Abstract
We hereby report studies that suggest a role for serum exosomes in the anchorage independent growth (AIG) of tumor cells. In AIG assays, fetal bovine serum is one of the critical ingredients. We therefore purified exosomes from fetal bovine serum and examined their potential to promote growth of breast carcinoma cells in soft agar and Matrigel after reconstituting them into growth medium (EEM). In all the assays, viable colonies were formed only in the presence of exosomes. Some of the exosomal proteins we identified, have been documented by others and could be considered exosomal markers. Labeled purified exosomes were up-taken by the tumor cells, a process that could be competed out with excess unlabeled vesicles. Our data also suggested that once endocytosed by a cell, the exosomes could be recycled back to the conditioned medium from where they can be up-taken by other cells. We also demonstrated that low concentrations of exosomes activate MAP kinases, suggesting a mechanism by which they maintain the growth of the tumor cells in soft agar. Taken together, our data demonstrate that serum exosomes form a growth promoting platform for AIG of tumor cells and may open a new vista into cancer cell growth in vivo.
Keywords: Breast, Carcinoma, Exosomes, Anchorage, Growth, MAP Kinase, Soft Agar, Matrigel
Introduction
The in vitro two dimensional growth of cultured cells including tumor cells is well characterized, particularly the growth signals emanating from the integrin/extracellular matrix interaction [1]. For a number of years, ideas have bounced back and forth as to whether the growth signaling mechanisms in two-dimensional tissue culture can be extrapolated to the growth cues in vivo [2]. A number of in vitro culture systems have been developed including organotypic culture system, that takes into account the three-dimensional in vivo growth of tumor cells [3]. Suffice to say, the in vivo growth of tumor cells is complex because not only is it three-dimensional in character, there are numerous variables including stromal factors that affect growth [4]. Apart from the limitations imposed on it, the anchorage independent growth of tumor cells in soft agar bears the closest resemblance to the in vivo growth when compared to the two-dimensional growth on plastic [5].
Serum, particularly fetal bovine serum is routinely used to supplement cell growth media. Serum has a number of adhesion or attachment factors which allow the cells to adhere and spread on the bottom of the culture flask to initiate growth signals. Apart from the well characterized adhesion proteins such as fibronectin, laminin, and collagen [6], serum also contains a myriad of factors whose role in cell growth is either defined or yet to be determined [7]. There are over 1,000 proteins in serum, where some are at nanomolar or lower concentrations and others at micro molar or higher concentrations. In studies designed to measure the contribution of a particular growth factor, it is customary to use serum free medium supplemented with graded doses of the protein or factor. Such studies have been done to determine the growth factor most suited to support anchorage independent growth [8]. Interestingly, in majority of such studies, in addition to the growth factor of interest, the medium was also supplemented with fetal bovine serum [9]. Serum proteins appear to be particularly important requirement for anchorage independent growth. We have made an effort in the present study to identify the protein or group of proteins “platform” associated with anchorage independent growth in serum.
We hypothesized that fetuin-A or a group of proteins which form a complex with or co-purify with it is/are responsible for anchorage independent growth in serum. We had demonstrated that fetuin-A is one of the serum proteins that could support anchorage independent growth in breast carcinoma cells [10]. Unfortunately fetuin-A easily associates with other serum proteins making it difficult to single it out as the principal driver of growth. Recent studies indicated that exosomes (nanovesicles) purified from HIV infected monocyte-derived macrophages and urine, contain fetuin-A among other serum proteins [11, 12]. We hereby present data that demonstrate that serum proteins or factors (including fetuin-A) that confer anchorage independent growth are associated with exosomes.
Materials and Methods
Cells: Breast carcinoma cell lines MDA-MB-435, and a sub-clone of BT-549 stably transfected with galectin-3 and re-named BT-549Gal-3, were kindly donated by Dr. Avraham Raz, Karmanos Cancer Research Institute. MCF-10A and MDA-MB-231, were purchased from ATCC. The cells were routinely cultured in DMEM/F12 supplemented with essential and non-essential amino acids, 100 μg/ml penicillin-streptomycin, 2.5 μg/ml Fungizone, 20 ng/ml epidermal growth factor, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, 98 ng/ml cholera toxin, and 10% heat inactivated fetal bovine serum at 37°C, 5% CO2, in a humidified incubator. GFP-annexin-II expression plasmid was kindly donated to us by Dr Carl E. Creutz (University of Virginia). All the other reagents unless otherwise stated were purchased from Sigma (St Louis, MO).
Exosome Purification Protocol
To purify exosomes from fetal bovine serum, we used one batch of FBS from Gemini Bioproducts (West Sacramento, CA), one from PAA Laboratories, Inc. (New Bedford, MA), and two batches from Atlanta biologicals, Lawrenceville, GA. The serum (100 ml) was mixed with RPMI-1640 medium at a ratio of 50:50. This was centrifuged at 20,000 × g to remove micro vesicles and other cellular debris and the resultant supernatant was centrifuged at 100,000 × g for 10 h. The supernatant (exosome free fraction) was carefully decanted and named exosome free medium (EFM) while the resulting precipitate was dissolved in PBS and centrifuged at 200,000 × g for 1 hr. The resulting precipitate was dissolved in RPM-1640 and constituted the exosome enriched medium (EEM). Both EEM and EFM were adjusted to a concentration of 6 mg/ml using serum free DMEM/F12 medium.
Anchorage Independent growth Assays
The anchorage independent growth assay (AIG) was modified for the 96-well culture plates. Aliquots of agar Noble (Difco) (0.8% w/v in water; 100 ml) were sterilized and stored at room temperature in conical flasks. The agar was melted, and kept in water bath at 50°C. To make the bottom layer, one part of the melted 0.8% agar (700 μl) was mixed with 560 μl of 2× DMEM/F12 serum free medium containing either 140 μl of exosome enriched medium (EEM) or exosome free medium (EFM). This final mixture (0.4% agar) was then added to wells at100 μl/well and allowed to gel at room temperature. To prepare the top layer (0.3% soft agar) 700 μl of 0.6% agar was mixed with 560 μl of 2× SFM containing the cells (500 cells/well) and either 140 μl of EEM or EFM. This mixture was then added to the bottom layer (100μl/well) and allowed to gel at room temperature. After three weeks of growth the colonies were photographed. Alamar Blue (Biosource, Camarillo, CA) was also added to the wells to measure the growth potential of the colonies by monitoring the release of reducing metabolites as a read-out. In some experiments we used agarose (Invitrogen) instead of soft agar. We also assayed for 3-dimensional growth of tumor cells in Matrigel. Growth factor reduced Matrigel (BD Biosciences, Bedford, MA) from frozen stock was kept overnight at 4°C to liquefy. This was then added to 96-well culture plate (100 μl/well) and allowed to solidify by incubating at 37°C for at least 20 min. Serum free medium (SFM), EFM, and EEM were added (200 μl/well) on top of Matrigel followed by BT-549Gal3 cells (500 cells/well). The plates were then incubated in a humidified CO2 incubator at 37°C for two weeks.
Proteomic Analysis of the proteins in Exosomes
In-Gel Digestion
Equal amounts of proteins from EEM and EFM were loaded into SDS-PAGE (4-12% gradient gel), resolved and stained with coomassie brilliant blue. Each lane was cut into 4 regions and each subjected to in-gel tryptic digestion [13]. Briefly, the gel regions were excised and washed with 100 mM ammonium bicarbonate for 15 minutes. The liquid was discarded and replaced with fresh 100 mM ammonium bicarbonate and the proteins reduced with 5 mM DTT for 20 minutes at 55° C. After cooling to room temperature, iodoacetamide was added to 10 mM final concentration and placed in the dark for 20 minutes at room temperature. The solution was discarded and the gel pieces washed with 50% acetonitrile/50 mM ammonium bicarbonate for 20 minutes, followed by dehydration with 100 % acetonitrile. The liquid was removed and the gel pieces were completely dried, re-swelled with 0.3 μg of modified trypsin (Promega) in 100 mM NH4HCO3, and digested overnight at 37°C. Peptides were extracted by three changes of 60% acetonitrile/0.1% TFA, and extracts from each region were combined separately, generating 4 samples per lane, and dried in vacuo. Samples were reconstituted in 30 μL 0.1 % formic acid for LC-MS-MS analysis.
LC-MS-MS Analysis and Protein Identification
Resulting peptides were analyzed using a Thermo Finnigan LTQ ion trap instrument equipped with a Thermo MicroAS autosampler and Thermo Surveyor HPLC pump, Nanospray source, and Xcalibur 2.0 SR2 instrument control. Peptides were separated on a packed capillary tip (Polymicro Technologies, 100 μm × 11 cm) with Jupiter C18 resin (5 μm, 300 Å, Phenomenex) using an in-line solid-phase extraction column (100 μm × 6 cm) packed with the same C18 resin [using a frit generated with liquid silicate] similar to that previously described [14]. The flow from the HPLC pump was split prior to the injection valve to achieve flow-rates of 700 nL-1000 μL min-1 at the column tip. Mobile phase A consisted of 0.1 % formic acid and Mobile phase B consisted of 0.1% formic acid in acetonitrile. A 95 min gradient was performed with a 15 min washing period (100 % A for the first 10 min followed by a gradient to 98% A at 15 minutes) to allow for solid-phase extraction and removal of any residual salts. Following the washing period, the gradient was increased to 25% B by 50 min, followed by an increase to 90 % B by 65 min and held for 9 min before returning to the initial conditions. Tandem spectra were acquired using a data dependent scanning mode in which one full MS scan (m/z 400-2000) was followed by 9 MS-MS scans. Tandem spectra were searched against the bovine subset of the UniRef100 database using the SEQUEST algorithm. The database was concatenated with the reverse sequences of all proteins in the database to allow for the determination of false positive rates. Protein matches were preliminarily filtered using the following criteria: cross-correlation (Xcorr) value of ≥ 1.0 for singly charged ions, ≥ 1.8 for doubly charged ions, and ≥ 2.5 for triply charged ions. A ranking of primary score (RSp) of ≤ 5 and a preliminary score (Sp) of ≥ 350 were also required for positive peptide identifications. Once filtered based on these scores, all proteins identified by less than two peptides were eliminated, resulting in false positive rates of <1%. The Sequest output was also filtered using IDPicker using a false positive ID threshold (default is 0.05 or 5% false positives) based on reverse sequence hits in the database.
