]
- Mallick, S. P., Carragher, B., Potter, C. S. & Kriegman, D. J. (2005). ace: automated CTF estimation. Ultramicroscopy 104, 8–29. [DOI] [PubMed] [Google Scholar]
- Plus, N. (1978). Endogenous viruses of Drosophila melanogaster cell lines: their frequency, identification and origin. In Vitro Cell Dev Biol Plant 14, 1015–1021. [Google Scholar]
- Plus, N., Croizier, G., Jousset, F. X. & David, J. (1975). Picornaviruses of laboratory and wild Drosophila melanogaster: geographical distribution and serotypic composition. Ann Microbiol (Paris) 126, 107–117. [PubMed] [Google Scholar]
- Plus, N., Croizier, G., Veyrunes, J. & David, J. (1976). A comparison of buoyant density and polypeptides of Drosophila P, C and A viruses. Intervirology 7, 346–350. [DOI] [PubMed] [Google Scholar]
- Poch, O., Sauvaget, I., Delarue, M. & Tordo, N. (1989). Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8, 3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle, F. M., Kalmakoff, J. & Ward, V. K. (2001). Analysis of the capsid processing strategy of Thosea asigna virus using baculovirus expression of virus-like particles. J Gen Virol 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Pringle, F. M., Johnson, K. N., Goodman, C. L., McIntosh, A. H. & Ball, L. A. (2003). Providence virus: a new member of the Tetraviridae that infects cultured insect cells. Virology 306, 359–370. [DOI] [PubMed] [Google Scholar]
- Roseman, A. M. (2003). Particle finding in electron micrographs using a fast local correlation algorithm. Ultramicroscopy 94, 225–236. [DOI] [PubMed] [Google Scholar]
- Roxstrom-Lindquist, K., Terenius, O. & Faye, I. (2004). Parasite-specific immune response in adult Drosophila melanogaster: a genomic study. EMBO Rep 5, 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatier, L., Jouanguy, E., Dostert, C., Zachary, D., Dimarcq, J. L., Bulet, P. & Imler, J. L. (2003). Pherokine-2 and -3. Eur J Biochem 270, 3398–3407. [DOI] [PubMed] [Google Scholar]
- Schneider, I. (1972). Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol 27, 353–365. [PubMed] [Google Scholar]
- Sousa, D. & Grigorieff, N. (2007). Ab initio resolution measurement for single particle structures. J Struct Biol 157, 201–210. [DOI] [PubMed] [Google Scholar]
- Suloway, C., Pulokas, J., Fellmann, D., Cheng, A., Guerra, F., Quispe, J., Stagg, S., Potter, C. S. & Carragher, B. (2005). Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60. [DOI] [PubMed] [Google Scholar]
- Tang, L., Johnson, K. N., Ball, L. A., Lin, T., Yeager, M. & Johnson, J. E. (2001). The structure of Pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat Struct Biol 8, 77–83. [DOI] [PubMed] [Google Scholar]
- Tang, L., Lin, C. S., Krishna, N. K., Yeager, M., Schneemann, A. & Johnson, J. E. (2002). Virus-like particles of a fish nodavirus display a capsid subunit domain organization different from that of insect nodaviruses. J Virol 76, 6370–6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate, J., Liljas, L., Scotti, P., Christian, P., Lin, T. & Johnson, J. E. (1999). The crystal structure of cricket paralysis virus: the first view of a new virus family. Nat Struct Biol 6, 765–774. [DOI] [PubMed] [Google Scholar]
- Teixeira, L., Ferreira, A. & Ashburner, M. (2008). The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol 6, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wilk, F., Dullemans, A. M., Verbeek, M. & Van den Heuvel, J. F. (1997). Nucleotide sequence and genomic organization of Acyrthosiphon pisum virus. Virology 238, 353–362. [DOI] [PubMed] [Google Scholar]
- van Rij, R. P., Saleh, M. C., Berry, B., Foo, C., Houk, A., Antoniewski, C. & Andino, R. (2006). The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 20, 2985–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, P. A., Marshall, D. & Schneemann, A. (2009). Dual roles for an arginine-rich motif in specific genome recognition and localization of viral coat protein to RNA replication sites in flock house virus-infected cells. J Virol 83, 2872–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon, R. A., Nandakumar, M., Vakharia, V. N. & Wu, L. P. (2005). The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci U S A 102, 7257–7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.