The use of sampling statistics has been shown to be a useful measure of relative protein abundance in label-free-LC-MS/MS shotgun proteomics. Recent studies have revealed a direct relationship between protein abundance and sampling statistics, such as percent sequence coverage, peptide count and spectral count. Most notably, the spectral count is the total number of tandem mass spectra (MS/MS) that are matched to the peptides of a protein. Liu et al. [15], demonstrated that spectral counts could be a reliable indicator of protein abundance with a linear range over 2 orders of magnitude by using a spectral count based measure; spectral sampling. In contrast to spectral counts, other sampling measures such as percent sequence coverage and number of peptides identified per protein did not have nearly as good of a linear correlation to actual protein abundance [15]. Moreover, spectral count has the highest technical reproducibility, followed by the less-reproducible peptide count and relatively non-reproducible sequence coverage [16]. Protein reassembly from identified peptide sequences is done with the aid of a parsimony method recently described by Zhang et al. [17] which identifies indiscernible proteins (protein groups) that can account for the identified peptides. Relative protein abundance (fold ratio changes) between exosome free medium (EFM) and exosome enriched medium (EEM), were compared based on spectral counts from two independent LC-MS/MS runs corresponding to two aliquots from the same preparation run in separate lanes of SDS-PAGE gels (Table 2).
Table 2. Relative Fold Change Ratios of proteins in Exosome Enriched Medium (EEM) versus Exosome Free Medium (EFM).
| Spectral Counts | ||||
|---|---|---|---|---|
| Accession Number | EFM | EEM | Ratio | Description |
| UniRef100_UPI00005BC629 | 5 | 95 | 19 | similar to Alpha-2-macroglobulin precursor |
| UniRef100_UPI00005BC5E5 | 1 | 15 | 15 | similar to pregnancy-zone protein |
| UniRef100_P07589 | 4 | 33 | 8.3 | Fibronectin |
| UniRef100_UPI00005BB909 | 1 | 8 | 8 | similar to Ceruloplasmin precursor (Ferroxidase) |
| UniRef100_UPI00006830EC | 5 | 35 | 7 | ANTITHROMBIN III |
| UniRef100_Q3SZV7 | 1 | 7 | 7 | Similar to hemopexin |
| UniRef100_P00735 | 5 | 32 | 6.4 | Prothrombin precursor (Coagulation factor II) |
| UniRef100_UPI00005B628A | 6 | 37 | 6.2 | hypothetical protein LOC540261 |
| UniRef100_P97280 | 2 | 12 | 6 | Inter-alpha-trypsin inhib. heavy chain H3 prec |
| UniRef100_Q2PQT6 | 1 | 6 | 6 | Vinculin |
| UniRef100_UPI0000110A3C | 5 | 27 | 5.4 | BETA-ACTIN |
| UniRef100_Q28194 | 3 | 16 | 5.3 | Thrombospondin-1 |
| UniRef100_Q3ZBS7 | 3 | 15 | 5 | Vitronectin |
| UniRef100_UPI00005BC623 | 185 | 861 | 4.7 | Similar:Alpha-2-macroglobulin precur. (A2-M) |
| UniRef100_UPI0000693DE0 | 66 | 298 | 4.5 | complement component 3 |
| UniRef100_Q2UVX4 | 74 | 325 | 4.4 | Complement C3 precursor |
| UniRef100_UPI00005C2942 | 2 | 8 | 4 | Similar:Ig lambda chain V-I region BL2 precur. |
| UniRef100_Q32T06 | 1 | 4 | 4 | Endopin 2C |
| UniRef100_Q0VCX1 | 1 | 4 | 4 | Complement component 1, s subcomponent |
| UniRef100_UPI00005C201D | 1 | 4 | 4 | Similar:C4b-binding prot alpha chain precur |
| UniRef100_UPI00005C27D5 | 1 | 4 | 4 | Similar:gamma-2a immunoglobulin heavy chain |
| UniRef100_Q06805 | 1 | 4 | 4 | Tyrosine-protein kinase receptor Tie-1 precursor |
| UniRef100_Q95121 | 4 | 14 | 3.5 | Pigment epithelium-derived factor precursor |
| UniRef100_Q03247 | 14 | 47 | 3.4 | Apolipoprotein E precursor |
| UniRef100_UPI00005BC731 | 3 | 10 | 3.3 | similar to Fibulin-1 precursor isoform 1 |
| UniRef100_P02584 | 3 | 10 | 3.3 | Profilin-1 |
| UniRef100_P17697 | 7 | 23 | 3.3 | Clusterin precursor (Glycoprotein III) |
| UniRef100_P01966 | 22 | 72 | 3.3 | Hemoglobin subunit alpha |
| UniRef100_Q28178 | 9 | 27 | 3 | Thrombospondin-1 precursor |
| UniRef100_UPI00005BE9E4 | 2 | 6 | 3 | similar to Heparin cofactor II precursor (HC-II) |
| UniRef100_Q9N2I2 | 2 | 6 | 3 | Plasma serine protease inhibitor precursor |
| UniRef100_P17690 | 1 | 3 | 3 | Beta-2-glycoprotein 1 precursor |
| UniRef100_P02081 | 50 | 138 | 2.8 | Hemoglobin fetal subunit beta |
| UniRef100_Q5E9B1 | 3 | 8 | 2.7 | L-lactate dehydrogenase B chain |
| UniRef100_P02662 | 3 | 8 | 2.7 | Alpha-S1-casein cursor |
| UniRef100_Q29RQ1 | 8 | 20 | 2.5 | Complement component C7 precursor |
| UniRef100_UPI00005C18A3 | 2 | 5 | 2.5 | similar to Complement factor H precursor |
| UniRef100_UPI00005C1B6C | 2 | 5 | 2.5 | Perlecan |
| UniRef100_Q05B55 | 5 | 12 | 2.4 | Hypothetical protein |
| UniRef100_UPI00005BFBFC | 15 | 34 | 2.3 | similar to Complement C4 precursor |
| UniRef100_UPI00005C211E | 13 | 28 | 2.2 | similar to Ig gamma-1 chain C region |
| UniRef100_Q0VCM5 | 15 | 31 | 2.1 | Similar:Inter-alpha-trypsin inhib. heavy chain H1 |
| UniRef100_P02070 | 31 | 62 | 2 | Hemoglobin subunit beta |
| UniRef100_P22226 | 5 | 10 | 2 | Cyclic dodecapeptide precursor |
| UniRef100_Q3SWW8 | 3 | 6 | 2 | Similar to thrombospondin 4 |
| UniRef100_Q2HJF0 | 3 | 6 | 2 | Similar to Serotransferrin |
| rUniRef100_UPI00005C24B2 | 2 | 4 | 2 | similar to Laminin alpha-3 chain precursor |
| rUniRef100_UPI00005C16C9 | 1 | 2 | 2 | similar to CG9164-PA, isoform A |
| UniRef100_UPI00005BDE5A | 1 | 2 | 2 | similar to Neuropilin-1 precursor |
| UniRef100_UPI00005BD502 | 1 | 2 | 2 | Similar:Cofactor for Sp1 trans activation sub 3 |
| UniRef100_Q5E956 | 1 | 2 | 2 | Triosephosphate isomerase |
| UniRef100_UPI00005BFBFF | 30 | 54 | 1.8 | similar to Complement C4 precursor, partial |
| UniRef100_Q3SZH5 | 10 | 18 | 1.8 | Similar to Angiotensinogen |
| UniRef100_P06868 | 32 | 57 | 1.8 | Plasminogen precursor (EC 3.4.21.7) |
| UniRef100_Q2KIF5 | 7 | 12 | 1.7 | Vl1a protein |
| UniRef100_P02672 | 15 | 25 | 1.7 | Fibrinogen alpha chain precursor |
| UniRef100_P01030 | 29 | 44 | 1.5 | Complement C4 precursor |
| UniRef100_Q9BGI3 | 2 | 3 | 1.5 | Peroxiredoxin-2 |
| UniRef100_Q5EA67 | 57 | 84 | 1.5 | Inter-alpha (Globulin) inhibitor H4 |
| UniRef100_Q28085 | 22 | 32 | 1.5 | Complement factor H precursor |
| UniRef100_Q3T052 | 58 | 84 | 1.5 | Inter-alpha (Globulin) inhibitor H4 |
| UniRef100_Q3SX14 | 24 | 33 | 1.4 | Similar to Gelsolin |
| UniRef100_Q0IIK2 | 289 | 390 | 1.4 | Transferrin |
| UniRef100_Q9GLY6 | 18 | 24 | 1.3 | Inter-alpha-trypsin inhibitor heavy chain2 |
| UniRef100_Q3SZ57 | 25 | 33 | 1.3 | Alpha-fetoprotein precursor |
| UniRef100_Q2KIU6 | 44 | 57 | 1.3 | Similar to Complement factor B |
| UniRef100_P28800 | 14 | 18 | 1.3 | Alpha-2-antiplasmin precursor |
| UniRef100_Q1RMN8 | 7 | 9 | 1.3 | Similar to Ig lambda-like polypep. 1 |
| UniRef100_P00978 | 27 | 34 | 1.3 | AMBP protein precursor |
| UniRef100_Q3SZZ4 | 26 | 32 | 1.2 | AMBP protein |
| UniRef100_UPI00005BDE33 | 82 | 99 | 1.2 | similar to inter-alpha globulin inhib. H2 polypep |
| UniRef100_Q2KIS7 | 5 | 6 | 1.2 | Tetranectin precursor |
| UniRef100_P02453 | 5 | 6 | 1.2 | Collagen alpha-1(I) chain precursor |
| UniRef100_Q3UZM0 | 23 | 27 | 1.2 | 8 days embryo whole body cDNA, RIKEN |
| UniRef100_P15497 | 30 | 35 | 1.2 | Apolipoprotein A-I precursor |
| UniRef100_UPI00001116D7 | 13 | 15 | 1.2 | TRYPSIN |
| UniRef100_Q0V8M9 | 62 | 69 | 1.1 | Inter-alpha (Globulin) inhibitor H3 |
| UniRef100_O46375 | 30 | 32 | 1.1 | Transthyretin precursor |
| UniRef100_UPI00000718F2 | 39 | 41 | 1.1 | keratin 10 |
| UniRef100_UPI00006830E5 | 17 | 17 | 1 | TRYPSIN |
| UniRef100_Q58CQ9 | 4 | 4 | 1 | Pantetheinase precursor |
| UniRef100_Q6LBN7 | 3 | 3 | 1 | Lactoferrin |
| r-UniRef100_Q2KI19 | 2 | 2 | 1 | Similar to CG10053-PA |
| UniRef100_Q2KIW5 | 1 | 1 | 1 | Centromere protein U |
| UniRef100_UPI00005C2368 | 1 | 1 | 1 | similar to HGF activator preproprotein |
| UniRef100_UPI00005C11DB | 1 | 1 | 1 | similar to Spectrin beta chain |
| UniRef100_UPI000016C268 | 1404 | 1389 | 1 | Albumin |
| UniRef100_UPI00001AE6F7 | 55 | 51 | 0.9 | keratin 9 |
| UniRef100_Q3MHN5 | 53 | 45 | 0.9 | Vitamin D-binding protein precursor |
| UniRef100_Q2KJF1 | 18 | 15 | 0.8 | Alpha-1-B glycoprotein |
| UniRef100_UPI0000E2470B | 65 | 54 | 0.8 | Cytokeratin-14 |
| UniRef100_Q58D62 | 47 | 39 | 0.8 | Fetuin-B precursor |
| UniRef100_P34955 | 158 | 130 | 0.8 | Alpha-1-antiproteinase precursor |
| UniRef100_P18902 | 13 | 10 | 0.8 | Plasma retinol-binding protein |
| UniRef100_UPI00001A389F | 13 | 10 | 0.8 | Plasma retinol-binding protein |
| UniRef100_Q2KIT0 | 4 | 3 | 0.8 | Similar to collagen, type X, alpha 1 |
| UniRef100_UPI0000D9CC8C | 48 | 35 | 0.7 | similar to keratin 1 isoform 7 |
| UniRef100_UPI0000167B80 | 62 | 45 | 0.7 | keratin 1 |
| UniRef100_UPI0000110894 | 7 | 5 | 0.7 | BETA-LACTOGLOBULIN |
| UniRef100_Q3ZEJ6 | 3 | 2 | 0.7 | Serpina 3-3 |
| UniRef100_Q4VAQ2 | 43 | 27 | 0.6 | Keratin 2 |
| UniRef100_UPI00005A1BEF | 8 | 5 | 0.6 | similar to keratin 25D |
| UniRef100_P12763 | 400 | 235 | 0.6 | Alpha-2-HS-glycoprotein precursor |
| UniRef100_Q3SZR3 | 12 | 7 | 0.6 | Alpha-1 acid glycoprotein |
| UniRef100_UPI0000D9CC7B | 15 | 8 | 0.5 | keratin 6A |
| UniRef100_P01044-2 | 22 | 11 | 0.5 | Isoform LMW of P01044 |
| UniRef100_UPI0000167F3F | 22 | 11 | 0.5 | kininogen 1 |
| UniRef100_Q9TTJ5 | 4 | 2 | 0.5 | Regucalcin |
| UniRef100_P02465 | 4 | 2 | 0.5 | Collagen alpha-2(I) chain precursor |
| UniRef100_UPI00006A12E2 | 4 | 2 | 0.5 | keratin 24 |
| rUniRef100_UPI00005C1B5E | 2 | 1 | 0.5 | similar to dynein, cytoplasmic, heavy polypep1 |
| UniRef100_Q08E14 | 2 | 1 | 0.5 | Similar to Collagen alpha 1(III) chain |
| UniRef100_P01035 | 2 | 1 | 0.5 | Cystatin C precursor |
| UniRef100_UPI00006D113E | 7 | 3 | 0.4 | keratin 5 isoform 20 |
| UniRef100_Q2KJ62 | 23 | 9 | 0.4 | KNG protein |
| UniRef100_UPI00005C202E | 13 | 5 | 0.4 | similar to Apolipoprotein B-100 precursor |
| UniRef100_P08779 | 9 | 3 | 0.3 | Keratin, type I cytoskeletal 16 |
| UniRef100_UPI00005BDB2A | 6 | 2 | 0.3 | similar to Apolipoprotein B-100 precursor |
| UniRef100_Q2KIQ9 | 6 | 2 | 0.3 | SERPINA3 protein |
| UniRef100_Q5DPW9 | 3 | 0 | 0 | Cystatin E/M |
| UniRef100_P35445 | 2 | 0 | 0 | Cartilage oligomeric matrix protein |
| UniRef100_P42916 | 2 | 0 | 0 | Collectin-43 precursor |
| UniRef100_UPI00005C2037 | 2 | 0 | 0 | similar to Collagen alpha 1(V) chain precursor |
Fold change ratio is calculated by the total number of spectral counts of a peptide/protein in the EEM divided by spectral count of EFM. Spectral counts are representative of two independent LC-MS/MS runs corresponding to two aliquots from the same preparation run in separate lanes of SDS-PAGE gels.
Transmission Electron Microscopy of the Exosomes
The exosomal sample was fixed in 2.5% paraformaldehyde for at least 1 hr, followed by 2 buffer washes (TBS) for 5 min each. The samples were then incubated with 50% ethanol for 15 min followed by 70% ethanol for 30 min. They were then mixed 1:1 with LR white resin in 100% ethanol for 30 min followed by 2:1 mix with LR white resin in 100% ethanol for 30 min. They were then immersed in pure LR white resin for 60 min and then another fresh LR white resin for 60 min followed by LR white resin overnight in a rotator. They were then placed in gelatin capsules with fresh LR white resin and cured at 50 degrees for 48 hr. The sections were cut to 100 nm thickness and within 30 min, staining process was started. The sections were washed in TBS buffer 3 times for 30 seconds each then the grids blocked using 2% BSA for 30 min. The sections were then fixed in 2.5 % glutaraldehyde for 30 seconds, washed in buffer and then 3 times in water for 30 seconds to remove salts and then stained for 30 seconds with 1% Uranyl acetate. The moisture was wicked away and then examined using a Phillips CM 12 TEM at 80 KV.
Up-take of Purified Serum Exosomes by Tumor Cells
Exosomes purified as described above were labeled with rhodamine isothiocynate (Sigma) as described [18] and incubated with adherent BT-549Gal3 cells. Briefly, the total exosomal proteins were determined by the Bradford assay and 100 μg/assay were equilibrated in 100 mM carbonate buffer pH 9.0 by centrifugation at 10,000 × g for 15 min in an ultra-filtration unit (MCO-100,000). The equilibrated exosomes were re-suspended in 100 μl of the same buffer and mixed with 100 μg/assay of rhodamine isothiocynate in DMSO. This was then incubated 2 h at room temperature and in the dark. After labeling, the exosomes were washed abundantly by centrifugation in the ultra-filtration unit to remove un-reacted rhodamine. Adherent breast tumor cells were grown on microscope cover slips, starved for 48 h by growth in DMEM supplemented with 0.5 % FBS, washed twice with Ca2+ and Mg2+-free Dulbecco PBS (Invitrogen) and once with Ca2+ and Mg2+-free HBSS (Invitrogen). Cells were then incubated for 30 min in HBSS containing 10 μg/ml rhodamine-labeled exosomes, 1 mM of divalent ions Ca2+/Mg2+, and the indicated concentrations of unlabeled exosomes. The cells were rinsed twice with cold PBS before and after fixation with -20°C cold methanol for 10 min and the cover slips mounted on slides using Prolong with DAPI (Invitrogen). Up-take was visualized by fluorescent microscopy using a Nikon Eclipse TE-2000-E inverted microscope equipped with NIS-Elements AR software (Nikon, Melville, NY, USA)
Recycling of Serum Exosomes by Tumor Cells
BT-549Gal-3 cells were transfected with GFP-annexin A2 or mock transfected using Fugene6 according to the manufacturer (Roche) and serum-starved for 48 h. The mock transfected cells were incubated with rhodamine-labeled exosomes for 30 min at 37°C, then washed twice with PBS at RT. The GFP-annexinA2 transfected and the rhodamine labeled cells were respectively harvested by treatment with 2 mM EDTA and washed in cold PBS and once in HBSS supplemented with 1 mM Mg2+ and 1 mM Ca2+. These were then mixed at a ratio of 1:3 (GFP-transfected cells to rhodamine-labeled cells), plated in six-well plates with cover slips and incubated for another 2 hr at 37°C. The cells were rinsed twice with cold PBS before and after fixation in -20°C cold methanol for 10 min and the cover slips mounted on slides using Prolong with DAPI (Invitrogen). Up-take/recycling were visualized by fluorescent microscopy using a Nikon Eclipse TE-2000-E inverted microscope equipped with NIS-Elements AR software (Nikon, Melville, NY, USA)
MAP kinase Signaling Mediated by Exosomes
Cells were plated in 10 cm dishes and serum-starved as above for 48h. The adherent cells were then washed twice with PBS and once with 10 mM HEPES pH 7.2, 140 mM NaCl, 4.7 mM KCl (HBS) and incubated in HBS containing 1 mM Mg2+ and 1 mM Ca2+ without or with 0.1 mg/ml of exosomes for the indicated time points at 37°C. The medium was rapidly discarded, replaced with ice-cold PBS and the dishes maintained on ice until the cells were harvested by scrapping. These were washed in the ice-cold PBS and the cell pellets stored at -80°C until needed. In parallel experiments, cell culture dishes were pre-treated with poly(HEMA) as previously described [19]. The poly(HEMA)-treated dishes were sterilized by UV then washed twice with PBS. About 2 ×106 cells/dish that have been serum-starved as above were plated on the poly(HEMA)-treated dishes in HBS containing 1 mM Mg2+, 1 mM Ca2+ without and with the indicated concentrations of exosomes (mg/ml) for 10 min or following a time course at 37°C. MAP kinase activation was visualized by the detection of phosphorylated ERK1/2 by western blotting using antibodies to the dually phosphorylated ERK1 and ERK2. Total ERK as the loading control was detected using antibodies to either ERK2 or ERK1/2.
Results
Role of Serum Exosomes on Anchorage dependent and independent growth of Breast Carcinoma Cells
Our objective here was to determine the serum fraction (exosome free or enriched) that supports anchorage independent growth of breast carcinoma cells. The breast carcinoma cells were suspended in soft agar and allowed to grow either in the absence or presence of serum exosomes for at least 3 weeks. The data clearly show that AIG is robust only in those wells containing exosomes (Fig. 1). This serum fraction which we have named exosome enriched medium (EEM) in addition to common serum proteins such as albumin, also contains a unique set of proteins that are absent in exosome free medium (EFM) (Table 1). The AIG assay was repeated at least 4 times with similar results. All the batches of fetal bovine serum we employed yielded exosomes that promoted AIG and the colonies grew in both soft agar and agarose at a similar rate. Not only did the cells growing in the presence of exosomes form large colonies with time, they were also healthy and actively released reducing metabolites into the conditioned medium, a hall mark of metabolic activity in growing cells (Fig. IB). It can be argued that MDA-MB-435 is not an authentic breast cancer cell line [20]. However, its growth patterns in soft agar being similar to the other breast carcinoma cell lines, demonstrate that the ability of exosomes to influence anchorage independent growth can be extrapolated to other tumor cells such as melanoma.
Figure 1. Bovine serum exosome mediated anchorage independent growth of breast cancer cells in soft agar.
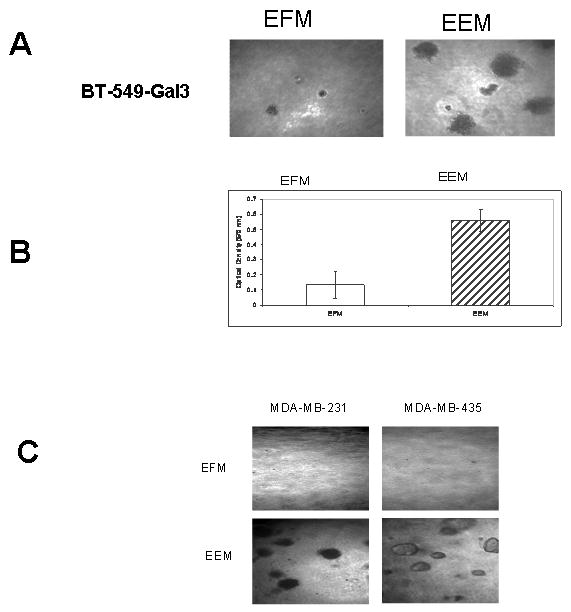
Soft agar growth assays were set up in 96-well micro titer plates as described in Materials and Methods. The cells were allowed to grow in soft agar either in the presence (EEM) or absence (EFM) of purified bovine exosomes. After 3 weeks the colonies were either photographed (panel A) or Alamar blue was added to the control wells and to the wells containing exosomes. After 4 hours of color change, the plates were read at 570 nm (panel B). The growth promoting properties of the exosomes were also determined using other breast cancer cell lines (panel C).
Table 1. Proteins Found only in Exosome Enriched Medium (EEM).
| Accession Number | spectral counts | Description | |
|---|---|---|---|
| EFM | EEM | ||
| UniRef100_UPI00005C0899 | 0 | 17 | similar to Filamin A |
| UniRef100_UPI00005C2264 | 0 | 8 | similar to myosin, heavy polypeptide 9, non-muscle, partial |
| UniRef100_P62935 | 0 | 6 | Peptidyl-prolyl cis-trans isomerase A |
| UniRef100_Q2KJF4 | 0 | 6 | Similar to Galectin-3 binding protein |
| UniRef100_Q76LV2 | 0 | 5 | Heat shock protein HSP 90-alpha |
| UniRef100_Q27967 | 0 | 5 | Secreted phosphoprotein 24 precursor |
| UniRef100_Q5E9F5 | 0 | 4 | Transgelin-2 |
| UniRef100_Q1RMK2 | 0 | 4 | IGHM protein |
| UniRef100_P01267 | 0 | 4 | Thyroglobulin precursor |
| UniRef100_UPI000051DE08 | 0 | 4 | Tubulin alpha chain |
| UniRef100_O02717 | 0 | 4 | Non-muscle myosin heavy chain |
| UniRef100_UPI00005BC732 | 0 | 4 | similar to fibulin 1 |
| UniRef100_O18787 | 0 | 4 | Elongation factor 1 alpha |
| UniRef100_UPI00005BFC16 | 0 | 3 | similar to Complement C4 precursor, partial |
| UniRef100_Q28107 | 0 | 3 | Coagulation factor V precursor |
| UniRef100_P10096 | 0 | 3 | Glyceraldehyde-3-phosphate dehydrogenase |
| UniRef100_UPI00005BC368 | 0 | 3 | similar to alpha 3 type VI collagen isoform 1 precursor |
| UniRef100_O46415 | 0 | 3 | Ferritin light chain |
| UniRef100_UPI00005BC752 | 0 | 3 | similar to Plexin B2 precursor (MM1) |
| UniRef100_Q3SZZ9 | 0 | 3 | FGG protein |
| UniRef100_UPI00005BEE04 | 0 | 3 | similar to Stem cell growth factor precursor |
| UniRef100_UPI00005BC072 | 0 | 3 | similar to extracellular matrix protein 1 |
| UniRef100_Q76LV1 | 0 | 2 | Heat shock protein HSP 90-beta |
| UniRef100_Q32LP0 | 0 | 2 | Unc-112-related protein 2 |
| UniRef100_Q05443 | 0 | 2 | Lumican precursor |
| UniRef100_UPI00005BDFA6 | 0 | 2 | similar to beta tubulin 1, class VI |
| UniRef100_UPI00005BE617 | 0 | 2 | similar to mannan-binding lectin serine protease 2 |
| UniRef100_UPI00005BD7A2 | 0 | 2 | similar to talin 2 |
| UniRef100_P28801 | 0 | 2 | Glutathione S-transferase P |
| UniRef100_UPI00005C03A6 | 0 | 2 | similar to ubiquitin-activating enzyme E1 isoform 3 |
| UniRef100_Q17QH6 | 0 | 2 | Similar to collectin sub-family member 11 isoform a |
| UniRef100_UPI00005C030B | 0 | 2 | similar to deleted in malignant brain tumors 1 |
| UniRef100_P33672 | 0 | 2 | Proteasome subunit beta type 3 |
| UniRef100_Q3MHN2 | 0 | 2 | Complement component C9 precursor |
| UniRef100_Q29451 | 0 | 2 | Lysosomal alpha-mannosidase A peptide |
| UniRef100_UPI00005A1C47 | 0 | 2 | similar to karyopherin beta 1 |
| UniRef100_P02676 | 0 | 1 | Fibrinogen beta chain precursor |
| UniRef100_Q28106 | 0 | 1 | Contactin-1 precursor |
| UniRef100_UPI00005C1679 | 0 | 1 | similar to Neogenin precursor, partial |
| UniRef100_UPI00005C18F6 | 0 | 1 | Similar:C1q and tumor necrosis factor related protein 3 |
| UniRef100_UPI00005C18F7 | 0 | 1 | similar to collagenous repeat-containing |
| UniRef100_Q3ZCJ2 | 0 | 1 | Aldo-keto reductase family 1, member A1 |
| UniRef100_Q2KJ63 | 0 | 1 | Kallikrein B, plasma (Fletcher factor) 1 |
| UniRef100_Q5E9B7 | 0 | 1 | Chloride intracellular channel protein 1 |
| UniRef100_Q3MHN0 | 0 | 1 | Proteasome subunit beta type 6 precursor |
| UniRef100_O02808 | 0 | 1 | Von Willebrand factor |
| UniRef100_UPI00005C164A | 0 | 1 | similar to ADAM metallopeptidase |
| UniRef100_UPI00005C052B | 0 | 1 | similar to Afamin precursor |
| UniRef100_UPI00005BB7FD | 0 | 1 | similar to Carboxypeptidase N subunit 2 precursor |
| UniRef100_Q3T149 | 0 | 1 | Heat-shock protein beta-1 |
| UniRef100_UPI0000562815 | 0 | 1 | heat shock 27kDa protein 1 |
Spectral counts are representative of two independent LC-MS/MS runs corresponding to two aliquots from the same preparation run in separate lanes of SDS-PAGE gels.
Tumor cells in serum free medium failed to grow in growth factor reduced Matrigel after two weeks of incubation (Fig.2A). However, in EFM, the cells grew mainly as a monolayer on top of the Matrigel with extensive spreading. In EEM on the other hand, there were distinctive three dimensional stellate colonies (Fig. 2C). These stellate colonies are typically observed when tumorigenic cells are grown in Matrigel [21]. Apart from anchorage independent growth in soft agar and Matrigel, we also examined the influence of exosomes on the growth of breast epithelial cells on plastic. In the presence of the vesicles, the tumor cells adhered and spread rapidly on plastic and with extended period of growth (seven days), the cells grew in tight aggregates on plastic (Fig 3B). This type of growth is not uncommon in tumor cells [22], further re-enforcing the potential role of these vesicles on AIG. Due to the inability of normal breast epithelial cells (MCF-10A) to grow in soft agar, we speculated that they would also fail to grow on plastic in the presence of exosomes. On the contrary, the cells proliferated in the presence of the vesicles and as expected in vesicle free medium (EFM). However, in serum free medium, growth was minimal (Fig 3A).
Figure 2. Bovine serum exosome mediated growth of stellate colonies of breast cancer cells in Matrigel.
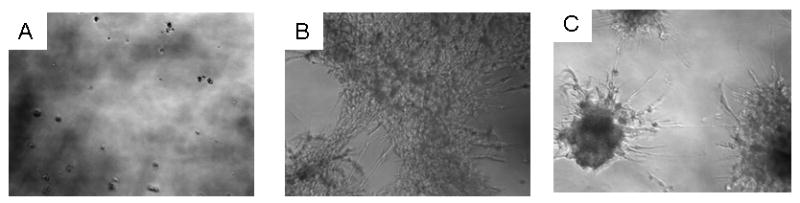
Liquid Matrigel (100μl/well) was allowed to solidify at 37°C in 96-well micro titer plate. The carcinoma cells were then added on the Matrigel (500 cells/well) in the presence of serum free medium (panel A), exosome free medium (panel B) and exosome enriched medium (panel C). The cells were then incubated in a humidified CO2 incubator for 14 days and then photographed.
Figure 3. Bovine serum exosome mediated growth of normal and breast cancer cells on plastic.
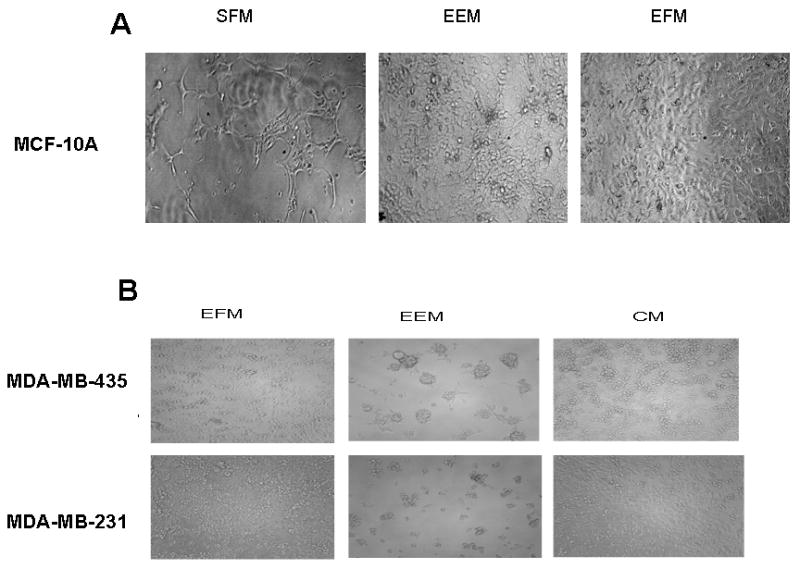
In panel A, MCF-10A were at 2×104 cells/well in 96-well micro titer plates in serum free medium, exosomal enriched medium (EEM) and exosomal free medium (EFM). The cells were allowed to grow for 5 days and then photographed. In panel B, one breast carcinoma cell line, MDA-MB-231 and a melanoma cell line, MDA-MB-435 were likewise plated at 2 × 104 cells/well in 96-well micro titer plates in EEM and EFM and in complete medium (CM) containing 10% fetal bovine serum.
Analysis of Proteins identified by mass-spectroscopy in both EEM and EFM
Transmission electron microscopy (TEM) revealed vesicles in EEM whose average diameter was approximately 150 nm (Fig. 4A). This average diameter was larger than that of bonafide cell derived exosomes with an average in diameter of between 60-100 nm [23]. Serum vesicles (micro-vesicles) precipitated at approximately 20,000 × g lacked any biological activity such as support of anchorage independent growth (data not shown). An examination of the distribution of two of the more abundant proteins in the EEM by sucrose gradient centrifugation revealed that while alpha 2-microglobulin was associated with the dense portion of the sucrose gradient, fetuin-A is associated with the less dense portion of the gradient (data not shown).
Figure 4. Proteins Associated with bovine serum exosomes.
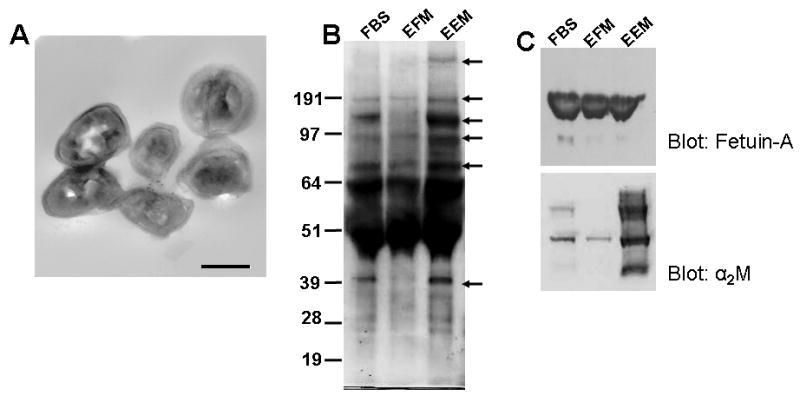
Panel A, transmission electron micrograph of exosomes purified from fetal bovine serum. The black line represents 100 nm. Panel B, aliquots of bovine serum, exosome free serum, and exosomes were resolved in gradient SDS-PAGE and stained with coomassie brilliant blue. The protein bands were photographed and then all the proteins in each lane cut and analyzed by mass spec as described in Materials and Methods. Panel C, proteins from bovine serum, exosome-free serum and exosomes were resolved in SDS-PAGE and then transferred to immobilon paper and probed for fetuin-A and alpha 2 macroglobulin.
When the proteins in EEM and EFM were resolved on SDS-PAGE, it was evident that whereas most of the serum proteins appeared in both fractions, some were more concentrated in EEM (Fig. 4B, arrows). Among the proteins recovered in the EEM lane, a number of them were found only in exosomes by mass spectroscopy (LC-MS/MS) (Table 1). The relative abundance of these proteins in either EFM or EEM was determined by use of spectral counts. The presence of at least two spectral counts was used as the threshold for peptide/protein identification. Given that the stringency during the LC-MS/MS was high and resulted in a measurable false positive rate of less than 1%, we conclude that a small spectral count is indicative of low abundance of the peptide, yet a high degree of confidence in the presence of that peptide in the sample. Interestingly, some of these proteins have also been reported in exosomes purified from other sources and are considered exosomal marker proteins. These include heat shock proteins HSP-90 [24], alpha tubulin [11], peptidyl prolyl trans isomerase [25, 26], elongation factor 1 alpha [11], galectin-3 binding proteins [27], glyceraldehyde-3-phosphate dehydrogenase [11], and chloride intracellular channel protein 1[11]. By far the most abundant protein associated with serum exosomes is alpha 2-macroglobulin (Fig. 4C and Table 2). Interestingly other acute phase proteins synthesized in the liver such as fetuin-A and heat labile complement factors were also associated with the exosomes (Table 2). The concentration of fetuin-A in EFM was more or less comparable to its concentration in EEM (Table 2 and Fig. 4C). It is likely that the high enrichment of serum proteins in bovine serum exosomes may account for their rather large size compared to exosomes purified from cells.
Up-take of Purified Exosomes by Tumor Cells
Exosomes can be up-taken or endocytosed by cells via a mechanism that is not well characterized in the literature [28]. We show here that the purified labeled bovine exosomes can be endocytosed by breast carcinoma cells. After about 2 hr of up-take, the exosomes appear to concentrate in the late endosomal compartment (Fig. 5A). This pattern of up-take is similar to that which has been observed by others [28]. Interestingly, when we incubated the cells with the labeled exosomes and excess unlabeled exosomes (Fig. 5B and 5C), the up-take of the former was blocked, suggesting that exosomes are endocytosed via a cell surface receptor/s.
Figure 5. Up-take of purified bovine serum exosomes by tumor cells.
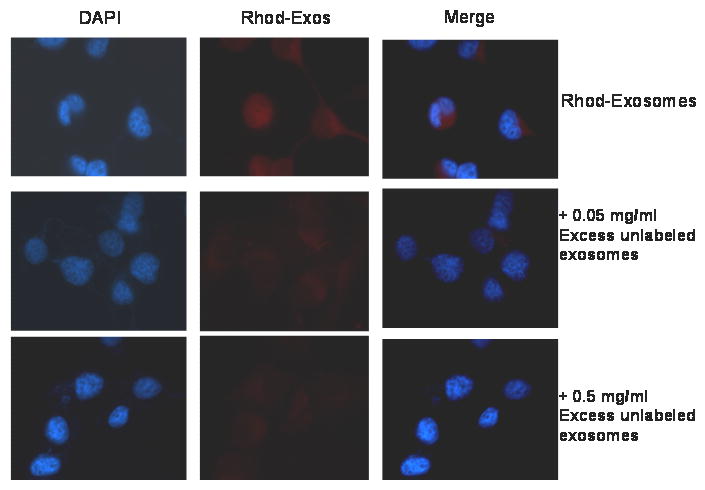
Purified bovine exosomes were labeled with rhodamine isothiocynate, purified again by ultra-filtration to remove excess unreacted rhodamine and then added to BT-549Gal-3 cells on cover-slips which had been serum starved for 48 h. The cells were incubated with labeled exosomes for 30 min in the absence and presence of excess unlabeled exosomes (0.05 mg/ml) and (0.5 mg/ml) respectively in HBSS containing divalent ions. The cells were rinsed twice with cold PBS and then fixed with -20°C cold methanol for 10 min, cover-slipped and visualized by fluorescent microscopy. The experiment was repeated with MDA-MB-231 and MCF-10A cells with similar results.
Re-cycling of Serum Exosomes by Tumor Cells
It was interesting to observe that by giving the tumor cells just enough exosomes ∼0.1 mg/ml, the cells were able to grow in soft agar for up to three weeks without the addition of more exosomes. This prompted us to question whether endocytosed exosomes can be re-cycled to maintain growth. To test this working hypothesis, we transfected BT-549-Gal3 cells with GFP-annexinA2 and incubated the cells with rhodamine-isothiocynate labeled exosomes. For the first 30 min. we could see only red and green cells (Fig 6A). However, after 3 hr of incubation, we mostly observed red and orange cells (Fig. 6B and 6C). This means that the cells that had endocytosed exosomes, released some back to the conditioned medium where they were up-taken by GFP-annnexinA2-transfected cells (Fig. 6B and 6C). In Fig. 6C, it is evident that the two GFP-annexinA2 transfected cells had endocytosed the released exosomes that were now concentrated in the late endosomal compartment, where they co-localized with annexinA2 which is also secreted from the cells via the re-cycling pathway of exosomes. We further demonstrated this by isolating and purifying exosomes from GFP-annexinA2 transfected cells and showing that GFP-annexinA2 is indeed recovered in the exosomal fraction (Fig. 6D). Members of annexin family of membrane proteins are routinely observed in cellular exosomes and can be considered markers of cellular exosomes [29]. In addition, galectin-3, another marker of cellular exosomes [29], was shown to be present in the exosomes purified from the tumor cells (Fig. 6E).
Figure 6. Recycling of bovine serum exosomes by tumor cells.
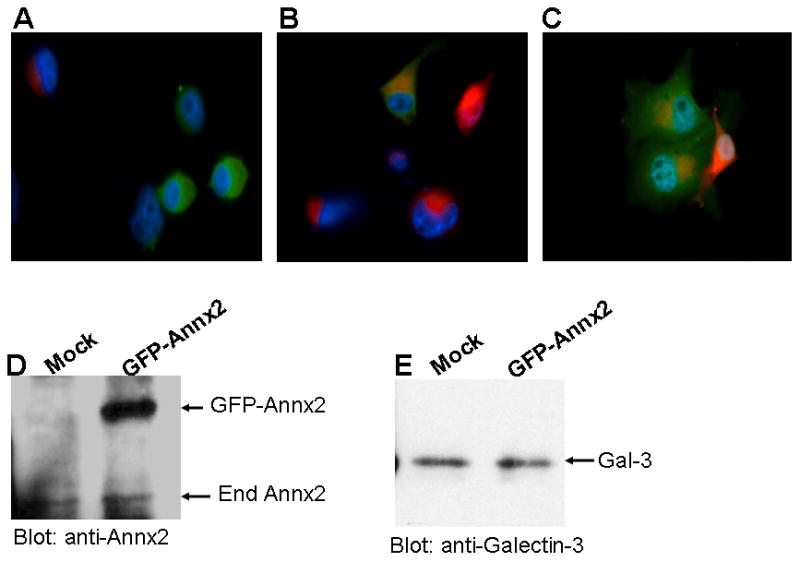
BT-549Gal-3 cells were transfected with GFP-annexin-A2 or mock transfected using Fugene6 and serum starved for 48 h. The mock transfected cells were then incubated with rhodamine-labeled exosomes for 30 min at 37°C followed by washing with PBS at room temperature. The GFP-annexin-A2 transfected cells (green cells) and rhodamine labeled (red) cells were harvested with 2 mM EDTA, washed in cold PBS and mixed at a ratio of 1:3 (green cells to red cells), plated in six-well plates on coverslips and incubated for 10 min (Panel A) and then for 2hr at 37°C (Panels B and C). The GFP-annexin-A2 and mock transfected cells were also detached with EDTA and incubated separately in HBSS in siliconized tubes for 2 h at 37°C. Exosomes were isolated and purified from the conditioned medium as described in Materials and Methods. The cellular exosomes were resolved in SDS-PAGE, transferred to immobilon membranes and probed with annexin-2 (Panel D) and galectin-3 (Panel E) antibodies.
Map Kinase Signaling Mediated by Exosomes
We next investigated the role of exosomes in transmitting growth signals to cells on plastic or non-adhered cells growing on poly-(HEMA) coated dishes. It was interesting to note that on plastic and in the presence of Ca2+/Mg2+ ions, ERK was activated (phospho-ERK) within 10 min of exosome addition and the activity diminished after 90 min. (Fig. 7A). This is the pattern of activation that is usually observed when one plots a time course of ERK activation [30]. However in the presence of a low concentration of exosomes (0.1 mg/ml), ERK was again activated within 5 min. but the signal remained strong for at least 240 minutes (Fig. 7A). This mode of activation is in line with adhesion and spreading of the cells on plastic in the presence of exosomes. In non-adherent cells, ERK activation was observed in the presence of low concentrations of exosomes (0.05 mg/ml) but at elevated concentrations approaching 0.5 mg/ml, activation of ERK was inhibited (Fig. 7B). This has been a consistent observation in at least 3 different experiments. Again as observed in adhered cells, in the presence of low concentrations of exosomes (0.1 mg/ml), the phosphorylated signal in ERK1/2 remained strong for at least 2 h (Fig 7C). Surprisingly the ERK1/2 activation signal in anchorage independent cells in the presence of exosomes (0.1 mg/ml) was evident even after 18 hours of growth (data not shown). A similar sustained activation of ERK in non-adhered cells was recently reported in ovarian cancer cells [31].
Figure 7. MAP Kinase signaling mediated by bovine serum exosomes.
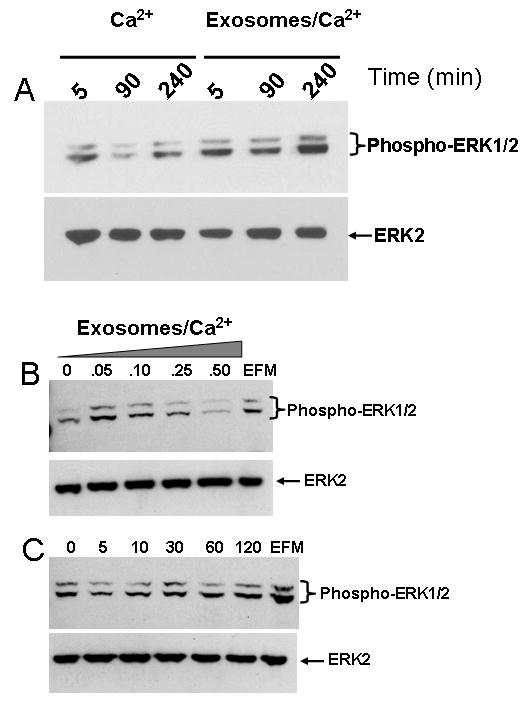
BT-549Gal3 cells were plated in 10 cm. dishes and serum starved for 48 h. The adherent cells were incubated in HBS containing 1 mM of the divalent ions (Mg2+/Ca2+) for the indicated time points (Panel A). The cells were scrapped, washed in ice-cold PBS and stored at -80°C until needed. In parallel experiments, cells (2 × 106 cells/dish) were plated on poly(HEMA)-treated dishes in HBS containing the divalent ions (Ca2+/Mg2+) without or with indicated concentrations of exosomes (mg/ml) for 10 min (Panel B). EFM (0.5 mg/ml) was also included as a positive control. In Panel C, the cells were incubated with 0.1 mg/ml of exosomes on poly(HEMA) coated wells for the indicated time points. The EFM (0.1 mg/ml) was also incubated with the cells for 120 min as a control. MAP kinase activation was visualized by detection of phosphorylated ERK1/2 by western blotting using antibodies to the dually phosphorylated ERK1 and ERK2. Total ERK (loading control) was detected using antibodies to ERK2.
Discussion
In the present studies, we have re-visited a long standing question regarding the identity of factor(s) in fetal bovine serum that mediates anchorage independent growth of tumor cells. A number of growth factors such as IGF, EGF, TGF-β, βFGF, PDGF have individually and as a group, been shown to play a role in anchorage independent growth of tumor cells [8, 32, 33]. However, even after all these growth factors were included as a cocktail in serum free medium and mixed with soft agar, colony formation by tumor cells was still less compared to growth in the medium containing serum [34]. It is in this background of looking for serum proteins that may influence anchorage independent growth that we questioned whether fetuin-A (a serum glycoprotein) plays a role in anchorage independent growth of tumor cells. In our earlier studies we determined that antibodies to this glycoprotein reduced anchorage independent growth potential in soft agar (data not shown). However, fetuin-A in addition to being secreted into the blood as a free protein, is also bound to other proteins [35] and has been shown to be associated with exosomes in the serum [11, 12].
In the present analyses, our goal was to identify the serum fraction (exosome free or exosome enriched) that supports anchorage independent growth (AIG) of breast carcinoma cells. The data clearly show that the membranous fraction of fetal bovine serum (contains exosomes) is the one that mediates AIG of the tumor cells. The in vitro growth of tumor cells in soft agar is regarded as an excellent predictor of tumor growth in vivo [5]. The ability of exosomes to mediate the growth of stellate colonies [21] in Matrigel further underscores the potential role of exosomes in the in vivo tumor growth. Whereas Matrigel also supported the growth of tumor cells in EFM, the cells mainly grew in 2-dimensions and not inwards or upwards in this medium.
Therefore in order for us to dissect in vivo tumor growth mechanisms, the interaction between these exosomes and carcinoma cells deserves a rigorous analysis. The exosomes are able to promote the growth of both normal (MCF-10A) and breast carcinoma cells. Growth signals for anchorage independent growth of tumor cells are well documented in the literature [2]. In normal cells (such as MCF-10A) that are attached or adhered to the substratum, the engagement of integrins by extracellular matrix proteins transmits growth signals that can also crosstalk with those emanating from growth factor/receptor interaction again resulting in cell growth [2]. However, in normal cells, lack of adhesion to the substratum results in anoikis or programmed cell death in case of epithelial and endothelial cells [2]. Transformed cells on the other hand are capable of transmitting growth signals whether or not they are attached to the substratum. However, the precise mechanism by which transformed cells coordinate the cross talks among the various growth signals while simultaneously inhibiting the apoptotic signals is poorly understood.
Despite the anchorage independence of transformed cells, they still require a number of growth factors, some of which the cells synthesize (autocrine) and some obtained from the serum. Our data suggest that in addition to providing the breast carcinoma cells with the growth factors such as TGF, the membranous portion of serum (contains exosomes) provide unique growth platforms. The vesicles could in theory concentrate growth factors on the surfaces of tumor cells. They could also promote cell to cell aggregation resulting in the enhancement of growth signals. The bovine vesicles just like the human exosomes, could also be derived from a variety of cell types including epithelial cells [36], macrophages [37], and platelets [38]. Some of the proteins could be imbedded in the vesicles while others such as fetuin-A could be on the surface. A number of serum proteins associated with exosomes have been implicated in anchorage independent growth of tumor cells in soft agar. Studies by Calhoun et al [39], have demonstrated that knockdown of cyclophilin A expression using RNAi in U2OS cells resulted in disruption of the F-actin structure and decreased anchorage independent growth, proliferation and migration. Interestingly, Emmprin (CD 147), a cell surface glycoprotein that plays a critical role in cellular transformation and anchorage independent growth, is a receptor for cyclophilin A [40]. The other protein that is abundantly expressed in the exosomes, HSP-90 alpha also plays a role in anchorage independent growth together with the other members of the family. The majority of known substrates for the 90 kDa heat shock protein (HSP-90) are signal intermediates of clinical importance such as kinases [41, 42]. For example, activated Cd42-associated kinase (AcK1) expression in LNCaP cells enhanced anchorage-independent growth in vitro and accelerated tumorigenicity in nude mice. However, its activity required the binding of HSP-90 beta [43]. Furthermore, the expression of the small heat shock protein (HSP-27) is associated with increased anchorage independent growth, increased invasiveness and resistance to chemotherapeutic drugs, poor prognosis and reduced disease free survival [44]. Studies by Anand et al. [45] have demonstrated that the gene encoding protein elongation factor (eEF-1 alpha) is amplified in 25% of primary ovarian tumors and is highly expressed in approximately 30% of ovarian tumors. They further showed that eEF-1alpha enhances focus formation, enhances anchorage independent growth and decreases doubling time of rodent fibroblasts.
Apart from the above mentioned serum proteins that are exclusively associated with exosomes, others such as alpha2-macroglobulin are concentrated in them but also occur in the exosome free fraction of serum. Another group of proteins which include fetuin-A, is more or less equally distributed in both EEM and EFM. Fetuin-A may after all, act as an opsonin to facilitate the up-take of the exosomes by the tumor cells [46]. Some of the exosomal associated serum proteins, particularly those that are loosely attached to their surfaces, may have specific receptors on the tumor cells through which they transmit their growth signals. However, the transit time of the proteins on the cell surface may be limited because by 10 min of adding exosomes to the cells, they are endocytosed. Once inside the cells, the exosomal proteins could directly promote anchorage independent growth and then channeled to the degradative pathway [47]. They could also be diverted into the re-cycling pathway of exosomes [48]. There are bovine serum proteins including fetuin-A that are routinely observed in exosomes purified from cells [11]. The assumption therefore is that these are bovine serum exosomal proteins that are part of the re-cycling exosomal pathway.
We have directly demonstrated that labeled bovine exosomes taken up by one breast tumor cell can eventually exit this cell and be up-taken by another. As the serum exosomes including their assortment of proteins are up-taken by cells on hand, the released exosomes on the other hand bring out with them a unique set of cellular proteins. For example, we have demonstrated that galectin-3, annexin-2GFP (in transfected cells) and endogenous annexin-2 are part of the repertoire of proteins secreted in exosomes from tumor cells. Both of these have been observed in cellular exosomes [29]. Galectin-3 is one of the cellular proteins that have been implicated in anchorage independent growth of tumor cells in soft agar [49]. It is thus tempting to speculate that exosomal galectin-3 is one of the key proteins with anchorage independent growth promoting properties. The up-take of serum exosomes by tumor cells could in theory speed up the release of the cellular exosomes (bearing these unique proteins) which in turn are up-taken by neighboring cells to influence anchorage independent growth.
In order to begin to understand mechanistic pathways by which exosomes influence anchorage independent growth in tumor cells, we analyzed MAP kinase signaling in the interaction between tumor cells and purified exosomes. This is one of the key signaling pathway in tumor cells and in particular breast carcinomas that is responsible for maintenance of AIG [50, 51]. Obviously there are cross-talks between MAP kinase pathways and other pathways that have also been heavily implicated in anchorage independent growth such as PI3 kinase/Akt [52]. The strong activation of ERK1/2 in the tumor cells by the bovine exosomes in the absence of growth factors such as EGF, suggest that these vesicles act in a synergistic manner with other growth factors to promote AIG. The ability of low concentrations of exosomes to maintain anchorage independent growth in the tumor cells argues for re-cycling of the exosomal derived growth factors and is consistent with sustained activation of ERK1/2.
In summary, we have demonstrated that serum factors necessary for anchorage independent growth of tumor cells in soft agar are concentrated in the exosomal compartment of serum and that these exosomes are most likely recycled to maintain growth. It is highly likely that human serum exosomes will also display a similar protein profile as the bovine vesicles, and with the potential to drive the in vivo growth of tumors particularly of the breast. Work is on going in our laboratory to identify cell surface receptor/s responsible for the endocytic up-take of the bovine exosomes. The identity of the key proteins that drive anchorage independent growth, whether originating from the exosomal compartment of tumor cells or bovine serum, is another focus area.
Acknowledgments
Data analysis was performed by the Meharry Medical College Microarray and Bioinformatics Core (MMBC), which is supported in part by NIH grants G12RR03032-19 and P20RR011792. (url: http://www.mmc.edu/bioinformatics). Mass spectrometry and experimental design was conducted by the Vanderbilt University Mass Spectrometry Core Lab (MSRC). (url: http://www.mc.vanderbilt.edu/msrc/massspec/index.php)
The work was supported by 1SC1CA134018-01 (J.O.); DOD W81XWH-07-1-0254 (J.O)
Abbreviations
- EEM
exosome enriched medium
- EFM
exosome free medium
- HEMA
2-hydroxyethylmethacrylate
- AIG
anchorage independent growth
References
- 1.Klein EA, Yung Y, Castagnino P, Kothapalli D, Assoian RK. Cell adhesion, cellular tension, and cell cycle control. Methods Enzymol. 2007;426:155–175. doi: 10.1016/S0076-6879(07)26008-2. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz MA. Integrins, oncogenes, and anchorage independence. J Cell Biol. 1997;139:575–578. doi: 10.1083/jcb.139.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reichelt J. Mechanotransduction of keratinocytes in culture and in the epidermis. Eur J Cell Biol. 2007;86:807–816. doi: 10.1016/j.ejcb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Shekhar MP, Pauley R, Heppner G. Host microenvironment in breast cancer development: extracellular matrix-stromal cell contribution to neoplastic phenotype of epithelial cells in the breast. Breast Cancer Res. 2003;5:130–135. doi: 10.1186/bcr580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedman VH, Shin SI. Cellular tumorigenicity in nude mice: correlation with cell growth in semi-solid medium. Cell. 1974;3:355–359. doi: 10.1016/0092-8674(74)90050-6. [DOI] [PubMed] [Google Scholar]
- 6.Haslam SZ, Woodward TL. Host microenvironment in breast cancer development: epithelial-cell-stromal-cell interactions and steroid hormone action in normal and cancerous mammary gland. Breast Cancer Res. 2003;5:208–215. doi: 10.1186/bcr615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knupfer H, Preiss R. Significance of interleukin-6 (IL-6) in breast cancer (review) Breast Cancer Res Treat. 2007;102:129–135. doi: 10.1007/s10549-006-9328-3. [DOI] [PubMed] [Google Scholar]
- 8.Rizzino A, Ruff E, Rizzino H. Induction and modulation of anchorage-independent growth by platelet-derived growth factor, fibroblast growth factor, and transforming growth factor-beta. Cancer Res. 1986;46:2816–2820. [PubMed] [Google Scholar]
- 9.Childs CB, Proper JA, Tucker RF, Moses HL. Serum contains a platelet-derived transforming growth factor. Proc Natl Acad Sci U S A. 1982;79:5312–5316. doi: 10.1073/pnas.79.17.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kundranda MN, Henderson M, Carter KJ, Gorden L, Binhazim A, Ray S, Baptiste T, Shokrani M, Leite-Browning ML, Jahnen-Dechent W, Matrisian LM, Ochieng J. The serum glycoprotein fetuin-A promotes Lewis lung carcinoma tumorigenesis via adhesive-dependent and adhesive-independent mechanisms. Cancer Res. 2005;65:499–506. [PubMed] [Google Scholar]
- 11.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou H, Pisitkun T, Aponte A, Yuen PS, Hoffert JD, Yasuda H, Hu X, Chawla L, Shen RF, Knepper MA, Star RA. Exosomal Fetuin-A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int. 2006;70:1847–1857. doi: 10.1038/sj.ki.5001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smales CM, Marchant RJ, Underhill MF. Characterization of therapeutic proteins by membrane and in-gel tryptic digestion. Methods Mol Biol. 2005;308:375–379. doi: 10.1385/1-59259-922-2:375. [DOI] [PubMed] [Google Scholar]
- 14.Licklider LJ, Thoreen CC, Peng J, Gygi SP. Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal Chem. 2002;74:3076–3083. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
- 15.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 16.Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, Samatova NF. Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res. 2006;5:2909–2918. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 17.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6:3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furtak V, Hatcher F, Ochieng J. Galectin-3 mediates the endocytosis of beta-1 integrins by breast carcinoma cells. Biochem Biophys Res Commun. 2001;289:845–850. doi: 10.1006/bbrc.2001.6064. [DOI] [PubMed] [Google Scholar]
- 19.Fukazawa H, Mizuno S, Uehara Y. A microplate assay for quantitation of anchorage-independent growth of transformed cells. Anal Biochem. 1995;228:83–90. doi: 10.1006/abio.1995.1318. [DOI] [PubMed] [Google Scholar]
- 20.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 melanoma cells--a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2007;104:13–19. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 21.Sharp JA, Waltham M, Williams ED, Henderson MA, Thompson EW. Transfection of MDA-MB-231 human breast carcinoma cells with bone sialoprotein (BSP) stimulates migration and invasion in vitro and growth of primary and secondary tumors in nude mice. Clin Exp Metastasis. 2004;21:19–29. doi: 10.1023/b:clin.0000017167.17065.61. [DOI] [PubMed] [Google Scholar]
- 22.Chun MH. Serum signaling factors and spheroids. Crit Rev Oncol Hematol. 2000;36:89–98. doi: 10.1016/s1040-8428(00)00079-2. [DOI] [PubMed] [Google Scholar]
- 23.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 24.Hegmans JP, Bard MP, Hemmes A, Luider TM, Kleijmeer MJ, Prins JB, Zitvogel L, Burgers SA, Hoogsteden HC, Lambrecht BN. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am J Pathol. 2004;164:1807–1815. doi: 10.1016/S0002-9440(10)63739-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaput N, Flament C, Viaud S, Taieb J, Roux S, Spatz A, Andre F, LePecq JB, Boussac M, Garin J, Amigorena S, Thery C, Zitvogel L. Dendritic cell derived-exosomes: biology and clinical implementations. J Leukoc Biol. 2006;80:471–478. doi: 10.1189/jlb.0206094. [DOI] [PubMed] [Google Scholar]
- 26.Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 27.Ristorcelli E, Beraud E, Verrando P, Villard C, Lafitte D, Sbarra V, Lombardo D, Verine A. Human tumor nanoparticles induce apoptosis of pancreatic cancer cells. FASEB J. 2008;22:3358–3369. doi: 10.1096/fj.07-102855. [DOI] [PubMed] [Google Scholar]
- 28.Morelli AE, Larregina AT, Shufesky WJ, Sullivan ML, Stolz DB, Papworth GD, Zahorchak AF, Logar AJ, Wang Z, Watkins SC, Falo LD, Jr, Thomson AW. Endocytosis intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 29.Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 30.Audet N, Paquin-Gobeil M, Landry-Paquet O, Schiller PW, Pineyro G. Internalization and Src activity regulate the time course of ERK activation by delta opioid receptor ligands. J Biol Chem. 2005;280:7808–7816. doi: 10.1074/jbc.M411695200. [DOI] [PubMed] [Google Scholar]
- 31.Al-Ayoubi A, Tarcsafalvi A, Zheng H, Sakati W, Eblen ST. ERK activation and nuclear signaling induced by the loss of cell/matrix adhesion stimulates anchorage-independent growth of ovarian cancer cells. J Cell Biochem. 2008;105:875–884. doi: 10.1002/jcb.21889. [DOI] [PubMed] [Google Scholar]
- 32.Anzano MA, Rieman D, Prichett W, Bowen-Pope DF, Greig R. Growth factor production by human colon carcinoma cell lines. Cancer Res. 1989;49:2898–2904. [PubMed] [Google Scholar]
- 33.Boyd DD, Levine AE, Brattain DE, McKnight MK, Brattain MG. Comparison of growth requirements of two human intratumoral colon carcinoma cell lines in monolayer and soft agarose. Cancer Res. 1988;48:2469–2474. [PubMed] [Google Scholar]
- 34.Chang TY, Tsai WJ, Chou CK, Chow NH, Leu TH, Liu HS. Identifying the factors and signal pathways necessary for anchorage-independent growth of Ha-ras oncogene-transformed NIH/3T3 cells. Life Sci. 2003;73:1265–1274. doi: 10.1016/s0024-3205(03)00428-4. [DOI] [PubMed] [Google Scholar]
- 35.Nie Z. Fetuin: its enigmatic property of growth promotion. Am J Physiol. 1992;263:C551–562. doi: 10.1152/ajpcell.1992.263.3.C551. [DOI] [PubMed] [Google Scholar]
- 36.van Niel G, Raposo G, Candalh C, Boussac M, Hershberg R, Cerf-Bensussan N, Heyman M. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology. 2001;121:337–349. doi: 10.1053/gast.2001.26263. [DOI] [PubMed] [Google Scholar]
- 37.Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007;110:3234–3244. doi: 10.1182/blood-2007-03-079152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–3799. [PubMed] [Google Scholar]
- 39.Calhoun CC, Lu YC, Song J, Chiu R. Knockdown endogenous CypA with siRNA in U2OS cells results in disruption of F-actin structure and alters tumor phenotype. Mol Cell Biochem. 2008 doi: 10.1007/s11010-008-9896-0. [DOI] [PubMed] [Google Scholar]
- 40.Nabeshima K, Iwasaki H, Koga K, Hojo H, Suzumiya J, Kikuchi M. Emmprin (basigin/CD147): matrix metalloproteinase modulator and multifunctional cell recognition molecule that plays a critical role in cancer progression. Pathol Int. 2006;56:359–367. doi: 10.1111/j.1440-1827.2006.01972.x. [DOI] [PubMed] [Google Scholar]
- 41.Pearl LH, Prodromou C. Structure and in vivo function of Hsp90. Curr Opin Struct Biol. 2000;10:46–51. doi: 10.1016/s0959-440x(99)00047-0. [DOI] [PubMed] [Google Scholar]
- 42.Smith DF, Whitesell L, Katsanis E. Molecular chaperones: biology and prospects for pharmacological intervention. Pharmacol Rev. 1998;50:493–514. [PubMed] [Google Scholar]
- 43.Mahajan NP, Whang YE, Mohler JL, Earp HS. Activated tyrosine kinase Ack1 promotes prostate tumorigenesis: role of Ack1 in polyubiquitination of tumor suppressor Wwox. Cancer Res. 2005;65:10514–10523. doi: 10.1158/0008-5472.CAN-05-1127. [DOI] [PubMed] [Google Scholar]
- 44.Lee SA, Ndisang D, Patel C, Dennis JH, Faulkes DJ, D'Arrigo C, Samady L, Farooqui-Kabir S, Heads RJ, Latchman DS, Budhram-Mahadeo VS. Expression of the Brn-3b transcription factor correlates with expression of HSP-27 in breast cancer biopsies and is required for maximal activation of the HSP-27 promoter. Cancer Res. 2005;65:3072–3080. doi: 10.1158/0008-5472.CAN-04-2865. [DOI] [PubMed] [Google Scholar]
- 45.Anand N, Murthy S, Amann G, Wernick M, Porter LA, Cukier IH, Collins C, Gray JW, Diebold J, Demetrick DJ, Lee JM. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat Genet. 2002;31:301–305. doi: 10.1038/ng904. [DOI] [PubMed] [Google Scholar]
- 46.Wang H, Zhang M, Bianchi M, Sherry B, Sama A, Tracey KJ. Fetuin (alpha2-HS-glycoprotein) opsonizes cationic macrophagedeactivating molecules. Proc Natl Acad Sci U S A. 1998;95:14429–14434. doi: 10.1073/pnas.95.24.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med. 2003;9:65–76. [PMC free article] [PubMed] [Google Scholar]
- 48.Mobius W, van Donselaar E, Ohno-Iwashita Y, Shimada Y, Heijnen HF, Slot JW, Geuze HJ. Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic. 2003;4:222–231. doi: 10.1034/j.1600-0854.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 49.Honjo Y, Nangia-Makker P, Inohara H, Raz A. Down-regulation of galectin-3 suppresses tumorigenicity of human breast carcinoma cells. Clin Cancer Res. 2001;7:661–668. [PubMed] [Google Scholar]
- 50.Zhang KX, Ward KR, Schrader JW. Multiple aspects of the phenotype of mammary epithelial cells transformed by expression of activated M-Ras depend on an autocrine mechanism mediated by hepatocyte growth factor/scatter factor. Mol Cancer Res. 2004;2:242–255. [PubMed] [Google Scholar]
- 51.Uehara N, Matsuoka Y, Tsubura A. Mesothelin promotes anchorage-independent growth and prevents anoikis via extracellular signal-regulated kinase signaling pathway in human breast cancer cells. Mol Cancer Res. 2008;6:186–193. doi: 10.1158/1541-7786.MCR-07-0254. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi K, Murakami M, Yamanaka S. Role of the phosphoinositide 3-kinase pathway in mouse embryonic stem (ES) cells. Biochem Soc Trans. 2005;33:1522–1525. doi: 10.1042/BST0331522. [DOI] [PubMed] [Google Scholar]


